Published online Feb 7, 2015. doi: 10.3748/wjg.v21.i5.1488
Peer-review started: July 13, 2014
First decision: August 15, 2014
Revised: August 28, 2014
Accepted: September 30, 2014
Article in press: September 30, 2014
Published online: February 7, 2015
AIM: To determine how the oncogene miR-21 regulates the RAS signaling pathways and affects colon cancer cell behaviors.
METHODS: RAS p21 GTPase activating protein 1 (RASA1) protein expression in six colon cancer cell lines was assessed by Western blot. Colon cancer RKO cells were chosen for transfection because they are KRAS wild type colon cancer cells whose RASA1 expression is significantly decreased. RKO cells were transfected with vectors overexpressing or down-regulating either miR-21 or RASA1. Furthermore, a luciferase reporter assay was used to determine whether RASA1 is a gene target of miR-21. Then, changes in mRNA and protein levels of RASA1, RAS-GTP, and other components of the RAS signaling pathways were assessed in transfected RKO cells by real-time quantitative reverse transcription-polymerase chain reaction, Western blot and immunoprecipitation. Finally, cell proliferation, apoptosis, invasion, and tumor formation ability were assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide dye assay, flow cytometry, transwell assay, and animal experiment, respectively.
RESULTS: RASA1 protein levels were significantly decreased in RKO cells compared with the other 5 colon cancer cell lines, and RASA1 was confirmed as a target gene of miR-21. Interestingly, RASA1 mRNA and protein levels in pre-miR-21-LV (up-regulation of miR-21) cells were lower than those in anti-miR-21-LV (down-regulation of miR-21) cells (P < 0.05). In addition, pre-miR-21-LV or siRASA1 (down-regulation of RASA1) cells showed higher cell proliferation, reduced apoptosis, increased expression of RAS-GTP, p-AKT, Raf-1, KRAS, and p-ERK1/2, and higher invasion and tumor formation ability, compared with control, anti-miR-21-LV or pcDNA3.1-RASA1 (up-regulation of RASA1) cells (P < 0.05).
CONCLUSION: RASA1 is a target gene of miR-21, which promotes malignant behaviors of RKO cells through regulation of RASA1 expression.
Core tip: In this work, we confirmed RAS p21 GTPase activating protein 1 (RASA1) as an miR-21 target and assessed the role of the miR-21/RASA1 axis in colon cancer cells. Our data suggested that miR-21 activated the RAS signaling pathways by down-regulating RASA1 expression, and it played an important role in promoting cell proliferation, anti-apoptosis and tumor cell formation.
-
Citation: Gong B, Liu WW, Nie WJ, Li DF, Xie ZJ, Liu C, Liu YH, Mei P, Li ZJ. MiR-21/RASA1 axis affects malignancy of colon cancer cells
via RAS pathways. World J Gastroenterol 2015; 21(5): 1488-1497 - URL: https://www.wjgnet.com/1007-9327/full/v21/i5/1488.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i5.1488
Colorectal cancer is one of the most common malignant tumors. In the United States, there are nearly 140000 newly diagnosed cases each year, and colorectal cancer constitutes the second cause of cancer-related death[1]. In China, colorectal cancer is the fifth most common tumor and the fourth cause of cancer-related death[2]. In newly diagnosed cases, distant metastasis has been detected in approximately 20% of patients, micro-metastasis in 50%, and about 50% of patients who received radical surgery eventually develop metastasis[3-5]. Various treatment strategies including surgical resection, chemotherapy, and radiation therapy have been used to improve patient survival, but incomplete treatment response, drug resistance, and tumor recurrence are common. Therefore, it is necessary to identify new prognostic markers and explore new therapeutic targets in colorectal cancer.
MicroRNAs (miRNAs) are small noncoding single-stranded RNAs that are 18-25 nucleotides in length[6]. They bind the 3’-untranslated region (UTR) of target mRNAs and block translation or trigger the degradation of the target mRNAs, eventually reducing the synthesis of the coding protein[7]. MiR-21 is highly expressed in 31 tumor types, including colon cancer[8-11], and is thus considered an oncogene.
The development of colon cancer is closely related to abnormal activation of the KRAS signaling pathway. The relationship between the RAS signaling pathways and colon cancer still attracts increasing attention. Interestingly, the EGFR monoclonal antibody was shown to mainly down-regulate the RAS signaling pathways in KRAS wild type (KRAS-wt) colon cancers; in combination with cytotoxic chemotherapy, cetuximab prolongs patient survival[12]. However, KRAS mutant (KRAS-mt) colon cancers are resistant to regulation by EGFR monoclonal antibody due to autonomous activation of the RAS signaling pathways[13]. Nevertheless, a further study showed that for 60%-70% of patients with KRAS-wt colorectal cancers, only about 60% of individuals benefited from EGFR monoclonal antibody[5,14]. This might be due to excessive RAS activation and dysregulation of the RAS signaling pathways, in relation to abnormal or missing expression of some molecules in the RAS signaling pathways such as BRAF mutation, PI3K mutations and phosphatase and tensin homolog (PTEN) or RAS-GAP deletion[15,16] .
RAS proteins are involved in the cycle between RAS-GDP (inactivated form) and RAS-GTP (activated form). When the RAS protein combines with RAS guanine nucleotide exchange factors (RAS-GEFs), RAS-GDP is converted to RAS-GTP, which results in activation of the RAS pathways. On the other hand, combination of RAS with RAS GTPase activating proteins (RAS-GAPs) increases by more than 100000 times the intrinsic GTPase activity of RAS, which leads to hydrolysis of RAS-GTP to RAS-GDP. As a result, the RAS protein is inactivated and the RAS signaling pathways are turned off[17]. Therefore, RAS-GAPs play an important role in KRAS-wt cells but not in mutant cells[18]. When the RAS gene is mutated (very common in cancers), the RAS protein loses its ability to combine with RAS-GAPs, and RAS-GTP continuously activates intracellular signaling pathways, including RAS/RAF/MEK/ERK and RAS/PI3K/AKT pathways, which promote cell proliferation, anti-apoptosis and malignant transformation.
In recent years, studies have demonstrated that RAS wild-type tumors exhibit excessive activation of the RAS protein and dysregulation of the RAS signaling pathways; the mechanisms may be related with down-regulation, deletion of RAS-GAPs or RAS suppressor[16,19]. RASA1 is a member of the RAS-GAP family. This study aimed to assess the role of the miR-21/RASA1 axis in colon cancer cells.
The human colon cancer cell lines RKO, HCT116, colo205, SW480, HT29, and Caco2 were purchased from the Shanghai Institute of Cellular Biology of the Chinese Academy of Sciences (China). The cells were cultured in appropriate medium (MEM–Alpha or RPMI 1640) supplemented with 10% fetal bovine serum (FBS) in a 5% CO2 atmosphere at 37 °C.
Sense and antisense oligonucleotides of human miR-21 lentivirus gene transfer vector harboring green fluorescent protein (GFP) sequence were purchased from Genechem (Shanghai, China). The recombinant lentivirus pre-miR-21 precursor (pre-miR-21-LV), pre-non-coding sequence (pre-NC-LV), anti-miR-21 inhibitor (anti-miR-21-LV), and anti-non-coding sequence (anti-NC-LV) were constructed, packaged and purified by South China Sea Marine Biotechnology Engineering Center Co., LTD (Guangdong, China). Colon cancer RKO cells were chosen for transfection because they are KRAS-wt colon cancer cells whose RASA1 expression is significantly decreased. RKO cells were seeded (2 × 105 cells per well) in a 6-well plate and incubated at 37 °C for 24 h. After 24 h of incubation, cells were counted. According to the multiplicity of infection (MOI) and cell number, the volume of virus was calculated to be added to the culture medium to replace the old medium. Control groups were prepared in parallel. After 12 h of incubation, the medium was refreshed and fluorescence was observed by fluorescence microscopy at 72 h.
The pcDNA3.1 (+) expression vector was purchased from Invitrogen (San Diego, CA). pcDNA3.1-RASA1-GFP (pcDNA3.1-RASA1) was constructed by South China Sea Marine Biotechnology Engineering Center Co., LTD (Guangdong, China). The pcDNA3.1-GFP vector was used as a negative control (RASA1 NC). The vector GV102 used for RASA1 down-regulation was purchased from Genechem (Shanghai, China). The siRASA1 and siRASA1 negative control (siRASA1 NC) vectors were constructed by South China Sea Marine Biotechnology Engineering Center Co., LTD (Guangdong, China). For transfection, RKO cells were seeded (2 × 105 cells per well) in a 6-well plate and incubated at 37 °C for 24 h. Plasmid vectors were then transfected into RKO cells using the liposome transfection method according to manufacturer’s instructions included in the Lipofectamine 2000 reagent kit (Invitrogen, CA, United States).
The expression levels of miR-21 and RASA1 were detected by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) using cells with up/down-regulated miR-21 and RASA1, negative control cells, and non-transfected control cells. Trizol (Invitrogen, CA, United States) was used to isolate total RNA according to the manufacturer’s instructions. RNA was eluted in 30 μL of preheated nuclease-free water (95 °C) and stored at -80 °C until use. First strand cDNA was synthesized by reverse transcription at 42 °C for 60 min followed by 70 °C for 10 min, according to the manufacturer’s instructions for the ReverTra Ace cDNA qPCR RT Kit (Toyobo, Japan). The RT product was used as template for qRT-PCR performed on an ABI PRISM 7500 Quantitative PCR system (Applied Biosystems, Foster City, CA, United States). The qRT-PCR reaction conditions were as follows: 95 °C for 30 s, followed by 45 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 30 s. Each sample was analyzed in triplicate, and expression levels of miR-21 and RASA1 were normalized to those of U6 and ACTB. Relative expression levels were calculated by the 2-ΔΔCt method.
The pGL3-promoter vector was purchased from Promega (Wisconsin, United States). The pGL3-promoter-RASA1 was constructed by South China Sea Marine Biotechnology Engineering Center Co., LTD (Guangdong, China). The pGL3-promoter vector was used as a positive control. The pre-miR-21-LV cells were seeded (3-4 × 105 cells per well) in a 6-well plate. When cells typically reached 90% confluence, they were transfected with the plasmid vectors (pGL3-promoter-RASA1 and pGL3-promoter) using Lipofectamine 2000. After 6 h of incubation, the complete medium was added and further incubated at 37 °C for 72 h. Afterwards, cells were collected and lysed. Then, 100 μL cell lysate and 100 μL luciferase detection reagent (Beyotime, Shanghai) for each sample were mixed to detect luciferase activity in RLU (relative light units), using a GLOMAX 20/20 luminometer (Promega, Madison, Wisconsin, United States). The cell lysis solution was used as a blank control.
Total protein was extracted from cells using the RIPA lysis buffer. After denaturation for 5-10 min at 95-100 °C, the proteins were loaded onto SDS-PAGGE gels for separation, and electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes. The proteins on the membranes were immuno-blotted with primary antibodies followed by goat anti-rabbit or rabbit anti-mouse IgG-HRP secondary antibodies. The primary antibodies included rabbit anti-human RASA1 monoclonal antibody (LifeSpan BioScience), rabbit anti-human Raf1 polyclonal and mouse anti-human KRAS monoclonal antibodies (Abcam, United States), rabbit anti-human AKT polyclonal, rabbit anti-human p-AKT monoclonal, rabbit anti-human ERk1/2 monoclonal, and rabbit anti-human p-ERk1/2 monoclonal antibodies (CST, United States), rabbit anti-human GADPH monoclonal antibody (Bioworld Technology, St. Louis, United States) and rabbit anti-human β-actin polyclonal antibody (Boster, Wuhan). Signals were detected using enhanced chemiluminescence reagents (Invitrogen, CA, United States).
The RAS activity assay was carried out using a RAS Activation Assay Kit (Millipore). Active RAS was pulled down with Raf-1 RBD agarose beads. The pull-down proteins were subjected to SDS-PAGE and immunoblotting analysis with anti-RAS antibody.
Cells with up/down-regulated miR-21 or RASA1, negative control and non-transfected control cells were seeded (1 × 104 cells per well) in a 96-well plate and incubated at 37 °C for 24 h. In parallel, a blank control group containing only medium without cells was established. Each sample was analyzed six times. Cells were incubated for 24, 48, 72, and 96 h, respectively. At the end of each incubation period, the MTT solution (5 mg/mL, 20 μL/well) was added to plates and further incubated at 37 °C for 4 h. After the medium was carefully aspirated, 150 μL of dimethyl sulfoxide was added. Following shaking for 10 min, optical density (OD) was determined at 490 nm using enzyme-labeled instrument (Thermo Scientific, United States) to analyze cell proliferation rates.
1-5 × 105 cells with up/down-regulated miR-21 or RASA1, negative control and non-transfected control cells were collected. Then, 5 μL 7-AAD was mixed with 50 μL Binding Buffer and added to the cells, followed by incubation in the dark at room temperature for 5-15 min. Finally, 450 μL Binding Buffer was added, followed by incubation with 1 μL Annexin V-PE in the dark at room temperature for 5-15 min. The stained cells were detected by flow cytometry on FACSCalibur (BD, New Jersey, United States) within 1 h.
Matrigel (BD) and serum free medium (Gibco, Carlsbad, CA, United States) were diluted at 1:5. The resulting solutions (50 μL) were placed into each transwell chamber (Corning, Guangzhou, China) and incubated at 37 °C. Then, 50 μL serum free medium containing 10 g/L BSA at 37 °C was added to each chamber; cell density was adjusted to 5 × 105/mL and 100 μL suspension was added into each transwell chamber. The medium containing 20% FBS was added to the lower chamber, and incubated at 37 °C. At the end of the incubation, cells were fixed at room temperature with 4% paraformaldehyde for 30 min. Subsequently, cells that crossed the membrane were counted under an optical microscope (ZEISS, Baden-Württemberg, Germany) after crystal violet staining.
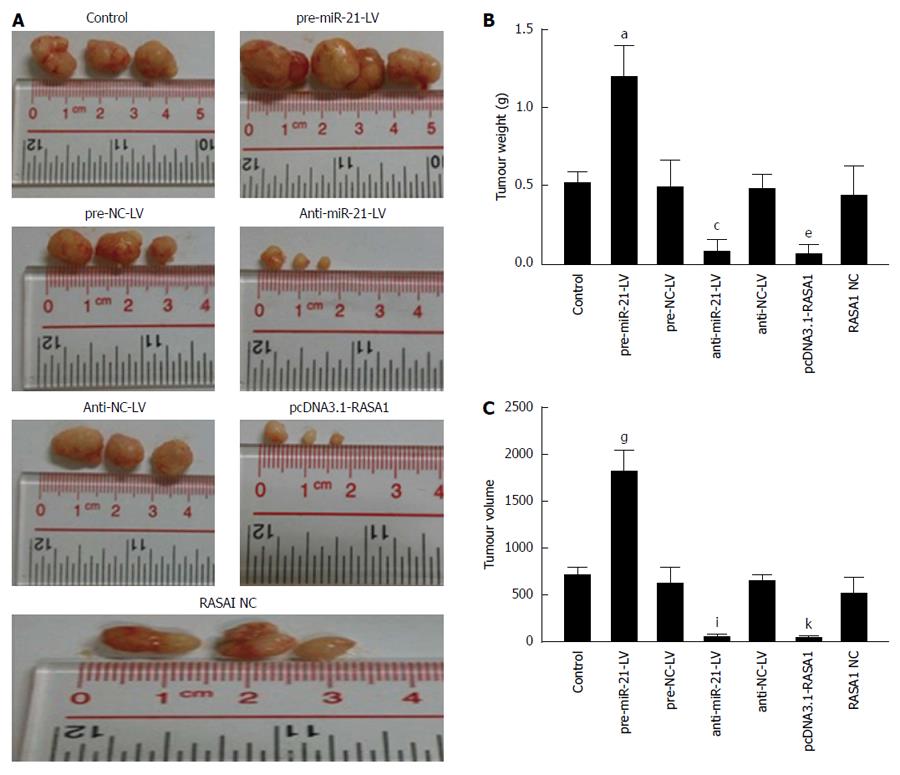
Four-week-old male BABL/c nude mice were obtained from Laboratory Animal Center of Sun Yat-sen University (Guangzhou, China), and divided into 7 groups (n = 3). Cells with up/down-regulated miR-21 or up-regulated RASA1 were resuspended at 2 × 107/mL in serum free medium, and 200 μL cell suspension was injected subcutaneously into BALB/C nude mice (n = 3). The animals were euthanized after 30 d, and tumors were excised and weighted. The tumor volume (mm3) = π/6* [(maximum diameter + minimum diameter)/2]. All animal experiments were approved by the animal care and use committee of Sun Yat-sen University. Institutional Review Boards or Ethics Committees from all participating institutes approved the study protocol.
Data are presented as mean ± standard deviation (SD). Statistical analysis was assessed by one-way or repeated measures analysis of variance (ANOVA), with LSD or Dunnett’s T3 selected for post hoc evaluation. P values < 0.05 were considered statistically significant. The SPSS13.0 software (SPSS Inc., United States) was used for statistical analyses, and all tests were two-sided.
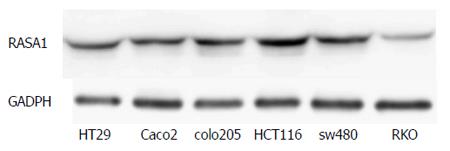
The RASA1 protein expression was assessed by Western blot in various colon cancer cell lines. Of the six colon cancer cell lines studied, the lowest RASA1 protein expression was observed in RKO cells (Figure 1). Therefore, RKO cells were selected for subsequent experiments.
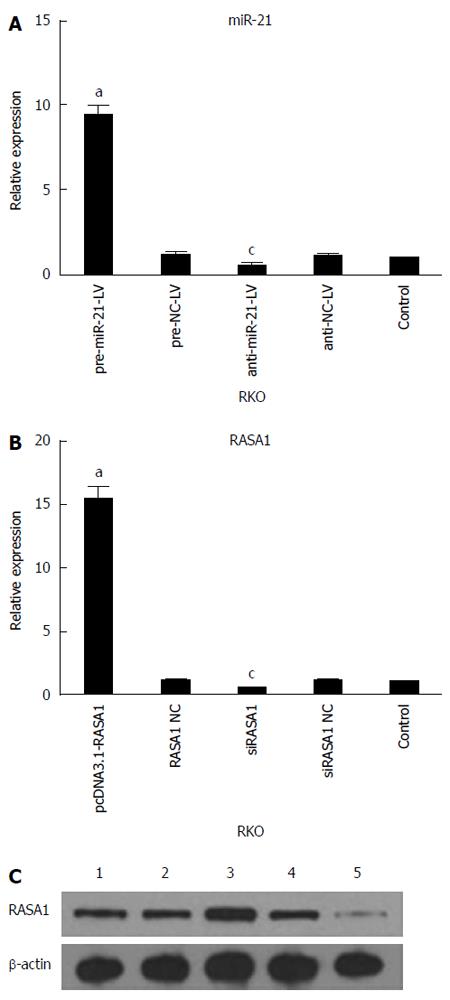
qRT-PCR was used to assess the expression of miR-21 and RASA1 mRNA levels in cells with up/down-regulated miR-21 or RASA1. As shown in Figure 2, miR-21 expression was higher in pre-miR-21-LV cells than in anti-miR-21-LV or control cells (P < 0.05), while it was lower in anti-miR-21-LV cells compared with control cells (P < 0.05). These findings indicated successful RKO cell transfection with lentivirus harboring pre-miR-21-LV and anti-miR-21-LV, which enhanced and inhibited the expression of miR-21, respectively.
The expression of RASA1 mRNA and protein in pcDNA3.1-RASA1 cells was higher than in siRASA1 and control cells (P < 0.05); however, RASA1 mRNA and protein levels in siRASA1 cells were lower than those in control cells (P < 0.05) as shown in Figure 2. These data indicated successful transfection with pcDNA3.1-RASA1 and siRASA1; cells with up/down-regulated RASA1 were harvested.
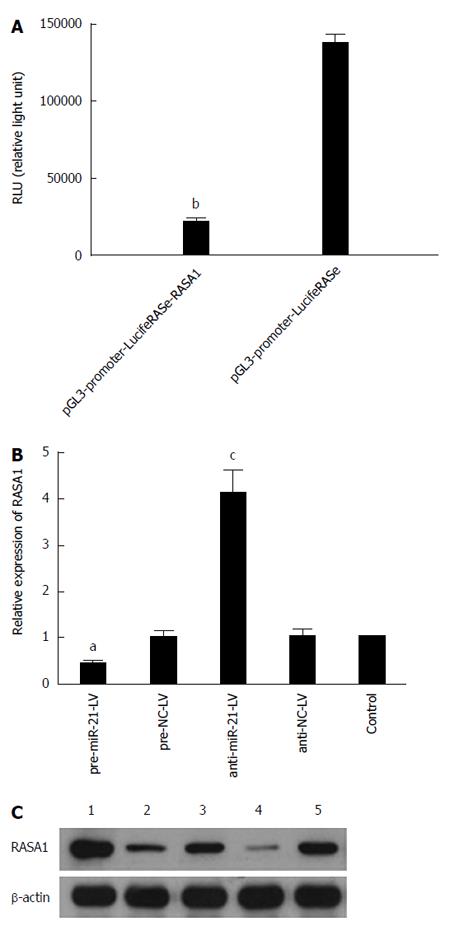
In order to unveil the molecular mechanism underlying the role of miR-21 in the development of cancer, bioinformatics software (PicTar, TargetScan and miRbase) was used to predict RASA1 as an miR-21 target gene; indeed, the 179-185 locus of 3’-UTR in RASA1 is complementary to miR-21. Based on this finding, luciferase reporter gene assay was performed to verify whether RASA1 is a target of miR-21. In pre-miR-21-LV cells co-transfected with pGL3-promoter-LucifeRASe-RASA1, lower RLU (relative light unit) values were obtained, compared with their counterparts co-transfected with pGL3-promoter-LucifeRASe (P < 0.01) (Figure 3A); these findings confirmed RASA1 as an miR-21 target.
To further verify these results, RASA1 mRNA and protein expression was detected in pre-miR-21-LV and anti-miR-21-LV cells. As shown in Figure 3B and C, RASA1 mRNA and protein levels in pre-miR-21-LV cells were significantly lower (P < 0.05), while significantly higher RASA1 mRNA and protein levels were observed in anti-miR-21-LV cells (P < 0.05).
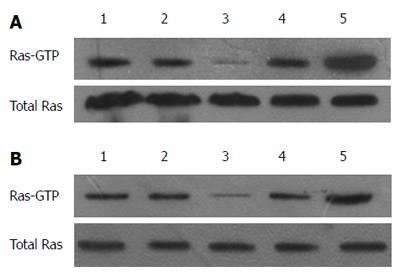
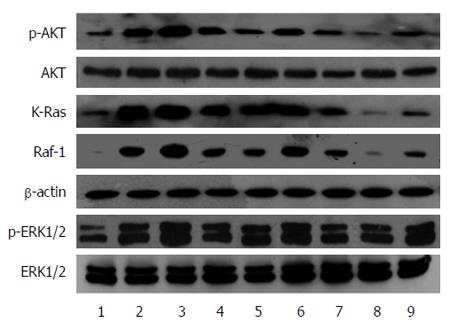
RAS activity is well known to be regulated by two types of molecules: RAS-GEFs and RAS-GAPs. RAS-GAPs can hydrolyze RAS-GTP into RAS-GDP, to inactivate RAS and switch off the RAS signaling pathways. RASA1 belongs to the RAS-GAP family, which is composed of 16 members. As shown above, miR-21 targets and regulates the expression of RASA1. RAS-GTP levels, which indicate RAS protein activity, were assessed in cells with up/down-regulated miR-21 and RASA1. As shown in Figure 4, overexpression of miR-21 enhanced RAS-GTP activity, so did down-regulation of RASA1; conversely, down-regulation of miR-21 or RASA1 overexpression reduced RAS-GTP activity.
The high levels of RAS-GTP suggested the activation of RAS proteins; RAS protein activation would amplify the related signaling pathways in cells, including RAS-Raf-MAPK and RAS-PI3K-AKT pathways, which promote cell proliferation, anti-apoptosis, and induce cell malignant transformation. We analyzed the changes in Raf-1, KRAS, AKT and p-AKT, ERK1/2 and p-ERK1/2 protein expression in cells with up/down-regulated miR-21 or RASA1. As shown in Figure 5, KRAS, Raf-1, p-AKT and p-ERK1/2 protein levels in cells with down-regulated miR-21 or overexpressed RASA1 were lower than those obtained after miR-21 overexpression or RASA1 down-regulation. These findings suggested that miR-21 might activate the RAS signaling pathways by down-regulating RASA1 expression, therefore playing an important role in promoting cell proliferation, anti-apoptosis and tumor cell formation.
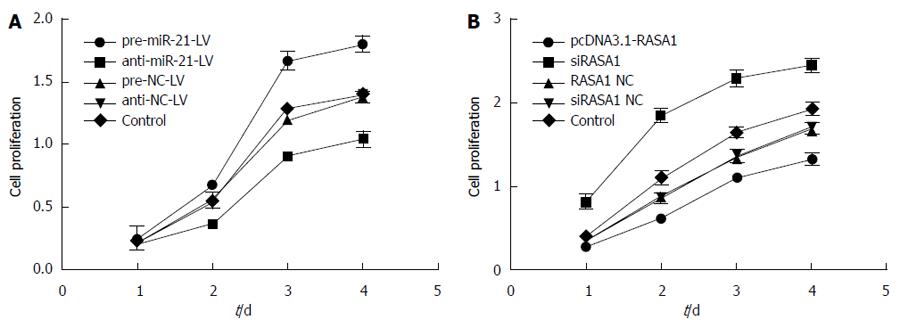
The effect of miR-21 and RASA1 on cell proliferation was detected by MTT assay. In pre-miR-21-LV cells, overtly higher OD values were obtained at different time points compared with those in anti-miR-21-LV and control cells (P < 0.01); meanwhile, transfection with pcDNA3.1-RASA1 also resulted in lower OD values, compared with those obtained in siRASA1 and control cells (P < 0.01) (Figure 6). These data indicated that low expression of miR-21 or RASA1 overexpression inhibited cell proliferation.
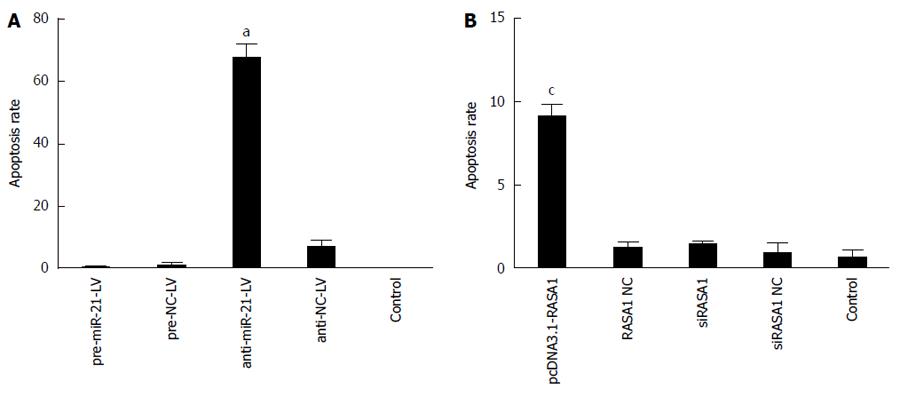
The role of miR-21 in RKO cell apoptosis via RASA1 regulation was analyzed after annexin V-PE/7-AAD double staining by flow cytometry. As shown in Figure 7, early apoptosis rate was higher in anti-miR-21-LV cells than in pre-miR-21-LV and control cells (P < 0.05); similar results were obtained for pcDNA3.1-RASA1 harboring cells (P < 0.05). These findings indicated that in RKO cells, overexpression of miR-21 inhibited cell apoptosis by down-regulating RASA1.
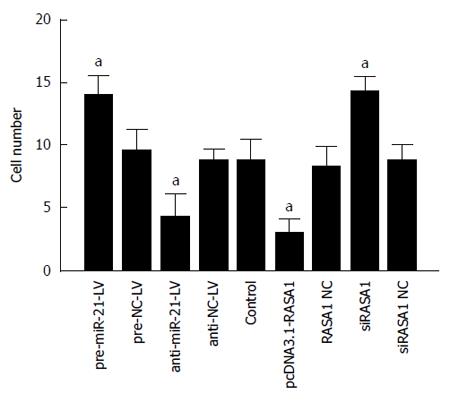
Malignant tumors are characterized by their invasive ability and metastasis. We confirmed that miR-21 and RASA1 affected RKO cell invasion ability by transwell assays. The numbers of cells that migrated through the membrane in anti-miR-21 and pcDNA3.1-RASA1 groups were significantly lower, compared with those obtained with pre-miR-21-LV, siRASA1 and control cells (P < 0.01) (Figure 8). These data suggested that miR-21, as an oncogene, promoted tumor cell invasion, likely through regulation of RASA1, which acts as a tumor suppressor and inhibits cell invasion.
In order to further verify the biological effects of miR-21 and RASA1 in vivo, a nude mouse tumor transplantation experiment was carried out. As shown in Figure 9, tumor weights in anti-miR-21-LV or pcDNA3.1-RASA1 groups were lower than those observed in pre-miR-21-LV animals (P < 0.05); in agreement with this, tumor volumes were smaller in anti-miR-21-LV or pcDNA3.1-RASA1 groups than in pre-miR-21-LV mice (P < 0.05) (Figure 9A-C). These data indicated that down-regulation of miR-21 resulted in decreased tumor formation, likely through up-regulation of RASA1.
The current clinical application of colon cancer targeted therapy such as cetuximab or panitumumab mainly aims at the activity of the RAS signaling pathways. It is known that RAS proteins often are in the recurrent states of activated RAS-GTP and inactivated RAS-GDP. RAS-GAPs function in KRAS-wt cells, not in the mutant cells. RAS gene mutations result in constitutively active state of RAS-GTP and open RAS signaling pathways, thereby facilitating cell proliferation, anti-apoptosis activity and malignant transformation.
So far, 16 members have been described in the RAS-GAP family, including p120GAP (RASA1), NF1, DAB2IP, GAP1m (or RASA2), GAP1IP4BP (orRASA3), CAPRI (orRASA4), RASAL1, SYNGAP, AF9Q34, FLJ21438, IQGAP1, IQGAP2, IQGAP3, PLXNB2, and TUBERIN[16,19]. The RASA1 gene is located on 5q13.3 and encodes the p120GAP protein, which is composed of 1047 amino acid residues[20].
Recent studies have found that in KRAS-wt tumors, because of down-regulation or inefficiency of the RAS-GAPs, there is activation of the RAS signaling pathways, which play a role in tumorigenesis. Interestingly, it was found that RASAL1 expression was significantly lower in KRAS-wt colon cancer cells than in mutant cells[16]. Calvisi et al[19] assessed the expression profile of RAS-GAPs in liver cancer tissue and found reduced expression of DAB2IP, RASAL1 and NF1; of note, the low expression of DAB2IP was significantly associated with poor prognosis in KRAS-wt liver cancer. These findings suggested RAS-GAPs to be tumor suppressor genes. RASA1 is an important member of the RAS-GAP family. It was demonstrated that RASA1 expression in colon cancer tissues was significantly lower compared with that in normal adjacent tissues (NAT)[21]. In addition, Hu et al[22] found markedly down-regulated RASA1 mRNA and protein expression in intrahepatic cholangiocarcinoma tissues and cells, compared with that in normal adjacent tissues .
Therefore, we hypothesized that RASA1, an important member of the RAS-GAP family, which is significantly down-regulated in colon cancer, may negatively regulate the RAS signaling pathways and act as a tumor suppressor gene. Indeed, we found that RASA1 up-regulation inhibited cell proliferation and turned off the RAS signaling pathways.
Many studies have demonstrated that miR-21 is overexpressed in most tumors, including colon cancer. It was proposed that miR-21 promotes tumor formation by down-regulating tumor suppressor genes, such as programmed cell death 4 (PDCD4), tissue inhibitors of metalloproteinase 3 (TIMP3)[23], and PTEN[24]. Herein, we have established RASA1 as an miR-21 target using a luciferase reporter gene assay (Figure 2A). Meanwhile, qRT-PCR and Western blot analyses confirmed that overexpression of miR-21 inhibited RASA1 expression (Figure 2B and C). In addition, we found that miR-21 was overexpressed in colon cancer tissues (unpublished data). As shown above, overexpression of miR-21 increased RAS-GTP activity in RKO cells; conversely, down-regulation of miR-21 inhibited the activity of RAS-GTP. Further analyses demonstrated that miR-21 mediated EGF/RAS signaling pathways in cancer cells through RASA1. There are mainly two pathways that promote tumor cell proliferation, anti-apoptosis and tumor formation: the RAS-Raf-1-MEK-MAPK and RAS-PI3K-AKT signaling pathways[25]. Our findings confirmed that high miR-21 levels inhibited the expression of RASA1 and enhanced the expression of Raf-1, p-AKT, and p-ERK, thus promoting RKO cell proliferation, invasion, and anti-apoptotic activity (Figures 4-7). In addition, using a nude mouse tumor transplantation model, we demonstrated that both low levels of miR-21 or RASA1 overexpression lead to in vivo inhibition of tumor formation.
In conclusion, miR-21 participates in the development of colon cancer by targeting RASA1. These data provide a basis for new targeted therapy in colon cancer. This is especially important in view of drug resistance observed with colon cancer targeted drug cetuximab or panitumumab, which is widely used clinically; this may also be associated with loss or down-regulation of RAS-GAPs. Therefore, finding valuable RAS-GAP candidates would provide the proof of principle that helps design new therapeutic agents to overcome cetuximab or panitumumab resistance.
The understanding of the pathogenesis of colon cancer and the successful application of targeted therapy, such as cetuximab or panitumumab, are mainly based on the research of the RAS signal pathways. However, due to the deregulation or deletion of some key molecules in the pathways, the RAS signaling pathways are activated persistently. As a result, drug resistance develops in cancer cells. Normally RAS-GAPs and RAS-GEFs are in a state of balance, and regulate the activation of the RAS signaling pathways. RAS p21 GTPase activating protein 1 (RASA1) is a member of RAS-GAP family. MiR-21 is the most common up-regulated miRNA in most cancers and thus is considered an oncogene. However, how the oncogene miR-21 regulates RASA1 and affects the cell behaviors of colon cancer is unknown.
RAS pathways play an important role in the development of colon cancer. At present, the targeted therapy drugs including cetuximab and panitumumab are targeted to the RAS signaling pathways. MiR-21 was overexpressed in colon cancer tissue and cells. However, few studies reported the relationship between miR-21 and RAS signaling pathways. The roles of RAS-GAP molecules regulating the RAS signaling pathway in carcinogenesis are hotspots of current research. However, the relationship between miR-21 and RASA1, and how they regulate the RAS signaling pathway and affects biological behaviors of colon cancer have not been reported.
Both miR-21 and RASA1 play an important role in the development of colon cancer. We demonstrated that RASA1 is a target of miR-21. This findings also confirmed that high miR-21 levels inhibited the expression of RASA1. Down-regulation of miR-21 or up-regulation of RASA1 promoted apoptosis, and inhibited the proliferation, invasion, and tumor formation of colon cancer cells.
MiR-21 and RASA1 may provide the proof of principle that helps design new targeted therapeutic agents to cure colon cancer and overcome cetuximab or panitumumab resistance.
This is a very interesting study comprehensively exploring the role of RASA1 and its inhibitor miR21 in colon cancer cells.
P- Reviewer: Formica V S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8283] [Cited by in F6Publishing: 8159] [Article Influence: 509.9] [Reference Citation Analysis (0)] |
| 2. | Zhang YL, Zhang ZS, Wu BP, Zhou DY. Early diagnosis for colorectal cancer in China. World J Gastroenterol. 2002;8:21-25. [PubMed] [Cited in This Article: ] |
| 3. | Goh S. Systematic review and meta-analysis of the evidence for flexible sigmoidoscopy as a screening method for the prevention of colorectal cancer (Br J Surg 2012; 99: 1488-1500). Br J Surg. 2013;100:1540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 4. | Wilke HJ, Van Cutsem E. Current treatments and future perspectives in colorectal and gastric cancer. Ann Oncol. 2003;14 Suppl 2:ii49-ii55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 36] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Lamas MJ, Duran G, Gallardo E. Anti-EGFR therapy in first-line colorectal cancer. Expert Rev Anticancer Ther. 2011;11:1499-1503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 6. | Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 946] [Cited by in F6Publishing: 961] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 7. | Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1750] [Cited by in F6Publishing: 1673] [Article Influence: 111.5] [Reference Citation Analysis (0)] |
| 8. | Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799-2803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1136] [Cited by in F6Publishing: 1170] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 9. | Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113-2129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 738] [Cited by in F6Publishing: 770] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 10. | Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915-1922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 220] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 11. | Ziyan W, Shuhua Y, Xiufang W, Xiaoyun L. MicroRNA-21 is involved in osteosarcoma cell invasion and migration. Med Oncol. 2011;28:1469-1474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 12. | Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757-1765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2724] [Cited by in F6Publishing: 2678] [Article Influence: 167.4] [Reference Citation Analysis (0)] |
| 13. | Troiani T, Zappavigna S, Martinelli E, Addeo SR, Stiuso P, Ciardiello F, Caraglia M. Optimizing treatment of metastatic colorectal cancer patients with anti-EGFR antibodies: overcoming the mechanisms of cancer cell resistance. Expert Opin Biol Ther. 2013;13:241-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | Asghar U, Hawkes E, Cunningham D. Predictive and prognostic biomarkers for targeted therapy in metastatic colorectal cancer. Clin Colorectal Cancer. 2010;9:274-281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, Byun DS, Nasser S, Arango D, Shin J, Klampfer L. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68:1953-1961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 354] [Cited by in F6Publishing: 378] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 16. | Ohta M, Seto M, Ijichi H, Miyabayashi K, Kudo Y, Mohri D, Asaoka Y, Tada M, Tanaka Y, Ikenoue T. Decreased expression of the RAS-GTPase activating protein RASAL1 is associated with colorectal tumor progression. Gastroenterology. 2009;136:206-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Wittinghofer A. Signal transduction via Ras. Biol Chem. 1998;379:933-937. [PubMed] [Cited in This Article: ] |
| 18. | Wittinghofer A, Scheffzek K, Ahmadian MR. The interaction of Ras with GTPase-activating proteins. FEBS Lett. 1997;410:63-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 87] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 19. | Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, Thorgeirsson SS. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol. 2011;54:311-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 20. | Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47-82. [PubMed] [Cited in This Article: ] |
| 21. | Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, Zhang CY, Chen J, Zhang J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol Chem. 2013;288:9508-9518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 145] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 22. | Hu C, Huang F, Deng G, Nie W, Huang W, Zeng X. miR-31 promotes oncogenesis in intrahepatic cholangiocarcinoma cells via the direct suppression of RASA1. Exp Ther Med. 2013;6:1265-1270. [PubMed] [Cited in This Article: ] |
| 23. | Selaru FM, Olaru AV, Kan T, David S, Cheng Y, Mori Y, Yang J, Paun B, Jin Z, Agarwal R. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009;49:1595-1601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 218] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 24. | Wang ZX, Lu BB, Wang H, Cheng ZX, Yin YM. MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch Med Res. 2011;42:281-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 25. | Kern HB, Niemeyer BF, Parrish JK, Kerr CA, Yaghi NK, Prescott JD, Gutierrez-Hartmann A, Jedlicka P. Control of MicroRNA-21 expression in colorectal cancer cells by oncogenic epidermal growth factor/Ras signaling and Ets transcription factors. DNA Cell Biol. 2012;31:1403-1411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |