Published online Sep 21, 2015. doi: 10.3748/wjg.v21.i35.10150
Peer-review started: January 27, 2015
First decision: February 10, 2015
Revised: April 1, 2015
Accepted: June 9, 2015
Article in press: June 9, 2015
Published online: September 21, 2015
AIM: To analyze the polymorphisms of CTLA-4 gene involved in the response against hepatitis C virus (HCV) infection.
METHODS: We recruited 500 hemodialysed patients from several hemodialysis centers, all HCV-antibody positive, spread over different regions of Tunisia, as part of a national survey in 2008 conducted in the laboratory of immunology at the Charles Nicolle hospital Tunisia, classified into two groups G1 (PCR+) and G2 (PCR-) according to the presence or absence of viral RNA. Of these patients, 307 were followed prospectively on a viral molecular level over a period from 2002 to 2008, divided into two groups based on the persistence and viral clearance. PCR-RFLP was performed for the analysis of SNPs (+49) A/G and (+6230) G/A CTLA-4 for these 500 patients and 358 healthy controls.
RESULTS: Analysis of clinical and virological characteristics of our cohort suggests a nosocomial infection in our hemodialysed patients with transfusion history as a primary risk factor and a predominance of genotype 1b. The haplotype analysis revealed an increase of frequencies of GG (+49)/(CT60) CTLA-4 in the entire patients group compared to controls (P = 0.0036 and OR = 1.42; 95%CI: 1.12-1.79, respectively). This haplotype is therefore associated with susceptibility to HCV infection.
CONCLUSION: Our study suggests a possible role of CTLA-4 polymorphisms in the outcome of HCV infection in the Tunisian hemodialysed population.
Core tip: Clinical and virological characteristics of our cohort suggest a nosocomial hepatitis C virus (HCV) infection in Tunisian hemodialysis patients with transfusion history as a primary risk factor and a predominance of genotype 1b. No significant association was found for the two CTLA-4 SNPs studied either to spontaneous clearance, persistence or protection against HCV infection. The GG (+49)/(CT60) CTLA-4 haplotype is therefore associated with susceptibility to HCV infection. The study of other susceptibility genes for HCV infection will certainly allow a better understanding of the molecular mechanisms of spontaneous viral clearance or persistence of HCV infection.
- Citation: Ksiaa Cheikhrouhou L, Lakhoua-Gorgi Y, Sfar I, Jendoubi-Ayed S, Aouadi H, Makhlouf M, Ayed K, Ben Abdallah T. Natural evolution of hepatitis C virus infection in hemodialysis Tunisian patients and CTLA-4 SNP's. World J Gastroenterol 2015; 21(35): 10150-10158
- URL: https://www.wjgnet.com/1007-9327/full/v21/i35/10150.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i35.10150
Hepatitis C is relatively a common disease. An estimated 3% of the world population is chronically infected, and hepatitis C virus (HCV) is responsible of about 70% of cases of chronic hepatitis, cirrhosis and a major cause of hepatocellular carcinoma (HCC)[1]. Only 20% of infected patients spontaneously clear the virus.
The reason for this variation in disease expression is unknown and has been correlated with a strong immune response. Both CD4+ T helper and CD8+ cytotoxic T lymphocyte (CTL) responses are important in the response to HCV infection[2].
In early HCV infection, a vigorous CD4+ T cell response is associated with viral clearance. In contrast, patients developing a chronic infection show a predominant Th2 response. These findings indicate that the ability to mount an efficient cellular immune response is the main mechanism responsible for HCV control, while a defect in this response leads to chronicity[3].
Host genetic factors that govern these responses may also modify the course of HCV infection. Polymorphisms in HLA molecules, as well as inflammatory molecules genes, appear to be associated with natural clearance and chronic progression of HCV infection[4,5]. We and others have recently described the association of polymorphisms in chemokine and cytokine genes with both clearance and progression of HCV infection[6,7].
Differences in chemokine and cytokine expression between Th1 and Th2 cells might explain the regulating T helper cell polarization and their selective recruitment to liver tissue.
The CTL antigen-4 (CTLA-4), encoded by a gene on chromosome 2q33, is expressed on activated CD4+ and CD8+ T cells. It binds to the ligands B7-1 (CD80) and B7-2 (CD86) and down regulates T cell function[8]. Mice deficient in CTLA-4 exhibit polyclonal T cell activation and proliferation[9]. CTLA-4 gene has several polymorphic markers and the most frequently studied are +49 A/G in exon 1 and CT60 at 3’ untranslated region[10]. Change of +49A for allele G causes threonine to alanine conversion in CTLA-4 protein. In vitro study demonstrated that the presence of the G allele results in inefficient CTLA-4 glycosylation and reduced cell surface expression. The CT60 polymorphism represents substitution of A to G at +6230 region leading to reduced mRNA expression[11]. These polymorphisms have been extensively studied with association to several autoimmune disorders and infectious diseases.
The aim of this study was to investigate the distribution of SNPs (+49) A/G and (CT60) G/A CTLA-4 in HCV infected hemodialysis Tunisian patients in comparison with healthy controls. Furthermore, we have analyzed the association of particular genotypes with outcomes of HCV infection, in terms of susceptibility to HCV infection, spontaneous clearance or viral persistence.
This retrospective study involved 500 HCV-infected individuals dialyzed with confirmed antibody positivity to HCV. Dialyzed patients were recruited from different hemodialysis centers spread over different regions of Tunisia, as part of the national survey conducted in 2008 in the Laboratory of Immunology at the Charles Nicolle hospital, Tunisia. They were negative for hepatitis B surface antigen (HBsAg) and HIV infection. Data from each patient included age at diagnosis, gender and possible risk factors for HCV (such as transfusion) were obtained at the hemodialysis centers. Total patients were matched in age and gender and were divided into two groups according to the presence or absence of viral RNA in 2008: Group 1 (G1) included 240 patients who were HCV-RNA positive; and Group 2 (G2) consisted of 260 subjects who were HCV-RNA negative. Among the 500 patients, 307 were prospectively followed at a serological and molecular level of HCV infection over a period of 6 years, from 2002 to 2008. These patients were classified into: “Persistence group” which included 159 patients with persistent HCV infection as assessed by two positive PCR tests for HCV-RNA, and “Clearance group” consisting of 148 subjects considered to have spontaneously recovered from HCV infection as suggested by two negative consecutive HCV-PCR detections one year apart. None of these patients had received treatment for HCV infection before entering the study. In addition, blood samples were obtained from 358 ethnically and geographically matched healthy individuals who tested negative for HBsAg, HIV-Ab and HCV-Ab. These 358 subjects served as a control group. The study was approved by the ethics committee of Charles Nicolle Hospital (Tunis, Tunisia) and all patients gave informed consent.
HCV RNA in serum was detected by RT-PCR (Inno-Lipa HCV II, Innogenetics, Belgium) according to the manufacturer’s instructions. Patients who were HCV-PCR positive on the initial assessment and became consistently HCV-PCR negative were classified HCV-PCR negative.
HCV genotypes were determined by inverse hybridization using a specific oligonucleotide probe assay (HCV Genotype Assay Lipa Innogenetics).
Blood samples were collected in EDTA, and DNA was isolated by the Salting-Out method reported by Miller et al[12].
Typing of exon 1 A/G transition at position +49 and 3’ UTR (+6230) G/A CTLA-4 gene polymorphisms was achieved by the PCR-RFLP (Restriction Fragment Length Polymorphism) method using a PCR System 2700 Thermal Cycler (Applied Biosystems, Gene Amp®). The PCR protocol and the primers used are listed in Table 1. PCR was carried out in a final volume of 25 μL containing 50 ng of genomic DNA, 1.5 mmol/L MgCl2, 0.2 mmol/L dNTP, 10 pmol of each primer and 0.5 U of Taq DNA polymerase (Promega United States). All PCR products were confirmed by 2% agarose gel electrophoresis. Three microliters of reaction mixture from each sample was digested with 5 U KpnI restriction enzyme for (+49) CTLA-4, 5 U NcoI restriction enzyme for CT60 CTLA-4 at 37 °C for 16 h. These products were loaded into 4% agarose gels and stained by ethidium bromide.
| Polymorphisms | Primers | Temperature, time and cycles for PCR |
| CTLA-4(+49) A/G | Forward: | Initial denaturation for 4min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 67 °C and 1 min at 72 °C and final elongation 5 min at 72 °C |
| 5’CAAGGCTCAGCTGAACCTGGGT3’ | ||
| Reverse: | ||
| 5’TACCTTTAACTTCTGGCTTTG3’ | ||
| CTLA-4(+6230) G/A | Forward | Initial denaturation for 5 min at 94 °C, 30 cycles of 40 s at 94 °C, 30 s at 61 °C and 50 s at 72 °C and final elongation 7 min at 72 °C |
| CT60 Sens: | ||
| 5’ CACCACTATTTGGGATATACC 3’ | ||
| Reverse | ||
| CT60 Antis: | ||
| 5’ AGCTCTATATTTCAGGAAGGC 3’ |
Genotype and allele frequencies were performed using the SPSS 17.0. Haplotype frequencies and Hardy-Weinberg equilibrium P values were estimated by the web site http://bioinfo.iconcologia.net/snpstats/start.html. Statistical comparisons were made between the different groups of patients and controls by the χ2 test calculated on 2 × 2 contingency tables. The Fisher’s exact test was used when expected cell values were less than 5. A P value less than 0.05 was considered statistically significant. The strength of the association between genotypes or alleles in each group was estimated by calculating the odds ratios (OR) and 95%CI using the same software. Logistic regression models were used to evaluate the relationships with the different factors (including confounders) and to estimate adjusted ORs (Exp(B)).
The clinical and virological characteristics of the total patient population are summarized in Table 2. Two hundred and forty eight men and 252 women with a mean age of 54.8 ± 14.17 years were recruited from 102 dialysis centers (23 from the public sector and 79 from the private sector) with a regional distribution as follows: 44% in Tunis, 26.2% in the South, 10% in the North West, 9.2% in Central, 6.6% in the North and 4% in the North East. One hundred and sixty nine patients (71%) had a history indicating risk of exposure to HCV due to blood transfusions and 53.8% due to surgical or medical invasive procedures. Further characteristics are listed in Table 2.
| Characteristics | Patients (n = 500) |
| Gender M/F (SR) | 248/252 (0.98) |
| Age (mean ± SD, yr) | 54.8 ± 14.17 |
| Regional distribution | |
| Tunis | 220 (44) |
| North west | 50 (10) |
| North | 33 (6.6) |
| North East | 20 (4) |
| Center | 46 (9.2) |
| South | 131 (26.2) |
| Risk factors | |
| Blood transfusions | 169 (71) |
| Invasive procedures | 128 (53.8) |
| Erythropoïetin | 103 (25) |
| Sector | |
| Private | 405 (80) |
| Public | 94 (20) |
| Hemodialysis average duration (mo) | 118.17 ± 71.74 |
G1 was composed of 121 males and 119 females (SR = 1.01), with a mean age of 54.7 ± 14.10 years. Of G2, 127 were males and 133 were females (SR = 0.95) with a mean age of 54.8 ± 14.6.
In the persistence group, 82 were males and 77 female (SR = 1.06, mean age = 54.7 ± 14.10 years). The clearance group were composed of 78 males and 70 females (SR = 1.11, mean age = 55 ± 14.01 years).
According to Table 3, we note that private dialysis centers are much more common than public centers. Nevertheless, this sectorization is not associated with viral replication. Indeed, the private sector was represented in 79.6% against 20.4% for the public sector in G1 and 82.3% vs 17.7% in G2.
| Groups | n | Region | No. of centers | n (%) | |||||||
| T | NW | N | NE | C | S | Pv | Pq | Pv | Pq | ||
| G1 | 240 | 105 (43.7) | 25 (10.4) | 1 6 (6.7) | 10 (4.2) | 22 (9.2) | 62 (25.8) | 65 | 19 | 191 (79.6) | 49 (20.4) |
| G2 | 260 | 115 (44.3) | 25 (9.6) | 17 (6.5) | 10 (3.8) | 24 (9.2) | 69 (26.6) | 72 | 24 | 214 (82.3) | 46 (17.7) |
| Persistence | 159 | 65 (40.9) | 13 (8.2) | 11 (6.9) | 10 (6.3) | 19 (11.9) | 41 (25.9) | 58 | 12 | 133 (83.6) | 26 (16.4) |
| Clearance | 148 | 52 (35.2) | 16 (10.8) | 6 (4) | 6 (4) | 17 (11,5) | 51 (34.5) | 53 | 10 | 132 (89.2) | 16 (10.8) |
However, while the frequency of the private sector was similar in the two groups of clearance (89.2%) and persistence (83.6%), an increase in the frequency of the public sector in the persistence group (16.4%) compared to that in the clearance group (10.8%) was observed.
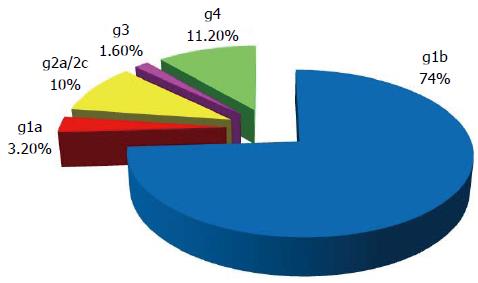
Overall the distribution of different genotypes in G1 reveals a predominance of genotype 1b (73.5%), followed by genotype 4 (11.2%), genotype 2a/2c (10%), then genotype 1a (3.2%) and genotype 3 (1.6%) (Figure 1) (HCV genotypes have been determined in 189/240 patients with active viral replication during the 2008 survey).
There is a great variability in the regional distribution of genotypes of HCV, indeed, genotype 1b is observed in all regions of Tunisia. It predominates in the center (90.4%), the South (86.7%) and North West (83.3%). This genotype is less common in Tunis (69.7%), North East (50%) and the North (45.5%). Apart from this genotype some areas are characterized by a relatively high frequency of a second genotype. This is the case for genotype 2a/2c in the Northern region (45.5%) and the South (13.3%) and also for HCV-4 with a frequency range from 40% in the North East to 9% in the North through the Tunis region (14.1%) and the North West (11.1%). The frequency of genotype 1a is relatively low, in the order of 6.1% in the Tunis region and 4.8% in the Center. HCV genotype-3 was observed in the North West (5.6%) and Tunis (2%).
HCV genotypes were analyzed in only 142/159 patients with persistent HCV infection and in 39/148 hemodialysis patients who spontaneously cleared the virus during the period 2002 to 2008 because the serial and annual harvest of these 39 patients were positive in the years 2002-2003 and consequently HCV-RNA could be genotyped. As shown in Table 4, if the frequencies of the genotypes 1b and 4 are similar, HCV 2a/2c is observed among patients with persistent viral replication compared to those having cleared the virus spontaneously. However, this difference is not statistically significant.
| Genotypes | Persistence (n = 142) | Clearance (n = 39) |
| 1b | 105 (73.9) | 30 (76.9) |
| 1a | 2 (1.4) | 0 |
| 1a/1b | - | 1 (2.6) |
| 2a/2c | 14 (9.9) | 1 (2.6) |
| 3 | 2 (1.4) | 1 (2.6) |
| 4 | 19 (13.4) | 5 (12.8) |
| 1b/4 | - | 1 (2.5) |
The genotype and allele frequencies of CTLA-4 gene polymorphisms in hemodialysis patients and in the control group are shown in Table 5.
| Haplotypes | Controls | Patients | G1 | G2 | Persistence | Clearance |
| (n = 358) | (n = 500) | (n = 240) | (n = 260) | (n = 159) | (n = 148) | |
| GA | 39% | 29.5% | 36% | 34.5% | 35.2% | 36% |
| GG | 29.9% | 35.3%1 | 26.7% | 32.2% | 28.3% | 32.9% |
| AG | 26.8% | 25.2% | 25.7% | 24.9% | 26.5% | 22.8% |
| AA | 4.2% | 10% | 11.6% | 8.4% | 10% | 8.3% |
Comparison of genotype and allele frequencies in all HCV-infected patients and controls and between other different groups of patients did not reveal a significant difference.
Haplotypes formed by variants (+49) A/G and (CT60) G/A were reconstructed in the cohort of 500 patients and the normal controls. A near-complete linkage disequilibrium (LD) exists between these two variants (D = 0.0627, D’ = 0.45, r = 0.27). The four haplotypes (GG, GA, AA and AG) were present in all groups studied (Table 6). The distribution of haplotypes differed significantly between the total HCV infected patients and healthy control groups. The (+49) G (CT60) G haplotype was present in more of the entire patient group compared to healthy controls (35.3% vs 29.9%, P = 0.0036; OR = 1.42; 95%CI: 1.12-1.79). No significant association was found according to the presence or absence of RNA HCV, or to the persistence or clearance of HCV infection.
| SNPs | Controls | Patients | G1 | G2 | Persistence | Clearance |
| (n = 358) | (n = 500) | (n = 240) | (n = 260) | (n = 159) | (n = 148) | |
| (+49) A/G CTLA4 | ||||||
| Genotypes | ||||||
| AA | 40 (11.2) | 77 (15.4) | 43 (17.9) | 34 (13.1) | 25 (15.7) | 19 (12.84) |
| AG | 142 (39.7) | 198 (39.6) | 93 (38.8) | 105 (40.4) | 66 (41.5) | 54 (36.5) |
| GG | 176 (49.2) | 225 (45) | 104 (43.3) | 121 (46.5) | 68 (42.8) | 75 (50.7) |
| Alleles | ||||||
| A | 0.31 | 0.352 | 179 (37.3) | 173 (33.3) | 116 (36.5) | 92 (31.1) |
| G | 0.69 | 0.648 | 301 (62.7) | 347 (66.7) | 202 (63.5) | 204 (68.9) |
| (+6230) G/A CTLA4 | ||||||
| Genotypes | ||||||
| GG | 124 (34.6) | 194 (38.8) | 97 (40.4) | 97 (37.3) | 65 (40.9) | 55 (37.2) |
| GA | 158 (44.1) | 217 (43.4) | 102 (42.5) | 115 (44.2) | 66 (41.5) | 64 (43.2) |
| AA | 76 (21.3) | 89 (17.8) | 41 (17.1) | 48 (18.5) | 28 (17.6) | 29 (19.6) |
| Alleles | ||||||
| G | 0.567 | 0.605 | 296 (61.7) | 309 (59.4) | 196 (61.6) | 174 (58.8) |
| A | 0.433 | 0.395 | 184 (38.3) | 211 (40.6) | 122 (38.4) | 124 (41.2) |
The mechanisms that determine spontaneous viral clearance following acute hepatitis or passage to the chronic stage have not yet been fully elucidated. A number of factors were studied: those related to the virus; the mode of transmission[13], genotype[14], viral load[15], its genetic variability or its coinfection with other viruses[16]. Other factors related to the host have also been associated with the evolution of HCV infection, such as sex, age, ethnicity[13,17] and genetic susceptibility to infection by this virus[5].
Data from the literature suggest that patient’s gender could predict the evolution of HCV infection. Indeed, several studies have shown that female patients have a statistically higher chance to spontaneously eliminate the virus[13,17]. The relationship between female gender and a better response to interferon therapy has also been shown. Our study shows an increase in the number of women in G2 compared to men (133 f/127 m). However, this difference is not statistically significant and confirms the results reported by Cox et al[18] (2010) which excluded the role of sex factor in the development of HCV infection.
Similarly, the age at infection was often involved in the natural history of HCV infection, although the role of this factor has been controversial in the literature[19,20]. At younger ages, hepatic fibrosis develops slowly[19], while patients infected at an older age have more severe histological lesions and more rapid progression to cirrhosis and HCC[21,22]. Therefore, young age was associated with spontaneous viral clearance of HCV[19,23]. In our study, the mean age of patients in the different groups and subgroups studied was similar, which is consistent with literature data that discriminate the role of this factor in the evolution of viral hepatitis C[18,24].
Hemodialysis patients are one of the high risk groups for viral hepatitis, particularly HCV. It is now well established in most epidemiological studies performed in different populations, including Tunisia, that HCV infection is statistically correlated to transfusion history[25-29]. In our study, despite the fact that 25% of hemodialysis patients have received treatment for their anemia (recombinant human erythropoietin) history of blood transfusions is the main risk factor for HCV infection (71%).
The distribution of genotypes identified among patients in our cohort revealed a predominance of genotype 1b (74%) compared to other genotypes: 1a, 2a/2c, 3 and 4 (27%). These results are consistent with those reported in the national survey of different dialysis centers in the Laboratory of Immunology, Charles Nicolle Hospital in 2001-2002, revealing a frequency of 70.8% of HCV-1b[26]. Also, the study of Djebbi et al[30] (2003) performed during the same period in the region of Tunis, on a sample of 93 patients infected with HCV, showed a predominance of genotype 1b (79%) compared to other genotypes [2a (7%), 1a (5%), 2b (3%), 3 (3%) and 4a (1%)].
The role of HCV genotype during the course of infection to the chronic stage was analyzed by several studies. Amoroso et al[31] (1998) reported a persistence rate of 92% in patients exposed to HCV genotype 1b infection against 33%-50% in patients exposed to other genotypes. Lehmann et al[14] (2004) suggest that infection with HCV-3 is associated, in the acute stage, with viral clearance compared to those with genotype 1. Moreover, it is now accepted that the HCV genotype appears to be a predictor of both the response to antiviral treatment and duration of this therapy.
In our cohort, the prevalence of HCV-1b was similar in patients with persistence of viral replication over a period of 6 years (73.9%) and those who spontaneously cleared the virus during the same period (76.9%). It is the same for genotype 4 (13.4% vs 12.8%). Although the frequency of HCV 3 was low compared to other genotypes detected, it was more frequently observed in the Clearance group compared to the Persistence group (2.6% vs 1.4%). Conversely, HCV-2a/2c genotype was more identified among patients with persistent viral replication (9.9%) compared to those who spontaneously cleared the virus (2.6%).
There are variations in the distribution of genotypes in different regions of Tunisia. Genotype 1b predominates in all regions, particularly in the center (90.4%), the South (86.7%) and North West (83.3%). Its frequency in the North West corroborates studies by Ben Alaya Bouafif et al[32] (2007) and Mejri et al[33] (2005). Outside this region genotype1b and each of the other regions is characterized by a high frequency of a second genotype. The variability of distribution of HCV genotypes among different regions suggests a nosocomial infection in our hemodialysis cohort. Phylogenetic analysis by Kchouk et al[34] (2013) for genotype 4 and Djebbi et al[35] (2008) for HCV-1b confirmed this hypothesis. This mode of transmission of HCV, especially among hemodialysis patients, has been well documented by epidemiological and phylogenetic studies in different Spanish, French, Swiss, Iranian and American populations[36-41].
The CD4+ T cell and cytotoxic CD8+ T cell responses implicated in HCV infection involve immuno-regulating costimulatory molecules, such as CTLA-4 that acts as a negative regulator of the cell-mediated immune response[8,9,42,43]. Thus, mutations in the gene CTLA-4 could reduce or interfere with its function as a negative regulator and even promote the overgrowth of the immune response. CTLA4 polymorphisms contribute to the development of autoimmune diseases[10,44-46], and viral infections[47], including HCV[48,49].
Polymorphism (+49) A/G in exon 1 of the gene of human CTLA-4 has been extensively studied as it affects the function of the inhibitory molecule CTLA-4. The A allele (49) has been identified as protective and the G allele is associated with increased susceptibility to autoimmune diseases by reducing the expression of CTLA-4 at the cell surface. The CT60 SNP is a functional polymorphism and substitution A to G at +6230 position in the 3’ NC of the CTLA-4 gene is associated with lower rates of mRNA isoform of the soluble molecule, resulting in an increase in T cell activation by a defect in upregulation via CTLA-4[11]. The G allele represents the risk allele for both loci. Thus, SNPs (49) A/G and the (+6230) G/A CT60 CTLA-4 have been the subject of our cohort. But the distribution of alleles and genotypes of these common SNPs did not differ among different studied groups.
According to http://www.ncbi.nlm.nih.gov/snp, the frequency of the allele (+49) G varies by ethnicity. It is more common among Asians (65%) than Caucasians (20%) or Africans (29%). Similarly, the G allele (CT60) is more common among Asians (70%) and Africans (80%) than Caucasians (50%).
However, the two polymorphisms show different statistical powers depending on the population studied. Tunisian studies Krichen et al[46] (2010) and Hadj Kacem et al[50] (2001) showed that these two risk alleles are also present at a high frequency among the controls. This frequency averaged 70% for the variant (+49) G and 54% for the G allele (CT60). These results are similar to those found in total HCV infected hemodialysis patients in our study [(49) G = 69% (CT60) G = 57%]. In addition, the frequency of the GG haplotype combination (+49) (CT60) of the CTLA-4 gene was significantly higher in the patients in our series compared to controls (35.3% vs 29.9%, P = 0.0036, OR = 1.42, 95%CI: 1.12-1.79).
An US study associated the (+49) G allele with a favorable response to combination therapy in Caucasian patients infected with HCV genotype 1[51]. In the study of Danilovic et al[49] (2012), the two G risk variants are associated with increased inflammation in chronic hepatitis C, which corroborates the role, suggested that the GG haplotype (+49) (CT60) CTLA-4 with susceptibility to HCV infection among the patients in our study.
Other CTLA-4 SNPs have also been studied in several autoimmune diseases[10,47]. This is, in particular, a substitution of a cytosine by a thymine (C/T) at position (-318) gene promoter[52], which is associated with more high promoter activity, and therefore to an increase in the expression of the molecule and its immuno-regulatory activity. Similarly, repetition of the base pairs of a microsatellite (AT)n in the 3’ UTR region of exon 3 of the CTLA-4 gene may be associated with instability of mRNA and a decrease in the transcription of the gene and the expression of CTLA-4[53,54]. In a German study haplotype (+49) A / (- 318) C CTLA-4 is associated with spontaneous resolution of HCV infection among male patients[48]. Danilovic et al[49] (2012) also reported that the polymorphism (AT) n is associated with a progression to the chronic stage of HCV infection. Although our study shows no association between the polymorphisms studied and spontaneous clearance or persistence of HCV infection, the analysis of other polymorphisms CTLA-4, [(AT)n and (-318) C/T], would be important to develop a profile of allelic variants of CTLA-4 susceptibility, progression and clearance of HCV infection.
In summary, we demonstrate that CTLA-4 SNPs could influence susceptibility to HCV infection. We confirm data from previous epidemiological studies that suggest a nosocomial infection in Tunisian hemodialysis patients with transfusion history as a primary risk factor for HCV infection and a predominance of genotype 1b.
We thank all medical staff and technicians of dialysis centers who agreed to participate in this study.
Differences in host susceptibility to infectious disease and disease severity cannot be attributed to the virulence of the microbial agent. Indeed, the outcome of hepatitis C virus (HCV) entry and of subsequent acute infection appears to be predeterminated by immune competence of the host in the moment of infection.
CTLA-4 polymorphisms have been extensively studied with association to several autoimmune disorders and infectious diseases. Most individuals infected with HCV develop chronic infection and suffer from T-cell mediated hepatitis. Thus, it is of interest whether factors that modify the cellular immune response influence the course of disease.
The authors confirm data of previous epidemiological studies that suggest a nosocomial infection in Tunisian hemodialysis patients with transfusion history as primary risk factor for HCV infection and a predominance of genotype 1b.
CTLA-4 SNP’s should be added to the spectrum of immunogenetic factors involved in chronic HCV disease in Tunisian population.
This is a good descriptive study in which the authors analyzed the natural course of HCV infection in Tunisian hemodialysed cohort between 2002 and 2008. CTLA-4 polymorphisms have been studied in all patients groups and controls. Study of other susceptibility genes for HCV infection will certainly allow a better understanding of the molecular mechanisms of spontaneous clearance or persistence of HCV infection.
P- Reviewer: Alam S, Dang SS, Wong GLH S- Editor: Wang JL L- Editor: O’Neill M E- Editor: Wang CH
| 1. | Maasoumy B, Wedemeyer H. Natural history of acute and chronic hepatitis C. Best Pract Res Clin Gastroenterol. 2012;26:401-412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 2. | Ishii S, Koziel MJ. Immune responses during acute and chronic infection with hepatitis C virus. Clin Immunol. 2008;128:133-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 3. | Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, Govindarajan S, Purcell RH, Chisari FV. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci USA. 2002;99:15661-15668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 479] [Cited by in F6Publishing: 469] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 4. | Ksiaa L, Ayed-Jendoubi S, Sfar I, Gorgi Y, Najjar HA, Abdallah TB, Ayed K. Clearance and persistence of hepatitis C virus in a Tunisian population: association with HLA class I and class II. Viral Immunol. 2007;20:312-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 5. | Thio CL, Gao X, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, O’Brien SJ, Karacki P, Astemborski J, Carrington M. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792-4797. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 146] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 6. | An P, Thio CL, Kirk GD, Donfield S, Goedert JJ, Winkler CA. Regulatory polymorphisms in the interleukin-18 promoter are associated with hepatitis C virus clearance. J Infect Dis. 2008;198:1159-1165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Goulding C, McManus R, Murphy A, MacDonald G, Barrett S, Crowe J, Hegarty J, McKiernan S, Kelleher D. The CCR5-delta32 mutation: impact on disease outcome in individuals with hepatitis C infection from a single source. Gut. 2005;54:1157-1161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Sansom DM, Walker LS. The role of CD28 and cytotoxic T-lymphocyte antigen-4 (CTLA-4) in regulatory T-cell biology. Immunol Rev. 2006;212:131-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 9. | Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, Thompson CB, Bluestone JA. CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994;1:405-413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 10. | Howard TD, Postma DS, Hawkins GA, Koppelman GH, Zheng SL, Wysong AK, Xu J, Meyers DA, Bleecker ER. Fine mapping of an IgE-controlling gene on chromosome 2q: Analysis of CTLA4 and CD28. J Allergy Clin Immunol. 2002;110:743-751. [PubMed] [Cited in This Article: ] |
| 11. | Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506-511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1595] [Cited by in F6Publishing: 1538] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 12. | Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13387] [Cited by in F6Publishing: 14165] [Article Influence: 393.5] [Reference Citation Analysis (0)] |
| 13. | Grebely J, Raffa JD, Lai C, Krajden M, Conway B, Tyndall MW. Factors associated with spontaneous clearance of hepatitis C virus among illicit drug users. Can J Gastroenterol. 2007;21:447-451. [PubMed] [Cited in This Article: ] |
| 14. | Lehmann M, Meyer MF, Monazahian M, Tillmann HL, Manns MP, Wedemeyer H. High rate of spontaneous clearance of acute hepatitis C virus genotype 3 infection. J Med Virol. 2004;73:387-391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 15. | Santantonio T, Wiegand J, Gerlach JT. Acute hepatitis C: current status and remaining challenges. J Hepatol. 2008;49:625-633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 109] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 16. | Soriano V, Mocroft A, Rockstroh J, Ledergerber B, Knysz B, Chaplinskas S, Peters L, Karlsson A, Katlama C, Toro C. Spontaneous viral clearance, viral load, and genotype distribution of hepatitis C virus (HCV) in HIV-infected patients with anti-HCV antibodies in Europe. J Infect Dis. 2008;198:1337-1344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 17. | Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat. 2006;13:34-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 18. | Cox AL, Netski DM, Mosbruger T, Sherman SG, Strathdee S, Ompad D, Vlahov D, Chien D, Shyamala V, Ray SC. Prospective evaluation of community-acquired acute-phase hepatitis C virus infection. Clin Infect Dis. 2005;40:951-958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 167] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 19. | Vogt M, Lang T, Frösner G, Klingler C, Sendl AF, Zeller A, Wiebecke B, Langer B, Meisner H, Hess J. Prevalence and clinical outcome of hepatitis C infection in children who underwent cardiac surgery before the implementation of blood-donor screening. N Engl J Med. 1999;341:866-870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 358] [Cited by in F6Publishing: 374] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 20. | Rerksuppaphol S, Hardikar W, Dore GJ. Long-term outcome of vertically acquired and post-transfusion hepatitis C infection in children. J Gastroenterol Hepatol. 2004;19:1357-1362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 21. | Pol S, Vallet-Pichard A, Corouge M, Mallet VO. Hepatitis C: epidemiology, diagnosis, natural history and therapy. Contrib Nephrol. 2012;176:1-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Veldt BJ, Chen W, Heathcote EJ, Wedemeyer H, Reichen J, Hofmann WP, de Knegt RJ, Zeuzem S, Manns MP, Hansen BE. Increased risk of hepatocellular carcinoma among patients with hepatitis C cirrhosis and diabetes mellitus. Hepatology. 2008;47:1856-1862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 206] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 23. | Yeung LT, To T, King SM, Roberts EA. Spontaneous clearance of childhood hepatitis C virus infection. J Viral Hepat. 2007;14:797-805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 24. | Wietzke-Braun P, Manhardt LB, Rosenberger A, Uy A, Ramadori G, Mihm S. Spontaneous elimination of hepatitis C virus infection: a retrospective study on demographic, clinical, and serological correlates. World J Gastroenterol. 2007;13:4224-4229. [PubMed] [Cited in This Article: ] |
| 25. | Eleftheriadis T, Liakopoulos V, Leivaditis K, Antoniadi G, Stefanidis I. Infections in hemodialysis: a concise review. Part II: blood transmitted viral infections. Hippokratia. 2011;15:120-126. [PubMed] [Cited in This Article: ] |
| 26. | Ayed K, Gorgi Y, Ben Abdallah T, Aouadi H, Jendoubi-Ayed S, Sfar I, Makni H. Hepatitis C virus infection in hemodialysis patients from Tunisia: national survey by serologic and molecular methods. Transplant Proc. 2003;35:2573-2575. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Rahnavardi M, Hosseini Moghaddam SM, Alavian SM. Hepatitis C in hemodialysis patients: current global magnitude, natural history, diagnostic difficulties, and preventive measures. Am J Nephrol. 2008;28:628-640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Ben Othman S, Bouzgarrou N, Achour A, Bourlet T, Pozzetto B, Trabelsi A. [High prevalence and incidence of hepatitis C virus infections among dialysis patients in the East-Centre of Tunisia]. Pathol Biol (Paris). 2004;52:323-327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Sekkat S, Kamal N, Benali B, Fellah H, Amazian K, Bourquia A, El Kholti A, Benslimane A. [Prevalence of anti-HCV antibodies and seroconversion incidence in five haemodialysis units in Morocco]. Nephrol Ther. 2008;4:105-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Djebbi A, Triki H, Bahri O, Cheikh I, Sadraoui A, Ben Ammar A, Dellagi K. Genotypes of hepatitis C virus circulating in Tunisia. Epidemiol Infect. 2003;130:501-505. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Amoroso P, Rapicetta M, Tosti ME, Mele A, Spada E, Buonocore S, Lettieri G, Pierri P, Chionne P, Ciccaglione AR. Correlation between virus genotype and chronicity rate in acute hepatitis C. J Hepatol. 1998;28:939-944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 90] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 32. | Ben Alaya Bouafif N, Triki H, Mejri S, Bahri O, Chlif S, Bettaib J, Héchmi S, Dellagi K, Ben Salah A. A case control study to assess risk factors for hepatitis C among a general population in a highly endemic area of northwest Tunisia. Arch Inst Pasteur Tunis. 2007;84:21-27. [PubMed] [Cited in This Article: ] |
| 33. | Mejri S, Salah AB, Triki H, Alaya NB, Djebbi A, Dellagi K. Contrasting patterns of hepatitis C virus infection in two regions from Tunisia. J Med Virol. 2005;76:185-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Kchouk FH, Gorgi Y, Bouslama L, Sfar I, Ayari R, Khiri H, Halfon P, Aouadi H, Jendoubi Ayed S, Ayed K. Phylogenetic analysis of isolated HCV strains from tunisian hemodialysis patients. Viral Immunol. 2013;26:40-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Djebbi A, Bahri O, Langar H, Sadraoui A, Mejri S, Triki H. Genetic variability of genotype 1 hepatitis C virus isolates from Tunisian haemophiliacs. New Microbiol. 2008;31:473-480. [PubMed] [Cited in This Article: ] |
| 36. | Shimokura G, Chai F, Weber DJ, Samsa GP, Xia GL, Nainan OV, Tobler LH, Busch MP, Alter MJ. Patient-care practices associated with an increased prevalence of hepatitis C virus infection among chronic hemodialysis patients. Infect Control Hosp Epidemiol. 2011;32:415-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 37. | Patel PR, Thompson ND, Kallen AJ, Arduino MJ. Epidemiology, surveillance, and prevention of hepatitis C virus infections in hemodialysis patients. Am J Kidney Dis. 2010;56:371-378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 38. | Gallego E, López A, Pérez J, Llamas F, Lorenzo I, López E, Illescas ML, Andrés E, Olivas E, Gómez-Roldan C. Effect of isolation measures on the incidence and prevalence of hepatitis C virus infection in hemodialysis. Nephron Clin Pract. 2006;104:c1-c6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Szyszymar B, Protas-Drozd F. [Electrothermonetric study of the skin at the site of white dermatographism in prurigo and other extensive allergic skin diseases]. Przegl Dermatol. 1977;64:31-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Halfon P, Roubicek C, Gerolami V, Quentin Y, Khiri H, Pepe G, Berland Y. Use of phylogenetic analysis of hepatitis C virus (HCV) hypervariable region 1 sequences to trace an outbreak of HCV in an autodialysis unit. J Clin Microbiol. 2002;40:1541-1545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Norder H, Bergström A, Uhnoo I, Aldén J, Weiss L, Czajkowski J, Magnius L. Confirmation of nosocomial transmission of hepatitis C virus by phylogenetic analysis of the NS5-B region. J Clin Microbiol. 1998;36:3066-3069. [PubMed] [Cited in This Article: ] |
| 42. | Gribben JG, Freeman GJ, Boussiotis VA, Rennert P, Jellis CL, Greenfield E, Barber M, Restivo VA, Ke X, Gray GS. CTLA4 mediates antigen-specific apoptosis of human T cells. Proc Natl Acad Sci USA. 1995;92:811-815. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 254] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 43. | McCoy KD, Le Gros G. The role of CTLA-4 in the regulation of T cell immune responses. Immunol Cell Biol. 1999;77:1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 44. | Mattner J. Genetic susceptibility to autoimmune liver disease. World J Hepatol. 2011;3:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 45. | Chang MC, Chang YT, Tien YW, Liang PC, Jan IS, Wei SC, Wong JM. T-cell regulatory gene CTLA-4 polymorphism/haplotype association with autoimmune pancreatitis. Clin Chem. 2007;53:1700-1705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 109] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 46. | Krichen H, Sfar I, Hadj Kacem H, Bardi R, Jendoubi-Ayed S, Makhlouf M, Ben Romdhane T, Besseghair F, Aouadi H, Ben Abdallah T. (AT) repeat in the 3’ untranslated region of the CTLA-4 gene and susceptibility to acute allograft rejection in Tunisian renal transplantation. Transplant Proc. 2010;42:4314-4317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 47. | Thio CL, Mosbruger TL, Kaslow RA, Karp CL, Strathdee SA, Vlahov D, O’Brien SJ, Astemborski J, Thomas DL. Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B virus infection. J Virol. 2004;78:11258-11262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 48. | Schott E, Witt H, Hinrichsen H, Neumann K, Weich V, Bergk A, Halangk J, Müller T, Tinjala S, Puhl G. Gender-dependent association of CTLA4 polymorphisms with resolution of hepatitis C virus infection. J Hepatol. 2007;46:372-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 49. | Danilovic DL, Mendes-Correa MC, Lima EU, Zambrini H, K Barros R, Marui S. Correlations of CTLA-4 gene polymorphisms and hepatitis C chronic infection. Liver Int. 2012;32:803-808. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 50. | Hadj Kacem H, Bellassoued M, Bougacha-Elleuch N, Abid M, Ayadi H. CTLA-4 gene polymorphisms in Tunisian patients with Graves’ disease. Clin Immunol. 2001;101:361-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 51. | Yee LJ, Perez KA, Tang J, van Leeuwen DJ, Kaslow RA. Association of CTLA4 polymorphisms with sustained response to interferon and ribavirin therapy for chronic hepatitis C virus infection. J Infect Dis. 2003;187:1264-1271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 52. | Deichmann K, Heinzmann A, Brüggenolte E, Forster J, Kuehr J. An Mse I RFLP in the human CTLA4 promotor. Biochem Biophys Res Commun. 1996;225:817-818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 110] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 53. | Polymeropoulos MH, Xiao H, Rath DS, Merril CR. Dinucleotide repeat polymorphism at the human CTLA4 gene. Nucleic Acids Res. 1991;19:4018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 54. | Wang XB, Kakoulidou M, Giscombe R, Qiu Q, Huang D, Pirskanen R, Lefvert AK. Abnormal expression of CTLA-4 by T cells from patients with myasthenia gravis: effect of an AT-rich gene sequence. J Neuroimmunol. 2002;130:224-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |