Published online Apr 14, 2018. doi: 10.3748/wjg.v24.i14.1531
Peer-review started: January 22, 2018
First decision: February 6, 2018
Revised: February 25, 2018
Accepted: March 18, 2018
Article in press: March 18, 2018
Published online: April 14, 2018
Processing time: 81 Days and 4.3 Hours
To characterize punctual mutations in 23S rRNA gene of clarithromycin-resistant Helicobacter pylori (H. pylori) and determine their association with therapeutic failure.
PCR products of 23S rRNA gene V domain of 74 H. pylori isolates; 34 resistant to clarithromycin (29 from a low-risk gastric cancer (GC) population: Tumaco-Colombia, and 5 from a high-risk population: Tuquerres-Colombia) and 40 from a susceptible population (28 from Tumaco and 12 from Túquerres) were sequenced using capillary electrophoresis. The concordance between mutations of V domain 23S rRNA gene of H. pylori and therapeutic failure was determined using the Kappa coefficient and McNemar’s test was performed to determine the relationship between H. pylori mutations and clarithromycin resistance.
23S rRNA gene from H. pylori was amplified in 56/74 isolates, of which 25 were resistant to clarithromycin (20 from Tumaco and 5 from Túquerres, respectively). In 17 resistant isolates (13 from Tumaco and 4 from Túquerres) the following mutations were found: A1593T1, A1653G2, C1770T, C1954T1, and G1827C in isolates from Tumaco, and A2144G from Túquerres. The mutations T2183C, A2144G and C2196T in H. pylori isolates resistant to clarithromycin from Colombia are reported for the first time. No association between the H. pylori mutations and in vitro clarithromycin resistance was found. However, therapeutic failure of eradication treatment was associated with mutations of 23S rRNA gene in clarithromycin-resistant H. pylori (κ = 0.71).
The therapeutic failure of eradication treatment in the two populations from Colombia was associated with mutations of the 23S rRNA gene in clarithromycin-resistant H. pylori.
Core tip: Mutations in 23S rRNA gene V domain of Helicobacter pylori (H. pylori) were studied in order to determine their association with therapeutic failure. In clarithromycin-resistant H. pylori isolated from individuals at high-risk of gastric cancer (GC) in Túquerres-Colombia and at low-risk of GC in Tumaco-Colombia, mutations A1593T1, A1653G2, C1770T, C1954T1, and G1827C in isolates from Tumaco, and A2144G from Túquerres were found. Mutations T2183C and C2196T from both cities were not associated with clarithromycin resistance. However, therapeutic failure of eradication treatment in the sampled Colombian populations was associated with mutations of 23S rRNA gene in clarithromycin-resistant H. pylori.
- Citation: Matta AJ, Zambrano DC, Pazos AJ. Punctual mutations in 23S rRNA gene of clarithromycin-resistant Helicobacter pylori in Colombian populations. World J Gastroenterol 2018; 24(14): 1531-1539
- URL: https://www.wjgnet.com/1007-9327/full/v24/i14/1531.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i14.1531
Eradication of Helicobacter pylori (H. pylori) from the gastric mucosa is the current treatment for conditions such as chronic gastritis, peptic ulcer, atrophic gastritis, dysplasia, and metaplasia[1]. The first line scheme for the eradication of H. pylori is triple therapy, which includes a proton pump inhibitor and two antibiotics such as amoxicillin and clarithromycin. This treatment aims to eradicate infection in at least 90% of patients. However, therapeutic failure is inherent and can be due to multiple factors (human and bacterial), including improper drug dose, short treatment duration, early treatment discontinuation, drug activity associated with the use of other substances, quick reinfection of successfully treated patients, and the presence of antibiotic-resistant strains[1-4]. Among the main causes of resistance to clarithromycin in H. pylori are mutations in the V domain of 23S rRNA gene, this domain is the binding site for macrolide-type antibiotics. The most frequent mutations are A2143G (69.8%), A2142G (11.7%), and A2142C (2.6%). In addition, mutations A2115G, G2141A, C2147G, T2190C, C2195T, A2223G and C2694A have also been reported, but their role in resistance to clarithromycin is not yet clear[3].
In Latin America and worldwide, H. pylori resistance to antibiotics has been documented, with eradication being negatively affected by clarithromycin resistance[2]. In Colombia, resistance to this macrolide is estimated to be 17.2%[5]. Geographical conditions have also been documented to influence the risk of gastric cancer (GC). Coastal regions such as Tumaco have a low risk of GC, while Andean regions such as Túquerres have a high risk of GC. Hence, these geographical differences offer unique opportunities for the study of mutations of 23S rRNA gene in H. pylori. This study characterized the mutations of 23S rRNA gene V domain in H. pylori and their association with clarithromycin resistance and with therapeutic failure in patients from two Colombian populations (Tumaco and Túquerres) who were at different risk of developing GC.
The subjects in this study included adult men and women with dyspepsia symptoms from Tumaco (n = 203) and from Túquerres (n = 206). Four gastric mucosal biopsies were obtained from each patient; two from the antrum and two from the gastric body, in order to isolate H. pylori, and determine in vitro susceptibility of the isolates to clarithromycin and amoxicillin using agar dilution and molecular biology procedures.
For H. pylori culture and genotyping, the gastric mucosa biopsies were preserved in 25% thioglycollate and glycerol. The biopsies were frozen in liquid nitrogen and later placed in dry ice and stored at -70 °C for analysis at the Microbiology Laboratory and Histopathology Laboratory of the Department of Pathology of the Universidad del Valle, in Cali, Colombia. This study was supported by the CIREH (Human Ethics Committee) of the Universidad del Valle. All study subjects signed an informed consent form.
After the antimicrobial susceptibility microbiological study, 74 H. pylori isolates were obtained, of which 34 (46%) were in vitro clarithromycin resistant and 40 (54%) were susceptible to the antibiotic. 39.2% (29/34) of the resistant isolates and 37.8% (30/42) of the susceptible isolates were taken from patients in Tumaco. In addition, the sequences of 23S rRNA gene V domain of strains ATCC 43502 and ATCC 700392 were amplified and used as positive controls. DNA extraction was carried out by salting out[6] and susceptibility tests were performed using the agar dilution method[7].
The amplification of 23S rRNA gene V domain of H. pylori by PCR was carried out using a thermal cycler (Swift MiniProTM, Esco, Cincinnati, OH, United States), and the following reagents were added to a 0.2 mL tube: buffer 1× (Buffer green 5× Promega®), MgCl2 1 μmol/L (Promega®), DMSO 10%, dNTPs 0.288 mmol/L (Promega®), 50 pmol/μL of each primer (starting position 1585, 5´-GATTGGAGGGAAGGCAAT-3´/3´-CTCCATAAGAGCCAAAGCCC-5´ final position 2247), 0.5 U of GoTaq DNA polymerase (Promega®); and 25 ng of H. pylori genomic DNA in a final volume of 50 μL. The thermal cycle consisted in an initial denaturation at 95 °C/2 min, followed by 35 cycles [95 °C/1 min, 54 °C/1 min, 59 °C/1 min and 72 °C/1 min] and a final extension at 72 °C/15 min[8].
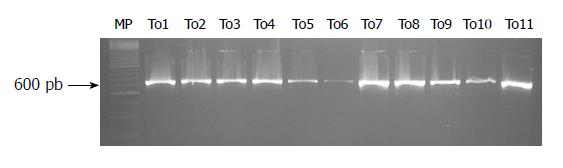
The amplification fragments were detected by 2% agarose gel electrophoresis (Sigma®), stained with 1 μL of ethidium bromide (Invitrogen, Carlsbad, CA, United States) (0.5 μg/mL), with an EC-105 power source (Thermo Fisher Scientific Inc., Asheville, NC, United States), at 75 V for 60 min, using a horizontal chamber (Spectroline bio-o-visión®). The DNA bands were visualized in UV light (260/280 nm), using a transilluminator (Spectroline bio-o-visión®). The size of the amplified fragment was approximately 662 pb (expected fragment by in silico analysis)[8].
The amplified fragments were sequenced in two directions (forward and reverse), using a genetic analyzer (ABI 3130 Applied Biosystem®) and the Big Dye Terminator methodology (Applied Biosystem®), following standardized conditions at Vanderbilt Genetic Institute Core Facilities, United States. The edition and alignment of the sequences was carried out using Bioedit software V 7.1.11® (Hall, 1999). Changes in sequences were matched by local alignment, with the reference sequence for 23S rRNA gene, code GenBank: U27270.1[8].
For categorical variables, McNemar’s Test was used for matching data, in order to identify significant differences between clarithromycin resistant and clarithromycin susceptible genotypes and the punctual mutations detected before treatment. The concordance correlation coefficient Kappa (k) was used to determine the concordance between the mutations of 23S rRNA gene V domain and in vitro clarithromycin resistance such as the concordance of mutations of 23S rRNA gene V domain with therapeutic failure in patients evaluated using the [13C]-Urea breath test (UBT), 45 d after completing H. pylori eradication treatment. The anti-H. pylori treatment included omeprazole (Genfar®) 20 mg, clarithromycin (Genfar®) 500 mg, and amoxicillin (Genfar®) 1000 mg, for 14 d in accordance with the recommendations of the Maastricht Consensus[9]. Therapeutic failure was considered in patients with a positive UBT. All data were analyzed using statistical software SPSS version 15.0 for Windows. Statistical significance was estimated at P < 0.05.
This study was approved by the Institutional Committee for Human Ethics Revision (CIREH) of the Faculty of Health of the Universidad del Valle, regulated by Resolution 008430 of October 4/1993, issued by the Colombian Ministry of Health.
The prevalence of H. pylori infection, which was diagnosed by histopathology, was higher in the low-risk GC population from Tumaco (88.77%), than in the high-risk GC population from Túquerres (85.4%), without a statistically significant difference. However, the prevalence of H. pylori resistance to clarithromycin and amoxicillin was significantly higher in the low-risk GC population from Tumaco, than in the high-risk GC population from Túquerres (20.5%, 22.8%) vs (3.4%, 5.4%), respectively, P < 0.05. Efficacy of the anti-H. pylori treatment was similar in both populations. Of 169 infected and treated patients from Tumaco, 130 (76.9%) were cured, and of 165 infected and treated patients from Tuquerres, infection was resolved in 123 (74.6%).
The amplification and sequencing of a fragment of 662 bp (Figure 1) between nucleotides 1585 and 2247 of 23S rRNA gene V domain of H. pylori, was carried out in 56 (76%) of the isolates, of which 39 (69.6%) were from Tumaco patients; of these, 20 (35.7%) were resistant and 19 (33.9%) were susceptible to clarithromycin under in vitro conditions. Five (8.9%) of the amplified isolates from Túquerres were resistant to clarithromycin and 12 (21.4%) were susceptible (Table 1).
| Helicobacter pylori isolates | Risk of gastric cancer | Total | |
| Low risk-Tumaco | High risk-Túquerres | ||
| Evaluated | |||
| Susceptible | 28 (37.8) | 12 (16.2) | 40 (54) |
| Resistant | 29 (39.2) | 5 (6.8) | 34 (46) |
| Total | 57 (77) | 17 (23) | 74 (100) |
| Amplified | 56 (76) | ||
| Susceptible | 19 (33.93) | 12 (21.43) | 31 (55.4) |
| Resistant | 20 (35.7) | 5 (8.93) | 25 (44.6) |
| Total | 39 (69.6) | 17 (30.4) | 56 (100) |
Table 1, shows the number of H. pylori isolates at baseline, which were susceptible and resistant to clarithromycin in vitro. The total number of H. pylori isolates from both populations and those used to amplify 23S rRNA gene V domain were evaluated; the number of H. pylori isolates amplified from both populations represents fragment amplification where possible. The total number of isolates is represented by bold typeface.
At least one mutation was identified in the sequences of 31 (55.3%) H. pylori isolates, with 17 (33.3%) resistant and 14 (25%) susceptible to clarithromycin. Of the resistant isolates, 13 (23.2%) were from Túmaco patients and 4 (7.1%) were from Túquerres patients. In addition, 9 (16.1%) of the resistant isolates did not show any mutations in their sequence; of these, 8 (14.3%) were isolated from Tumaco patients and 1 (1.8%) was isolated from Túquerres patients. The Kappa coefficients (κ = 0.17) and (κ = 0.23) for the low risk and high risk GC populations, respectively, suggest that there was no relationship between the presence of mutations and in vitro resistance to clarithromycin. Similarly, there was no association between the lack of mutations in 23S rRNA gene and in vitro susceptibility to clarithromycin in both populations, P > 0.05 (Table 2).
| Susceptibility | Risk of gastric cancer | |||||||
| Low risk n = 39 | High risk n = 17 | |||||||
| Mutant | Non mutant | Mutant | Non mutant | |||||
| Resistant | 13 (23.2) | 8 (14.3) | 4 (7.1) | 1 (1.8) | ||||
| Susceptible | 8 (14.3) | 10 (17.8) | 6 (10.7) | 6 (10.7) | ||||
| Kappa-P | k = 0.17 | P = 0.28 | k = 0.23 | P = 0.25 | ||||
| Total | 21 | 37.5 | 18 | 32.1 | 10 | 17.8 | 7 | 12.5 |
Twenty different mutations were characterized in 33 sequences of H. pylori evaluated. Mutations T2183C and C2196T were present only in resistant isolates in both populations; the first mutation was observed in 2 isolates from the low risk GC population (Tumaco) and in 1 isolate from the high risk GC population (Túquerres). The second mutation was observed in 1 isolate in each population. Similarly, mutations A1593T, A1653G, C1770T, C1954T, and G1827C, were observed only in resistant isolates in Tumaco patients. Conversely, mutation A2144G was present only in 1 isolate from Túquerres (Tables 3 and 4).
| Resistant n = 13 | Susceptible n = 8 | ||||
| Patient ID | Mutations | MIC | Patient ID | Mutations | MIC |
| 138 | A1593G 1T2183C | 1 | 17 | A1822G/G1827A/G1941A/T1831C | < 0.25 |
| 64 60 | A1653G | 2 4 | 94 | T1645C | < 0.25 |
| 4 | A1739G 1C1954T/G1695A | 4 | 96 | A1739G | < 0.25 |
| 65 | A1739G 1C2196T 1G1827C | 1 | 97 | T1645C | < 0.25 |
| 42 102 174 | A1822G/G1827A/T1831C | 1 2 1 | 98 | C1632T | < 0.25 |
| 88 | C1632T | > 4 | 101 | A1822G/G1827A/T1645C/T1831C | < 0.25 |
| 107 | 1C1770T | 1 | 103 | C1632T | < 0.25 |
| 38 36 | T1645C | 1 2 | 107 | A1667G/T1668C | < 0.25 |
| 6 | 1T2183C/A1593T/A1822G/G1827A/T1831C | 4 | |||
| ATCC 700392 | A1593G | ATCC 43504 | A1667G/T1668C | ||
| Resistant n = 4 | Susceptible n = 6 | ||||
| Patient ID | Mutations | MIC | Patient ID | Mutations | MIC |
| 323 | A1593G/A1822G/G1827A/T1645C/T1831C/1T2183C | 1 | 351 | A1822G/G1827A/T1831C | < 0.25 |
| 336 | A1593G/1C2196T | 2 | 377 | A1822G/G1827A/G2221A/T1645C/T1831C | < 0.25 |
| 339 | 1A2144G/G1827A | 4 | 394 | A1593G | < 0.25 |
| 440 | A1822G/G1827A/G2221A/T1831C | 4 | 457 | A1822G/G1827A/G2221A/T1831C | < 0.25 |
| 467 | A1739G/G1695A | < 0.25 | |||
| 513 | A1822G/G1827A/T1831C | < 0.25 | |||
| ATCC 700392 | A1593G | ATCC 43504 | A1667G/T1668C | ||
Tables 3 and 4 show the changes in the sequences of 23S rRNA gene V domain of H. pylori in high-risk and low-risk GC patients according to susceptibility or resistance to clarithromycin. Column MIC shows the minimum inhibitory concentration at μg/mL, which was evaluated using the agar dilution method. In Column mutations, the punctual changes in the nucleotides of 23S rRNA gene observed in the sequence of each isolate are shown.
It was found that the mutations of H. pylori susceptible to clarithromycin were located in domain IV of 23S rRNA gene, nucleotides 1562-1931, except for mutation G2221A which was located in domain V of an isolate susceptible to clarithromycin. In contrast, mutations in domain V, nucleotides 1932-2541, were mainly present in resistant isolates, except for changes C1770T, A1593T and G1827C, which were associated with mutations in domain IV (Table 5).
| Domain-Region | Tumaco | Túquerres | ||
| Resistant position | Susceptible position | Resistant position | Susceptible position | |
| Domain IV 1562-1931 | C1770T | A1593G | A1593G | |
| A1593T | A1667G | A1667G | ||
| G1827C | A1739G | A1739G | ||
| A1822G | A1822G | |||
| C1632T | C1632T | |||
| G1695A | G1695A | |||
| G1827A | G1827A | |||
| G1941A | G1941A | |||
| T1645C | T1645C | |||
| T1668C | T1668C | |||
| T1831C | T1831C | |||
| Domain V 1932-2541 | C1954T | G2221A | C2196T | G2221A |
| T2183C | T2183C | |||
| C2196T | A2144G | |||
| A1653G | ||||
| C2196T | ||||
Although the mutations in isolates resistant to clarithromycin were observed mainly in 23S rRNA gene V domain of H. pylori, no relationship was found between them and in vitro resistance to clarithromycin (P > 0.05, Tables 2-5). Punctual mutations in domain IV of the target gene were found in susceptible isolates (Table 5). However, the Kappa coefficient κ = 0.64 and κ = 0.69 shows that there was a good level of concordance between the mutations in 23S rRNA gene and therapeutic failure in patients unsuccessfully treated, both in the high-risk and low-risk GC populations, respectively, and the two populations together, κ = 0.71, as shown by the positive UBT, which was performed 45 d after the end of H. pylori eradication treatment (Table 6).
| Breath test[13C]-urea | Population at risk of gastric cancer | Total | ||||
| Low risk n = 39 | High risk n = 17 | |||||
| Mutant | No mutant | Mutant | No mutant | Mutant | No mutant | |
| Positive | ||||||
| Therapeutic failure | 18 | 3 | 8 | 1 | 26 | 4 |
| Negative | ||||||
| Therapeutic Success | 3 | 15 | 2 | 6 | 5 | 21 |
| Total | 21 | 18 | 10 | 7 | 31 | 25 |
| Kappa | k = 0.69 | k = 0.64 | k = 0.71 | |||
Research on the prevalence of clarithromycin resistance and characterization of the mutations of 23S rRNA gene, which may be associated with in vitro resistance in H. pylori, is scarce in Colombia. In general, research has focused on evaluating the frequency of mutations already reported and the most frequently observed mutations , such as mutations A2142G, A2143G y A2142C[3].
In Colombia, studies carried out in Risaralda, Quindío, and Cauca have reported frequencies between 1.85% and 7.3% for mutation A2142G, and between 2.2% and 2.46% for mutation A2143G in H. pylori isolates resistant to in vitro clarithromycin[10-12]. In our study, no H. pylori isolate which was resistant or susceptible to clarithromycin in vitro and exhibited these mutations was detected.
Among the mutations studied in H. pylori isolates resistant to clarithromycin was C2196T with a frequency of 0.05% (1/21) and 0.2% (1/5) in isolates from Tumaco and Túquerres patients, respectively. This change was reported in a study carried out in the Province of Guiyang (China), which found resistance of 30% (13/42) to in vitro clarithromycin, this study also reported mutation C2196T in a resistant and in a susceptible isolate, and mutation A2143G in susceptible isolates[13]. In contrast to this, mutation C2196T was found only in resistant isolates in our study, with a similar frequency. However, it was not linked to other mutations with such resistance, but it is important to consider the proximity of a nucleotide to mutation C2195T, associated with resistance[3].
Mutation T2183C exhibited frequencies of 0.09 (2/21) and 0.2 (1/5) in resistant isolates from high-risk and low-risk GC patients from Túquerres and Tumaco, respectively. Similar results were reported in studies carried out in H. pylori isolates from Korean dyspepsia patients, where the frequency of this mutation was between 0.25 (1/4)[14] and 0.35 (5/14)[15]. Although this mutation is found in domain V and occurred only in isolates resistant to in vitro clarithromycin, some researchers believe that its relationship with clarithromycin resistance is not yet clear, as it may be found in isolates both resistant and susceptible to this drug[16,17]. However, its presence in isolates growing at MIC ≥ 1 μg/mL of clarithromycin, suggests its capability to inhibit the effect of the antibiotic, at least as reported in this study.
Mutation A2144G was found in an H. pylori isolate from Túquerres, with a frequency of 0.25 (1/4), which corroborates findings which suggest that the mutation is clearly associated with in vitro clarithromycin resistance[18-20]. It was found that the frequency in the sampled population in this study, is in line with the frequencies reported in other regions, 0.01 (1/73)[21] and 0.81(9/11)[20-23]. This mutation was first reported in H. pylori isolates resistant to clarithromycin in Colombia, which indicates that it may be associated with the inclusion of strains from high frequency countries such as South Korea (frequency of 0.57)[15]; Japan (frequency of 0.7)[24] and Turkey (frequency between 0.29 and 0.81[20,22].
The mutations associated with clarithromycin resistance in the H. pylori isolates described in this study (A2144G, C2196T, and T2183C), are located in 23S rRNA gene V domain, as reported in the current literature[3]. Inhibition of the action of the macrolide may be due to spatial alterations in the V domain of 23S rRNA gene, which inhibit the target, as seen in transversion mutations A2143G, A2142G, A2142C[3], A2144G[18,19,22], where a nitrogenous base with two H groups (Adenine) is changed for another with three H groups (Guanine and Cytosine), with the inherent spatial alteration of the molecular structure, a phenomenon similar in transitions C2196T and T2183C[17].
This study found that there was no concordance between the presence of punctual mutations of H. pylori and in vitro resistance to clarithromycin and no association between the absence of mutations in the 23S rRNA gene and in vitro susceptibility to clarithromycin in both populations. These findings and the absence of mutations in 36% of the isolates resistant to in vitro clarithromycin may be explained by the occurrence of mutations outside the amplified region, a fragment located between positions 1585-2224. Among the changes associated with clarithromycin resistance, which are located outside this fragment, are A2223G, C2694A[3], T2711C[21], T2288C[24], and T2289C[25], and these mutations may explain the discrepancy of the results on the presence of punctual mutations in the amplified region, the in vitro resistance to clarithromycin and the good level of concordance between punctual mutations in the 23S rRNA gene of H. pylori with therapeutic failure in patients with unsuccessful eradication treatment. Clarithromycin resistance may be mediated by flow pumps that help H. pylori resist concentrations higher than 1 μg/mL of clarithromycin[23,26]. The presence of these mechanisms in H. pylori isolates in the high-risk and low-risk GC populations in Colombia was not evaluated in this study.
H. pylori resistance to clarithromycin is the main cause of failed eradication treatment; thus, the characterization of resistance is fundamental to validate gold standard methodology, such as the microbiological method of dilution in agar; however, this is a technically difficult and time-consuming method. It is worth mentioning that in our study, the sequencing method of the amplified H. pylori fragments of 23S rRNA gene by PCR and the detection of their punctual mutations were consistent with the UBT, a method used to diagnose therapeutic failure in patients with unsuccessful treatment, (κ = 0.64, κ = 0.69), both for high-risk and low-risk GC populations (κ = 0.71). These results may be reproducible in future studies, improve H. pylori infection eradication regimens and and may be applicable in clinical practice in Colombia. However the UBT is used to evaluate the follow-up of H. pylori treatments and its effectiveness should be an additional test in clinical practice and in the programs and policies for the prevention of GC in Colombia.
Although two first-line antibiotics were used in the anti-H. pylori treatment regimen, the results of resistance mechanisms in H. pylori to amoxicillin were not reported in this study. It is important to emphasize that H. pylori resistance to clarithromycin is mainly due to mutations in 23S rRNA gene V domain and is the main cause of first-line eradication treatment failure[2].
Other techniques that require less time for the identification of resistance include the E-test (sensibility of 45% and specificity of 95%) and DNA-based techniques, such as FISH (sensibility of 97% and specificity of 94%), PNA-FISH (sensibility of 80% and specificity of 93%), Line Probe Test (sensibility of 100% and specificity of 82.2%), and PCR (sensibility of 98% and specificity of 92%)[3], which require specific methods for each mutation (FISH; PNA-FISH, Line Probe Test) or sequencing of the amplified fragment (PCR). The efficiency of these tests is subject to knowledge of the mutations associated with clarithromycin resistance in H. pylori strains.
This study demonstrated that the resistant isolates from these two contrasting populations involved in the development of GC, mutations A2143G, A2142G, and A2142C, which are usually reported as the most frequent, were not found in the isolates evaluated. With regard to the design of these tests, the changes A2144G, T2183C and C2196T found in these populations should be considered for use in fast-diagnostic methods of clarithromycin resistance in clinical practice. These mutations associated with H. pylori resistance to clarithromycin are the first to be reported in Colombia.
It may be concluded that in H. pylori isolates resistant to clarithromycin in patients from both Colombian populations, no high-frequency mutation was observed in 23S rRNA gene V domain, but there was high genotypic variation among the isolates.
No relationship between the mutations in 23S rRNA gene V domain of H. pylori and in vitro resistance was found, contrary to that seen in other H. pylori non-mutant isolates resistant to clarithromycin, which may be explained by mutations outside the evaluated fragment or by the existence of flow pumps. However, the failure of eradication treatment in the Colombian populations in this study was associated with punctual mutations in 23S rRNA gene of H. pylori resistant to clarithromycin.
In the Colombian populations studied, it was difficult to use a fast-resistance detection test for specific mutations, as information is scarce and the mutations reported exhibited a low frequency.
Infection by Helicobacter pylori (H. pylori) is the leading risk factor for the development of gastric adenocarcinoma, especially in individuals infected with strains resistant to antibiotics used in primary treatment regimens. The eradication of H. pylori infection is a valid primary prevention strategy for gastric lesions, atrophy, and gastric cancer (GC). However, resistance of this microorganism to clarithromycin is associated with therapeutic failure and a major risk of GC in Colombia. Thus, although significant improvements in the efficacy of treatment regimens have been made, none of these regimens successfully eradicate the infection. A few studies have focused on the evaluation of clarithromycin-resistance mechanisms, particularly mutations of 23S rRNA gene of the infecting strains in Colombia, which are associated with treatment failure and early subsequent prevention of GC.
Taking into account that GC prevention programs are focused on the eradication of H. pylori, it is important to know the specific treatment regimens for each country seeking to apply this strategy. In Colombia, the efficacy of standard triple therapy which includes clarithromycin, amoxicillin, and a proton pump inhibitor is currently being questioned. However, there are insufficient multicenter studies suggesting alternative regimens and basic studies on antibiotic resistance mechanisms in H. pylori. Mutations in H. pylori 23S rRNA gene V domain were studied to evaluate in vitro resistance to clarithromycin. This study identified mutations not documented in the current literature, which although are not associated with in vitro resistance to clarithromycin, they are linked to the therapeutic failure of triple therapy. Punctual mutations in the Colombian strains could be useful in future studies focusing on diagnostic methods for antibiotic susceptibility and in the therapeutic efficacy of GC prevention schemes in Colombia.
In this study, the researchers characterized mutations in domain V of 23S rRNA gene in clarithromycin-resistant H. pylori and determined their association with therapeutic failure in a high-risk gastric cancer population from Tuquerres, Colombia, and in a low-risk gastric cancer population from Tumaco, Colombia. A very interesting basic study clearly showed that therapeutic failure of eradication treatment in the sampled Colombian populations was associated with mutations of 23S rRNA gene in clarithromycin-resistant H. pylori. Hopefully, these findings will help to further improve treatment success and may be applied in the future for the fast diagnosis of therapeutic failure. This study found no concordance between the presence of punctual mutations in H. pylori and in vitro resistance to clarithromycin and there was no association between the absence of mutations in the 23S rRNA gene and in vitro susceptibility to clarithromycin in both populations. These findings and the absence of mutations in 36% of the isolates resistant to in vitro clarithromycin may be explained by the occurrence of mutations outside the amplified region, a fragment located between positions 1585-2224. Among the changes associated with clarithromycin resistance, which are located outside this fragment, are A2223G, C2694A T2711C, T2288C, and T2289C, mutations that may explain the discrepancy between the presence of punctual mutations in the amplified region and in vitro resistance to clarithromycin
To achieve the objectives of this study, we used the capillary electrophoresis sequencing method of the amplified DNA fragments of the H. pylori 23S rRNA gene and the detection of its punctual mutations, which were concordant with the [13C]-Urea breath test. This method was used in a novel way to diagnose the therapeutic failure of anti-H. pylori treatment in vivo. The [13C]-Urea breath test was used during the follow-up period to evaluate the effectiveness of H. pylori treatments.
This study demonstrated that the resistant isolates from these two contrasting populations involved in the development of GC, mutations A2143G, A2142G, and A2142C, which are usually reported as the most frequent, were not found in the isolates evaluated. With regard to the design of tests, the changes A2144G, T2183C and C2196T found in these populations should be considered for use in fast-diagnostic methods of clarithromycin resistance in clinical practice.
These results are important in the definition of treatments for gastro-duodenal diseases caused by H. pylori. They suggest that the failure of anti-H. pylori treatment is mainly due to mutations in 23S rRNA gene V domain. The application of these findings could be complemented by studies on the genetics and virulence of the microorganism, as individuals with similar ancestry may not require anti-H. pylori treatment. In contrast individuals infected with strains of different evolutionary origins than their host, would benefit from additional studies on antibiotic susceptibility. These advances in basic studies tend to elucidate the African enigma, and indicate that human-H. pylori coevolution and virulence of the bacterium could explain the contrast in risk of disease observed in our study populations. These findings may contribute to the future identification of individuals at higher risk of GC and require antibiotic susceptibility studies prior to treatment of the infection and early GC prevention.
In this investigation, mutations A2144G, C2196T and T2183C were observed in 23S rRNA gene V domain of H. pylori resistant to clarithromycin and were associated with failure of eradication treatment. The mutations T2183C, A2144G and C2196T in 23S rRNA gene V domain are reported for the first time in clarithromycin-resistant isolates of H. pylori in Colombia. This study demonstrated that the therapeutic failure of H. pylori eradication treatment in high and low risk GC populations from Colombia was associated with mutations of the 23S rRNA gene of clarithromycin-resistant H. pylori. The sequencing method for the detection of punctual mutations of DNA amplified 23S rRNA gene fragments is proposed to predict therapeutic failure induced by clarithromycin-resistant H. pylori. This new knowledge allows us to propose the design of a rapid detection test for H. pylori resistance to clarithromycin where mutations A2144G, T2183C and C2196T should be considered and can be applied in clinical practice to predict therapeutic failure of anti-H. pylori treatment.
Following therapeutic failure, reinfection may occur in patients as well as medication with antagonistic drugs or others such as proton pump inhibitors, which allow the appearance of false positives. In this study, adherence to treatment and self-medication were taken into account during the follow-up period. Characterization of the mutations in the 23S rRNA gene in a larger number of Colombian populations is required, in order to confirm the mutations associated with clarithromycin resistance in H. pylori and to determine, from multicenter studies, the optimal treatment regimen in Colombia. The molecular analysis of 23S rRNA gene V domain of H. pylori and other candidate genes is required, in order predict therapeutic failure. It is possible to reproduce the method in future investigations using total DNA from gastric mucosa biopsies and validate the presence of mutations found in this study. The [13C]-Urea breath test is recommended during follow-up to evaluate the effectiveness of anti-H. pylori treatment.
We would like to thank the Microbiology and Molecular Biology Laboratory, and the Histopathology Laboratory of the Pathology Department of Universidad del Valle, for use of their facilities during this study. We are also grateful to Hospital San Andres de Tumaco and the Hospital San Jose de Túquerres, for use of their facilities for clinical sampling and isolation of the H. pylori fragments used in this study.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Colombia
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chuah SK, Tarnawski AS S- Editor: Ma YJ L- Editor: Webster JR E- Editor: Huang Y
| 1. | Wu W, Yang Y, Sun G. Recent Insights into Antibiotic Resistance in Helicobacter pylori Eradication. Gastroenterol Res Pract. 2012;2012:723183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 2. | Camargo MC, García A, Riquelme A, Otero W, Camargo CA, Hernandez-García T, Candia R, Bruce MG, Rabkin CS. The problem of Helicobacter pylori resistance to antibiotics: a systematic review in Latin America. Am J Gastroenterol. 2014;109:485-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 3. | Thung I, Aramin H, Vavinskaya V, Gupta S, Park JY, Crowe SE, Valasek MA. Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 2016;43:514-533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 410] [Cited by in RCA: 548] [Article Influence: 60.9] [Reference Citation Analysis (2)] |
| 4. | Ghotaslou R, Leylabadlo HE, Asl YM. Prevalence of antibiotic resistance in Helicobacter pylori: A recent literature review. World J Methodol. 2015;5:164-174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 155] [Cited by in RCA: 150] [Article Influence: 15.0] [Reference Citation Analysis (1)] |
| 5. | Trespalacios AA, Otero W, Marcela M. Helicobacter pylori resistance to metronidazole, clarithromycin and amoxicillin in Colombian patients. Rev Colomb Gastroenterol. 2010;25:31-38. |
| 6. | Ho SA, Hoyle JA, Lewis FA, Secker AD, Cross D, Mapstone NP, Dixon MF, Wyatt JI, Tompkins DS, Taylor GR. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29:2543-2549. [PubMed] |
| 7. | Bustamante-Rengifo JA, Matta AJ, Pazos A, Bravo LE. In vitro effect of amoxicillin and clarithromycin on the 3' region of cagA gene in Helicobacter pylori isolates. World J Gastroenterol. 2013;19:6044-6054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 8. | Taylor DE, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621-2628. [PubMed] |
| 9. | Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2220] [Cited by in RCA: 1967] [Article Influence: 245.9] [Reference Citation Analysis (1)] |
| 10. | Isaza MC, Henao BJ, Alvarez A, Moncayo JI, Santacruz JJ, Meisel E, Salazar F, Giraldo D. Comparación de dos protocolos de erradicación de Helicobacter pylori. Rev Médica Risaralda. 2007;13:1-8. |
| 11. | Alvarez A, Moncayo JI, Santacruz JJ, Corredor LF, Reinosa E, Martínez JW, Beltrán L. [Antimicrobial susceptibility of Helicobacter pylori strains isolated in Colombia]. Rev Med Chil. 2009;137:1309-1314. [PubMed] |
| 12. | Acosta CP, Hurtado FA, Trespalacios AA. [Determination of single nucleotide mutations in the 23S rRNA gene of Helicobacter pylori related to clarithromycin resistance in a population from Cauca, Colombia]. Biomedica. 2014;34 Suppl 1:156-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Boyanova L, Markovska R, Yordanov D, Gergova G, Mitov I. Clarithromycin Resistance Mutations in Helicobacter pylori in Association with Virulence Factors and Antibiotic Susceptibility of the Strains. Microb Drug Resist. 2016;22:227-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 14. | Yoon KH, Park SW, Lee SW, Kim BJ, Kim JG. Clarithromycin-based standard triple therapy can still be effective for Helicobacter pylori eradication in some parts of the Korea. J Korean Med Sci. 2014;29:1240-1246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Sung J, Kim N, Park YH, Hwang YJ, Kwon S, Na G, Choi JY, Kang JB, Kim HR, Kim JW. Rifabutin-based Fourth and Fifth-line Rescue Therapy in Patients with for Helicobacter pylori Eradication Failure. Korean J Gastroenterol. 2017;69:109-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 16. | Hwang TJ, Kim N, Kim HB, Lee BH, Nam RH, Park JH, Lee MK, Park YS, Lee DH, Jung HC. Change in antibiotic resistance of Helicobacter pylori strains and the effect of A2143G point mutation of 23S rRNA on the eradication of H. pylori in a single center of Korea. J Clin Gastroenterol. 2010;44:536-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 110] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 17. | Teh X, Khosravi Y, Lee WC, Leow AH, Loke MF, Vadivelu J, Goh KL. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLoS One. 2014;9:e101481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Momynaliev KT, Selezneva OV, Kozlova AA, Vereshchagin VA, Il'ina EN, Govorun VM. [A2144G is the main mutation in the 23S rRNA gene of Helicobacter pylori associated with clarithromycin resistance]. Genetika. 2005;41:1338-1344. [PubMed] |
| 19. | Sezgin O, Aslan G, Altintaş E, Tezcan S, Serin MS, Emekdaş G. Detection of point mutations on 23S rRNA of Helicobacter pylori and resistance to clarithromycin with PCR-RFLP in gastric biopsy specimens in Mersin, Turkey. Turk J Gastroenterol. 2008;19:163-167. [PubMed] |
| 20. | Caliskan R, Tokman HB, Erzin Y, Saribas S, Yuksel P, Bolek BK, Sevuk EO, Demirci M, Yılmazli O, Akgul O. Antimicrobial resistance of Helicobacter pylori strains to five antibiotics, including levofloxacin, in Northwestern Turkey. Rev Soc Bras Med Trop. 2015;48:278-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 21. | Toracchio S, Aceto GM, Mariani-Costantini R, Battista P, Marzio L. Identification of a novel mutation affecting domain V of the 23S rRNA gene in Helicobacter pylori. Helicobacter. 2004;9:396-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Tajbakhsh S, Falahi J, Motamed N, Tabib SM, Bahador A, Gharibi S. Prevalence of A2143G and A2144G point mutations responsible for clarithromycin resistance among Helicobacter pylori strains in Bushehr, Iran. Avicenna J Clin Microb Infec. 2016;3:e36521. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | Kim JM, Kim JS, Jung HC, Kim N, Kim YJ, Song IS. Distribution of antibiotic MICs for Helicobacter pylori strains over a 16-year period in patients from Seoul, South Korea. Antimicrob Agents Chemother. 2004;48:4843-4847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 136] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 24. | Rimbara E, Noguchi N, Kijima H, Yamaguchi T, Kawai T, Sasatsu M. Mutations in the 23S rRNA gene of clarithromycin-resistant Helicobacter pylori from Japan. Int J Antimicrob Agents. 2007;30:250-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Hao Q, Li Y, Zhang ZJ, Liu Y, Gao H. New mutation points in 23S rRNA gene associated with Helicobacter pylori resistance to clarithromycin in northeast China. World J Gastroenterol. 2004;10:1075-1077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 34] [Article Influence: 1.6] [Reference Citation Analysis (1)] |
| 26. | Hirata K, Suzuki H, Nishizawa T, Tsugawa H, Muraoka H, Saito Y, Matsuzaki J, Hibi T. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J Gastroenterol Hepatol. 2010;25 Suppl 1:S75-S79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |