Published online Sep 14, 2015. doi: 10.3748/wjg.v21.i34.9916
Peer-review started: February 26, 2015
First decision: March 26, 2015
Revised: April 13, 2015
Accepted: July 15, 2015
Article in press: July 15, 2015
Published online: September 14, 2015
AIM: To investigate macrophage migration inhibitory factor (MIF) expression and its clinical relevance in gastric cancer, and effects of MIF knockdown on proliferation of gastric cancer cells.
METHODS: Tissue microarray containing 117 samples of gastric cancer and adjacent non-cancer normal tissues was studied for MIF expression by immunohistochemistry (IHC) semiquantitatively, and the association of MIF expression with clinical parameters was analyzed. MIF expression in gastric cancer cell lines was detected by reverse transcription-polymerase chain reaction (RT-PCR) and Western blot. Two pairs of siRNA targeting the MIF gene (MIF si-1 and MIF si-2) and one pair of scrambled siRNA as a negative control (NC) were designed and chemically synthesized. All siRNAs were transiently transfected in AGS cells with OligofectamineTM to knock down the MIF expression, with the NC group and mock group (OligofectamineTM alone) as controls. At 24, 48, and 72 h after transfection, MIF mRNA was analyzed by RT-PCR, and MIF and proliferating cell nuclear antigen (PCNA) proteins were detected by Western blot. The proliferative rate of AGS cells was assessed by methylthiazolyl tetrazolium (MTT) assay and colony forming assay.
RESULTS: The tissue microarray was informative for IHC staining, in which the MIF expression in gastric cancer tissues was higher than that in adjacent non-cancer normal tissues (P < 0.001), and high level of MIF was related to poor tumor differentiation, advanced T stage, advanced tumor stage, lymph node metastasis, and poor patient survival (P < 0.05 for all). After siRNA transfection, MIF mRNA was measured by real-time PCR, and MIF protein and PCNA were assessed by Western blot analysis. We found that compared to the NC group and mock group, MIF expression was knocked down successfully in gastric cancer cells, and PCNA expression was downregulated with MIF knockdown as well. The cell counts and the doubling times were assayed by MTT 4 d after transfection, and colonies formed were assayed by colony forming assay 10 d after transfection; all these showed significant changes in gastric cancer cells transfected with specific siRNA compared with the control siRNA and mock groups (P < 0.001 for all).
CONCLUSION: MIF could be of prognostic value in gastric cancer and might be a potential target for small-molecule therapy.
Core tip: Macrophage migration inhibitory factor (MIF) is a multifunctional cytokine, which plays a significant role in the tumor development. The objective of this study was to investigate the expression of MIF in gastric cancer and the inhibitory effect of its knockdown on gastric cancer cell proliferation. The results show that MIF is expressed highly in gastric cancer, and its expression was related to clinical stage. The proliferation of gastric cancer cells was inhibited by MIF knockdown, suggesting that MIF is a potential target in molecular therapy for gastric cancer.
- Citation: He LJ, Xie D, Hu PJ, Liao YJ, Deng HX, Kung HF, Zhu SL. Macrophage migration inhibitory factor as a potential prognostic factor in gastric cancer. World J Gastroenterol 2015; 21(34): 9916-9926
- URL: https://www.wjgnet.com/1007-9327/full/v21/i34/9916.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i34.9916
Macrophage migration inhibitory factor (MIF) is a pluripotent cytokine that can inhibit macrophage migration in vitro and promote the accumulation of macrophages in delayed skin hypersensitivity[1,2]. Autocrine MIF signaling stimulates macrophage production of tumor necrosis factor-α, interleukin-1, nitric oxide, hydrogen peroxide and other inflammatory factors[3]. MIF is an important mediator in the pathogenesis of gastric inflammation in rats, and its expression increases significantly in Helicobacter pylori (H. pylori)-induced inflammation in humans[4,5].
Previously, we have shown that MIF knockout mice are protected against H. pylori-induced gastritis[6]. Furthermore, MIF is highly expressed in various tumors, including lung[7], liver[8], breast[9], gastric[10], colon[11], and prostate[12] cancers, and overexpression of MIF stimulates proliferation and inhibits apoptosis in cancer cells via the paracrine pathway[13,14]. These findings suggest that MIF plays an essential part in the development of chronic inflammation and cancer, and possibly in carcinogenesis related to chronic inflammation.
MIF has been suggested as a potential molecular target for cancer therapy. In an in vitro study, proliferation of colon cancer cells was inhibited when MIF was downregulated by transduction of MIF antisense plasmids[15]. In another report, the growth and invasion of DU-145 prostate cancer cells were attenuated by inhibition of MIF and its receptor, CD74[16]. However, whether MIF silencing has any effects on the development of gastric cancer and whether MIF is a potential therapeutic molecular target in this disease are not understood.
In this report, we demonstrate that MIF is substantially expressed in gastric cancer cells in vitro and in vivo, MIF expression is associated with patient survival in gastric cancer and thus may be of prognostic value, and MIF may act as a potential therapeutic target for gastric cancer.
In this study, we retrospectively recruited formalin-fixed, paraffin-embedded tissues from 117 cases of gastric cancer, which were resected initially between March 2001 and August 2003. Samples were collected from the archives of the Department of Pathology, the First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China. Tumor differentiation was defined based on the criteria proposed by the World Health Organization (WHO) Classification of Tumors (2003). Tumor stages were defined according to the American Joint Committee on Cancer (AJCC) TNM Staging Classification for Carcinoma of the Stomach (7th Ed, 2010). This study was approved by the Ethics Committee of the Sun Yat-sen University Cancer Center, and written informed consent was obtained from each patient.
Tissue microarrays (TMA) were constructed according to a method described previously[17]. Briefly, the individual block of tissue and the corresponding histological slides stained with hematoxylin and eosin were overlaid for microarray sampling, which was performed with a tissue arraying instrument (Beecher Instruments, Sun Prairie, WI, United States). Cylinders of tissue with a 0.6 mm diameter were removed from the samples-three from each primary tumor sample and three from each healthy tissue sample-and re-embedded in paraffin in a predetermined position. Multiple 5 μm sections were cut from the microarray block and mounted on microscope slides.
TMA sections were first deparaffinized in xylene, rehydrated through a series of graded alcohol, immersed in 3% hydrogen peroxide in phosphate-buffered saline (PBS) for 10 min to block endogenous peroxidase activity, and antigen-retrieved in a pressure cooker for 3 min in citrate buffer (pH 6.0). TMA sections were then blocked for nonspecific binding by incubation in 10% normal goat serum at room temperature for 30 min. Subsequently, the slides were incubated with a primary monoclonal antibody to MIF (Santa Cruz Biotechnology, Santa Cruz, CA, United States; 1:800 in PBS) at 4 °C overnight in a moist chamber. The next day, the sections were sequentially incubated with biotinylated goat anti-rabbit immunoglobulin (Envision, Dako, Denmark) as the secondary antibody, at a concentration of 1:100 for 40 min at 37 °C. Slides were then reacted with streptavidin-peroxidase conjugate for 30 min at 37 °C, followed by incubation with 3,3’-diaminobenzidine as a chromogen substrate. Cell nuclei were counterstained with Meyer’s hematoxylin. A negative control was achieved by replacing the primary antibody with normal murine IgG.
All immunohistochemistry slides were semiquantitively evaluated by the same observer who had no knowledge of the previous biochemical test results. Assessments were performed according to the procedure widely used to calculate the Remmele score[18]. The level of staining intensity (SI) was subdivided into four groups: 0 (negative); 1 (weak); 2 (moderate); and 3 (strong). The percentage of positive cells (PP) was regarded as 0 (none), 1 (< 10%), 2 (10%-50%), 3 (51%-80%) and 4 (> 80% positive tumor cells), respectively. The Remmele score is produced by multiplying the score of intensity and percentage (0-12). In this study, a score of 0-2 was regarded as negative or low expression, while > 2 as high expression.
The gastric cancer-derived cell lines used in this study were AGS, MKN-28, MKN-45, SGC-7901 and BCG-823. Cancer cells were grown in RPMI-1640 medium (Sigma-Aldrich, St Louis, MO, United States) supplemented with 10% fetal bovine serum. The cells were grown at 37 °C in a humidified atmosphere containing 5% CO2.
To achieve MIF knockdown, cells were transfected with small interfering RNA (siRNA). Cells were cultured in Opti-MEM reduced serum medium (Invitrogen, Carlsbad, CA, United States) in accordance with the manufacturer’s instructions. We used the online GenScript siRNA Target Finder Tool (GenScript, Piscatawy, NJ, United States) to design siRNA that would target the mRNA sequence of the human MIF gene. The siRNA duplexes (si-1 and si-2) consisted of 27 bp with a base deoxynucleotide overhang[19], and were synthesized commercially (TaKaRa Bio Inc., Otsu, Shiga, Japan) with the following sequences (sense strand): si-1, ACAUCAACUAUUACGACAUGAACGCGGdTdT and si-2, CAUCAUGCCGAUGUUCAUCGUAAACACdTdT. A scrambled sequence siRNA (GUUGCGCCCGCGAAUGAUAUUUAUAAUCdTdT; sense strand) was used as a negative control. All sequences were confirmed to have no homology with RIF mRNA by Blast searches.
All siRNAs were transfected using Oligofectamine reagent (Invitrogen, Carlsbad, CA, United States), according to the manufacturer’s instructions. Additionally, a mock group was transfected with reagent alone. Cells were harvested 24, 48 and 72 h after transfection and the levels of MIF mRNA were assessed by real-time polymerase chain reaction (RT-PCR). Western blot analysis was used to measure MIF and proliferating cell nuclear antigen (PCNA) protein expression.
Three independent experiments were done with each AGS and MKN-45 cells and similar results obtained in the following study, and as representatives, the showed results are of three independent experiments with AGS cells.
RT-PCR was carried out with SYBR Green mixture (TaKaRa, Japan), according to the manufacturer’s protocol. The following primers were used: MIF forward, 5’-CAGTGGTGTCCGAGAAGTCAG-3’ and reverse, 5’-TAGGCGAAGGTGGAGTTGTT-3’; GAPDH forward, 5’-TTTGGTATCGTGGAAGGAC-3’ and reverse, 5’-AAAGGTGGAGGAGTGGGT-3’. The data were analyzed with standard-curve analysis and charts created in Quantity One software (Bio-Rad, Hercules, CA, United States).
Cells were harvested in ready-to-use radioimmunoprecipitation assay buffer (Sigma-Aldrich) that contains 150 mmol/L sodium chloride, 1.0% igepal CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate and 50 mmol/L Tris (pH 8.0), supplemented with protease-inhibitor and phosphatase-inhibitor cocktails. The protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL, United States). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate 20 μg protein samples that were transferred on to polyvinylidene fluoride membranes (Perkin Elmer, Fremont, CA, United States) and then immunoblotted with MIF and PCNA polyclonal antibodies (Santa Cruz Biotechnology) and α-tubulin polyclonal antibody (Cell Signaling Technology, Danvers, MA, United States) or GAPDH polyclonal antibody (Abcam, Cambridge, United Kingdom) as internal controls. Antibodies conjugated with horseradish peroxidase and an enhanced chemiluminescence Western blotting system (Amersham Bioscience, Little Chalfont, United Kingdom) were used for detection. Gray values of the bands were analyzed with Quantity One software.
Cell growth was measured with the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, 2 d after transfection with siRNAs, AGS cells were seeded at 2000 cells per well in 96-well plates and incubated for 24, 48 or 72 h. To each well, 5 μL of 5 mg/mL MTT was added and the plates were incubated for an additional 4 h at 37 °C. Finally, formed formazan complex was solubilized with 100 μL dimethyl sulfoxide. Absorbance was measured 4 d after transfection with a microplate reader (Bio-Rad) at a wavelength of 492 nm.
Two days after transfection with MIF or negative-control siRNA, 500 cells were evenly spread on six-well plates and cultured for 10 d at 37 °C. At the end of the incubation period, the cells were stained with Giemsa solution (Sigma-Aldrich) and photographed with a digital camera. The number of colonies that contained more than 50 cells was counted with an inverted microscope.
Survival was estimated using the Mantel-Cox log-rank test. Association of MIF protein expression levels with clinicopathological data was analyzed using the χ2 test, stage-match univariate survival analysis, and a multiple Cox proportional hazards regression analysis.
All analyses were performed using SPSS statistical software package (version 13.0, SPSS Inc., Chicago, IL, United States). Differences were deemed to be significant when P values from two-tailed tests were less than 0.05.
This cohort consisted of 72 (61.5%) men and 45 (38.5%) women, with a median age of 61 years old. The average follow-up duration was 28.5 mo, and the median follow-up duration was 23.0 mo (range, 5.0 to 80.0 mo). Clinicopathological characteristics, including patient age and gender, tumor differentiation, stage, and relapse, were collected (Table 1).
| Variable | MIF | |||
| All cases | Low expression | High expression | P value1 | |
| Age (yr)2 | 0.819 | |||
| ≤ 61 | 55 | 16 (29.09) | 39 (70.91) | |
| > 61 | 61 | 20 (32.79) | 41 (67.21) | |
| Sex | 0.598 | |||
| Male | 74 | 21 (28.38) | 53 (71.62) | |
| Female | 43 | 15 (34.88) | 28 (65.12) | |
| Tumor differentiation | 0.049 | |||
| Well | 5 | 1 (20.00) | 4 (80.00) | |
| Moderate | 34 | 16 (47.06) | 18 (52.94) | |
| Poor | 78 | 19 (24.36) | 59 (75.64) | |
| Tumor stage | 0.001 | |||
| I and II | 27 | 16 (59.26) | 11 (40.74) | |
| III | 51 | 14 (27.45) | 37 (72.55) | |
| IV | 39 | 6 (15.38) | 33 (84.62) | |
| T stage | 0.034 | |||
| T1 and T2 | 14 | 8 (57.14) | 6 (42.86) | |
| T3 | 75 | 23 (30.67) | 52 (69.33) | |
| T4 | 28 | 5 (17.86) | 23 (82.14) | |
| Lymph node metastasis | 0.001 | |||
| No (N0) | 24 | 16 (66.67) | 8 (33.33) | |
| Yes (Nx) | 93 | 20 (21.51) | 73 (78.49) | |
| Distant metastasis | 0.564 | |||
| M0 | 99 | 32 (32.32) | 67 (67.68) | |
| M1 | 18 | 4 (22.22) | 14 (77.78) | |
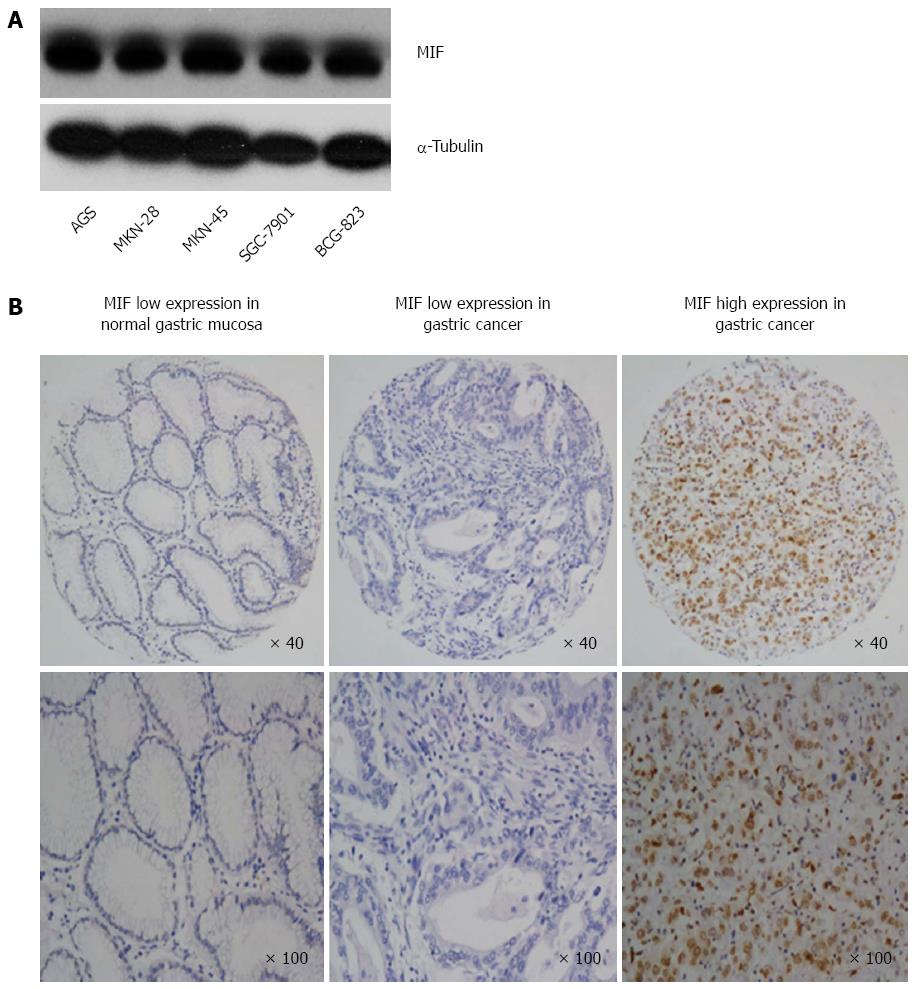
Western blot analysis showed that MIF was expressed in all five gastric cancer cell lines (AGS, MKN-28, MKN-45, SGC-7901 and BCG-823; Figure 1A). Immunochemistry showed that MIF protein was mainly expressed in the cell nuclei and cytoplasm of gastric mucosa cells (range of expression, 0%-100%; Figure 1B). This finding indicates that MIF is overexpressed in gastric cancer, compared to the adjacent normal mucosa. High MIF protein expression was associated with poor tumor differentiation, advanced T-stage, advanced tumor stage and lymph node metastasis in gastric cancer, but not with age, sex or distant metastasis (Table 1).
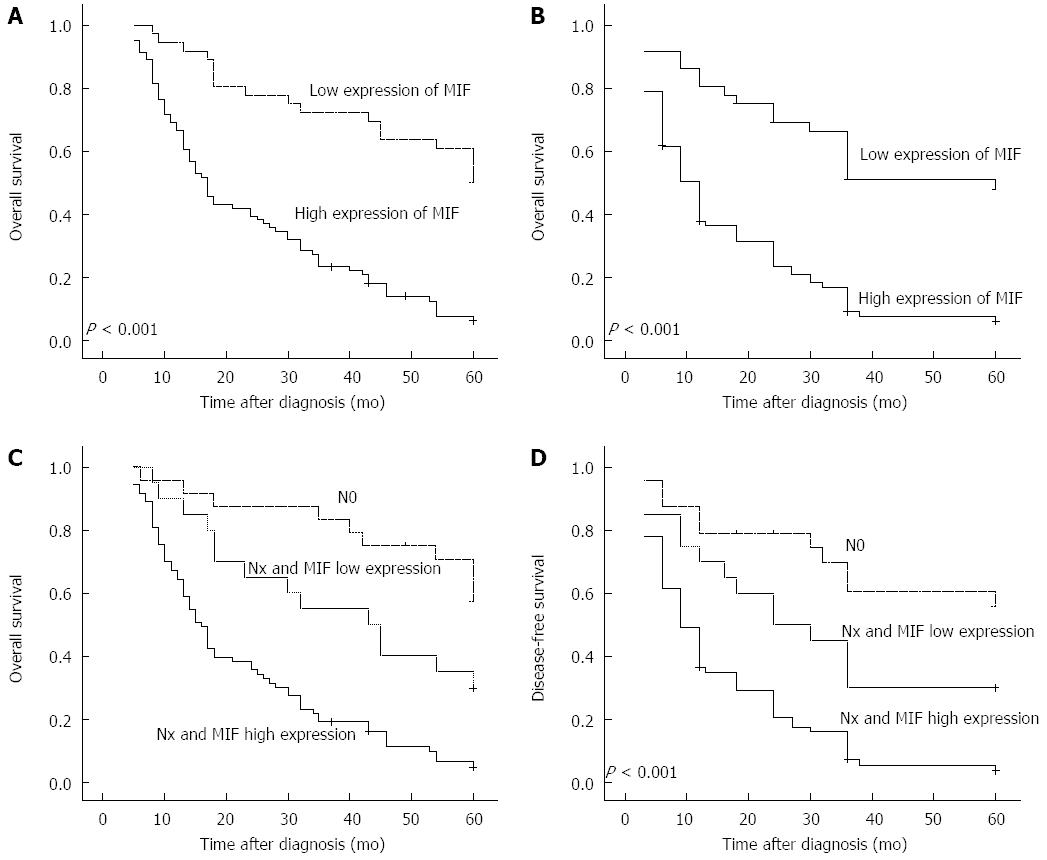
Multivariate regression analysis showed that MIF expression was an independent prognostic factor for poor patient survival (HR = 1.784; 95%CI: 1.338-2.380, P < 0.001), as well as advanced clinical stage (Table 2). Associations were observed between the degree of MIF protein expression, and overall survival and disease-free survival. In patients with high MIF expression, mean overall survival was 24.2 mo (SD = 1.96, 95%CI: 20.4-28.1), compared with 47.0 mo (SD = 3.18, 95%CI: 40.8-53.3) in those with low levels of MIF expression (P = 0.001; Figure 2A). Likewise, mean disease-free survival was significantly shorter in patients with high MIF expression (17.0 mo, SD = 1.83, 95%CI: 13.4-20.6) than those with low MIF expression (40.7 mo, SD = 1.833.72, 95%CI: 33.5-48.1) (P < 0.001; Figure 2B). In patients with lymph node metastasis (93/117; 79.49%), mean overall survival was shorter in those with high MIF expression (73/93; 78.5%, 22.7 mo, SD = 1.77, 95%CI: 18.9-22.6) than those with low MIF expression (20/93, 21.5%; 38.8 mo, SD = 4.55, 95%CI: 29.8-47.7). Mean overall survival was significantly longer in patients without lymph node metastasis (24/117, 20.51%; 51.2 mo, SD = 3.51, 95%CI: 44.3-58.0) than any patients with lymph node metastasis (P < 0.001; Figure 2C). The pattern was similar for disease-free survival (high MIF expression plus lymph node metastasis, 15.9 mo, SD = 1.80, 95%CI: 12.5-19.5; MIF expression plus lymph node metastasis, 30.9 mo, SD = 4.82, 95%CI: 21.5-40.4; no lymph node metastasis, 44.2 mo, SD = 4.64, 95%CI: 35.1-53.3. P < 0.001; Figure 2D).
| Characteristic | Hazards ratio | 95%CI | P value |
| Age ( ≤ 61 yr vs > 61 yr) | 1.099 | 0.719-1.680 | 0.663 |
| Sex (male vs female) | 1.202 | 0.779-1.855 | 0.406 |
| Tumor size ( ≤ 3 cm vs > 3 cm) | 1.606 | 0.964-2.677 | 0.069 |
| Stage (I-II vs III-V) | 1.885 | 1.171-3.305 | 0.009 |
| Differentiation (well to moderate vs poor) | 1.258 | 0.777-2.039 | 0.351 |
| MIF expression (low vs high) | 1.784 | 1.338-2.380 | < 0.001 |
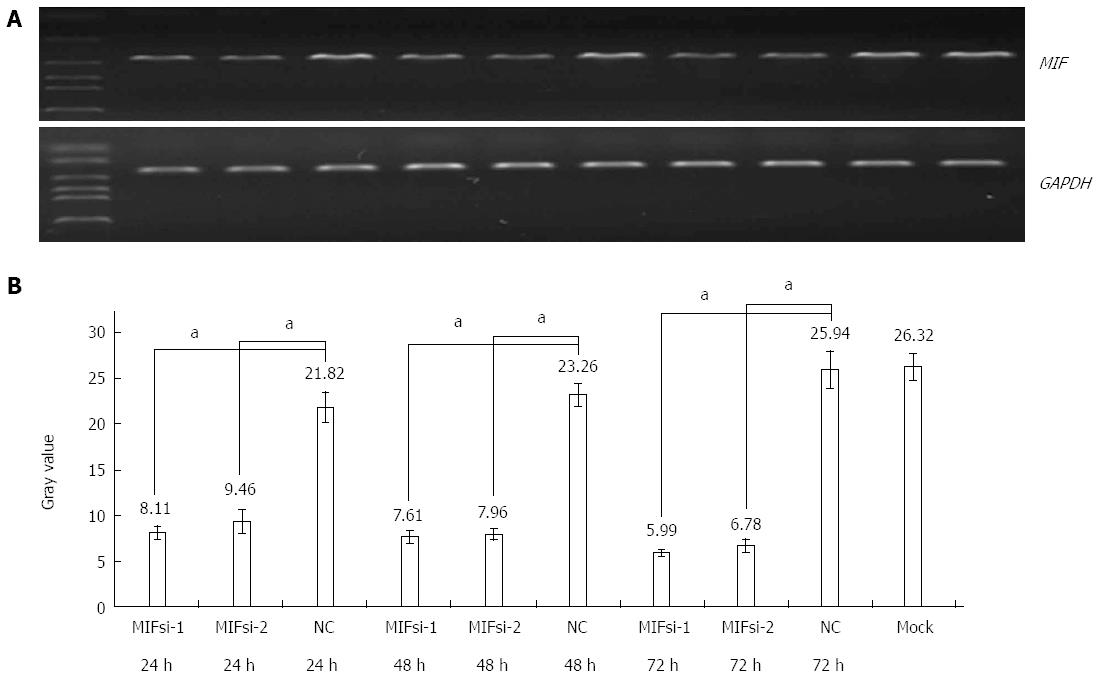
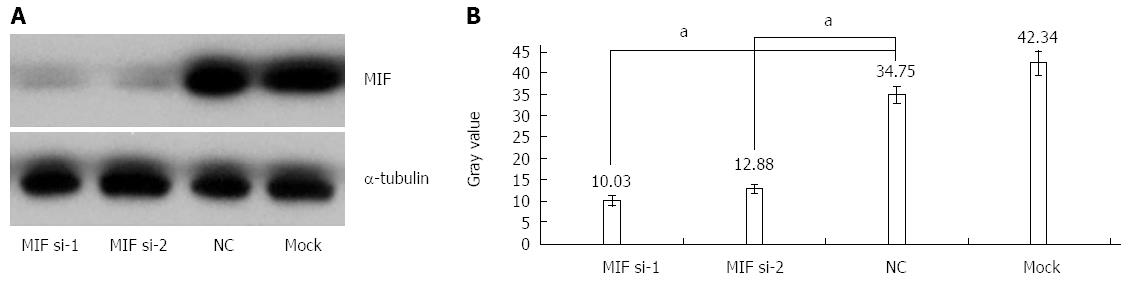
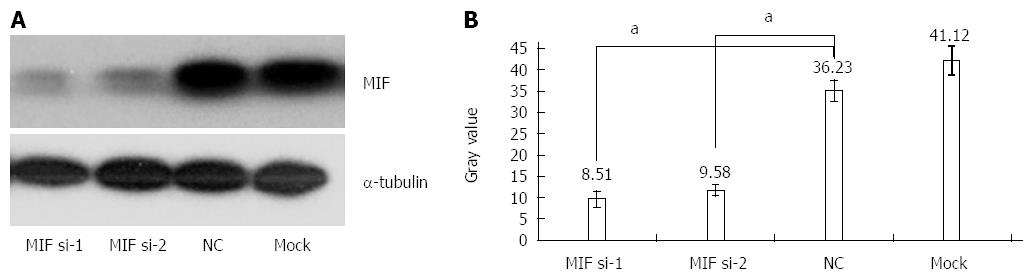
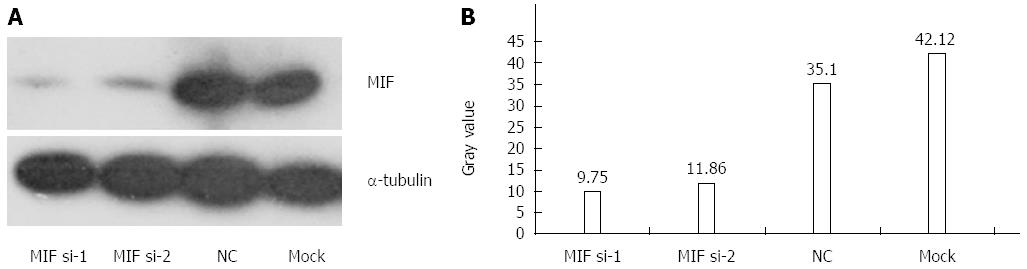
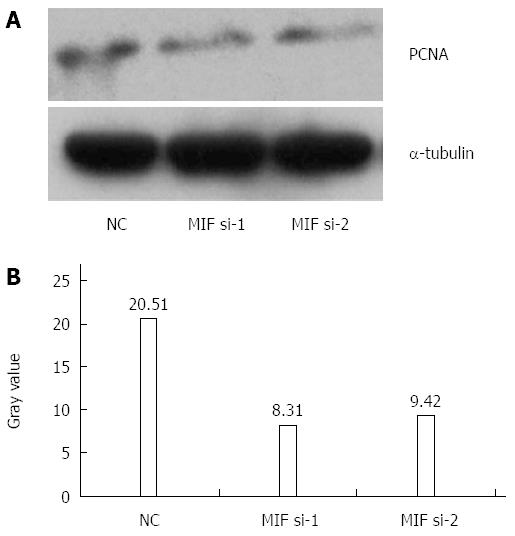
In comparison with the negative and mock groups, MIF mRNA levels decreased significantly in AGS cells transfected with MIF si-1 or si-2, compared with the internal control (Figure 3). The inhibition ratios of MIF si-1 in the experimental group were 62.83%, 67.28% and 76.91% at 24, 48 and 72 h after transfection, respectively, and those of MIF si-2 were 56.65%, 65.78% and 73.86% (P < 0.001 for all; Figure 3). At 24, 48 and 72 h after transfection, protein inhibition ratios for the MIF si-1 group were 71.00%, 72.73% and 76.51%, respectively, and for the MIF si-2 group were 62.94%, 66.21% and 73.55%, compared with the control (P < 0.001 for all; Figure 4, Figure 5 and Figure 6).
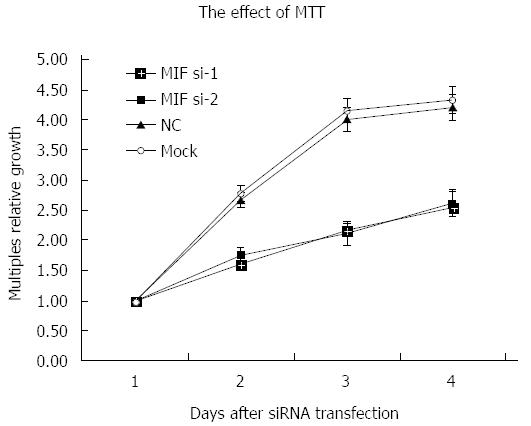
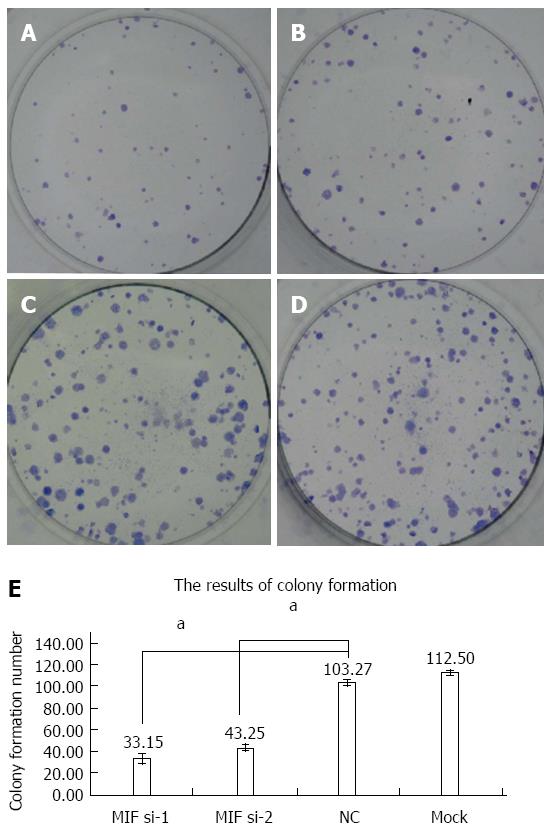
Four days after transfection, the mean cell counts in the MIF si-1 and si-2 groups were, respectively, 2.55 ± 0.13 times and 2.62 ± 0.15 times the original counts, compared with 4.21 ± 0.32 times and 4.33 ± 0.48 times, respectively, in the negative and mock groups (Figure 7). Mean doubling times were 2.58 ± 0.14 d and 2.42 ± 0.17 d, compared with 1.25 ± 0.06 d and 1.08 ± 0.14 d, respectively, in the two controls. The colony formation assay showed that 10 d after transfection with si-1 and si-2, mean colony numbers in AGS cells were 33.15 ± 4.12 and 43.25 ± 2.63, respectively, which were significantly lower than 103.27 ± 2.80 in the negative control and 112.50 ± 2.10 in the mock group (P < 0.001 for all; Figure 8). Gray values of the PCNA expression levels in the MIF si-1 and si-2 and control siRNA groups were 8.31, 9.42 and 20.51, respectively (Figure 9), all of which suggest that MIF siRNA transfection significantly inhibited the proliferation capacity of AGS cells.
MIF was highly expressed in gastric cancer compared with in adjacent healthy tissue, and was associated with poor differentiation, advanced disease and tumor stages, lymph node metastasis, and poor overall and disease-free survival. Moreover, the proliferation of gastric cancer cells was inhibited by MIF RNA interference. Our findings suggest that high expression of MIF has an important role and is clinically relevant in gastric cancer. Substantial MIF expression was also observed in all five gastric cancer cell lines studied (AGS, MKN-28, MKN-45, SGC-7901 and BCG-823).
He et al[10] found that MIF expression rates were 52.12%, 66.11% and 95.51% in mucosa samples from patients with chronic antral gastritis, gastric intestinal metaplasia, and gastric adenocarcinoma, respectively, compared with 12.19% in normal mucosa samples. Concentrations of MIF have been found to raise in the sera of patients with gastric cancer[20]. In this study, overexpression of MIF protein in gastric cancer tissues was significantly associated with poor overall and disease-free survival. Cox’s multivariate regression analysis showed that MIF expression, clinical stage and large tumor size were independent prognostic factors for patient survival. This finding is supported by previous studies, which have shown that MIF expression was related to poor prognosis and poor survival in patients with primary nasopharyngeal carcinoma or osteosarcoma[19,21]. However, in this study expression did not differ significantly between moderately to highly differentiated and poorly differentiated adenocarcinoma, which is not consistent with previous findings for neuroblastoma[22]. Moreover, Nabizadeh Marvast et al[23] recently showed that MIF expression statistically has no correlation with histological subtype, distant metastasis, lymph node involvement, tumor stage and grade in gastric cancer. Although it is not consistent with our results, the sample size in their report was smaller than that in our present study, and their analysis for immunostaining was qualitative, while our results for IHC were based on semiquantitative analysis. Anyway, further study is needed to clarify relationships between MIF expression and the degree of differentiation and tumor progression in gastric cancer.
RNA interference is a tool to identify mechanisms of action for target proteins and to assess therapeutic potential for small-molecule therapy. The underlying mechanism of RNA interference is unclear, but siRNA might be a substrate for Dicer function; Dicer and RNA-induced silencing complex work together to complete the processing of siRNA[24]. The results in our experimental group showed that 27 bp siRNA targeting MIF had excellent gene-silencing effects. Compared with the levels observed in control cells, MIF expression was significantly inhibited and detectable at both the transcription and translation levels. The silencing effect of MIF si-1 was slightly stronger than that of MIF si-2 at 24, 48 and 72 h. This finding suggests that MIF siRNAs targeting these two sites could quickly, effectively and specifically lower MIF expression. siRNAs of 21-23 bp in length are reasonably specific and do not activate the double-stranded-RNA-dependent protein kinase pathway, and were thought to yield optimum RNA interference. Various studies have shown, however, that 27 bp siRNAs are more potent inducers of RNA interference than 21 bp siRNAs, and are effective at lower concentrations[25]. The double-stranded-RNA-dependent protein kinase pathway is not activated by 27 bp siRNAs and, therefore, they are of appropriate length for knockdown of target genes.
The slight differences in the silencing effects of MIF si-1 and si-2 were probably because due to sequence differences between target sites. Not all sites in siRNAs play the same roles in the process of target sequence recognition[26] and siRNAs targeting different sites in the same gene sequence trigger different silencing effects[27]. In this study, MIF si-1 corresponded to the MIF mRNA sequence initiated at base 384, whereas MIF si-2 corresponded to that at base 94. Although the paired siRNAs had good interference efficiencies, the effect of MIF si-1 was slightly better than that of MIF si-2. As expected, the scrambled siRNA negative control did not trigger any silencing effect.
Cell growth was significantly inhibited after MIF silencing by MTT assay. The inhibitory effect began 24 h after transfection and persisted during the 4 d of observation, with the optimum effect seen at 48 h. The colony formation assay showed that gastric cancer cell proliferation was also significantly inhibited. Accordingly, PCNA expression decreased in gastric cancer cells after transfection. MIF seems, therefore, to promote proliferation of gastric cancer, and may be a suitable molecular therapeutic target for gene silencing or attenuation of MIF activity. In agreement with our results, Dessein et al[28] found that MIF had a role in the proliferation of established 5-FU-resistant colon cancer cells. The underlying mechanism remains elusive, but might be through promotion of the proliferation of cancer stem cells.
Besides cell proliferation, MIF expression might have a role in cancer cell apoptosis and tumor angiogenesis, invasion and metastasis. High MIF expression in human glioblastoma and melanoma has been correlated with tumor angiogenesis[29,30] and has been suggested to enhance invasion and metastasis in nasopharyngeal carcinoma and prostatic adenocarcinoma[31,32]. That MIF high expression has similar roles in gastric cancer progression seems feasible, and supports the possibility that MIF silencing could influence these characteristics. This hypothesis and its underlying mechanisms remain to be clarified.
In summary, MIF is substantially expressed in gastric cancer tissues and cell lines. High MIF expression was associated with poor tumor differentiation, advanced disease and tumor stages, lymph node metastasis, and poor patient survival. Thus, measurement of MIF expression has prognostic value in patients with gastric cancer. The proliferation of gastric cancer cells was inhibited by MIF knockdown, which suggests that MIF could be a suitable target for small-molecule therapy.
Migration inhibitory factor (MIF) is a pluripotent cytokine that can inhibit macrophage migration in vitro and promote the accumulation of macrophages in delayed skin hypersensitivity, which is highly expressed in a variety of tumors. Previous study by He et al demonstrated that MIF knockout mice are protected against Helicobacter pylori induced gastritis. Moreover, MIF stimulates the proliferation of cancer cells, as well as inhibits the apoptosis of cancer cells by paracrine pathway. These findings would imply that MIF might play an essential role in the development of chronic inflammation and cancer, and even carcinogenesis related to chronic inflammation.
The authors investigated MIF expression and its clinical relevance in gastric cancer, and effects of MIF knockdown on proliferation of gastric cancer cells. MIF expression was assessed by immunohistochemistry in a tissue microarray containing samples taken from 117 patients and its prognostic association was analysed. After siRNA transfection, MIF mRNA was measured by real-time PCR, MIF protein and proliferating cell nuclear antigen proliferating cell nuclear antigen were assessed by Western blot analysis, and the MTT assay, colony forming assay and cell counting were also performed.
This study demonstrates for the first time that MIF was highly expressed in gastric cancer compared with in adjacent healthy tissue, and was associated with poor differentiation, advanced disease and tumor stages, lymph node metastasis, and poor overall and disease-free survival. Moreover, the proliferation of gastric cancer cells was inhibited by MIF RNA interference.
Measurement of MIF expression has prognostic value in patients with gastric cancer. The proliferation of gastric cancer cells was inhibited by MIF knockdown, which suggests that MIF could be a suitable target for small-molecule therapy.
Macrophage migration inhibitory factor is a multi-functional cytokine, which is associated with inflammation and plays a significant role in tumorigenesis.
This study, consisting of clinical and experimental data, further explored the role of MIF in gastric carcinogenesis. Whereas the clinical data confirmed the previous findings, the experimental data in AGS cells using siRNAs provided novel evidence that MIF plays a critical role in the development of gastric cancer and MIF may be a potential therapeutic target.
P- Reviewer: Carulli L, Matsui H S- Editor: Yu J L- Editor: Wang TQ E- Editor: Liu XM
| 1. | David JR. Delayed hypersensitivity in vitro: its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc Natl Acad Sci USA. 1966;56:72-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 914] [Cited by in F6Publishing: 958] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 2. | Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153:80-82. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1173] [Cited by in F6Publishing: 1119] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 3. | Yu T, Morita I, Shimokado K, Iwai T, Yoshida M. Amlodipine modulates THP-1 cell adhesion to vascular endothelium via inhibition of protein kinase C signal transduction. Hypertension. 2003;42:329-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Huang XR, Chun Hui CW, Chen YX, Wong BC, Fung PC, Metz C, Cho CH, Hui WM, Bucala R, Lam SK. Macrophage migration inhibitory factor is an important mediator in the pathogenesis of gastric inflammation in rats. Gastroenterology. 2001;121:619-630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 56] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Xia HH, Lam SK, Huang XR, Wong WM, Leung SY, Yuen ST, Lan HY, Wong BC. Helicobacter pylori infection is associated with increased expression of macrophage migratory inhibitory factor--by epithelial cells, T cells, and macrophages--in gastric mucosa. J Infect Dis. 2004;190:293-302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 24] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 6. | Wong BL, Zhu SL, Huang XR, Ma J, Xia HH, Bucala R, Wong BC, Lan HY. Essential role for macrophage migration inhibitory factor in gastritis induced by Helicobacter pylori. Am J Pathol. 2009;174:1319-1328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | White ES, Flaherty KR, Carskadon S, Brant A, Iannettoni MD, Yee J, Orringer MB, Arenberg DA. Macrophage migration inhibitory factor and CXC chemokine expression in non-small cell lung cancer: role in angiogenesis and prognosis. Clin Cancer Res. 2003;9:853-860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Akbar SM, Abe M, Murakami H, Tanimoto K, Kumagi T, Yamashita Y, Michitaka K, Horiike N, Onji M. Macrophage migration inhibitory factor in hepatocellular carcinoma and liver cirrhosis; relevance to pathogenesis. Cancer Lett. 2001;171:125-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 9. | Bando H, Matsumoto G, Bando M, Muta M, Ogawa T, Funata N, Nishihira J, Koike M, Toi M. Expression of macrophage migration inhibitory factor in human breast cancer: association with nodal spread. Jpn J Cancer Res. 2002;93:389-396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 88] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | He XX, Yang J, Ding YW, Liu W, Shen QY, Xia HH. Increased epithelial and serum expression of macrophage migration inhibitory factor (MIF) in gastric cancer: potential role of MIF in gastric carcinogenesis. Gut. 2006;55:797-802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Legendre H, Decaestecker C, Nagy N, Hendlisz A, Schüring MP, Salmon I, Gabius HJ, Pector JC, Kiss R. Prognostic values of galectin-3 and the macrophage migration inhibitory factor (MIF) in human colorectal cancers. Mod Pathol. 2003;16:491-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 77] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | del Vecchio MT, Tripodi SA, Arcuri F, Pergola L, Hako L, Vatti R, Cintorino M. Macrophage migration inhibitory factor in prostatic adenocarcinoma: correlation with tumor grading and combination endocrine treatment-related changes. Prostate. 2000;45:51-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 13. | Mitchell RA, Bucala R. Tumor growth-promoting properties of macrophage migration inhibitory factor (MIF). Semin Cancer Biol. 2000;10:359-366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 114] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 14. | Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190:1375-1382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 474] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 15. | Takahashi N, Nishihira J, Sato Y, Kondo M, Ogawa H, Ohshima T, Une Y, Todo S. Involvement of macrophage migration inhibitory factor (MIF) in the mechanism of tumor cell growth. Mol Med. 1998;4:707-714. [PubMed] [Cited in This Article: ] |
| 16. | Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J Immunol. 2006;177:8730-8739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 190] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 17. | He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu DZ, Luo GY, Zhu SL, Xie D. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac J Cancer Prev. 2012;13:3173-3178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 18. | Boltze C, Riecke A, Ruf CG, Port M, Nizze H, Kügler C, Miethke C, Wiest N, Abend M. Sporadic and radiation-associated papillary thyroid cancers can be distinguished using routine immunohistochemistry. Oncol Rep. 2009;22:459-467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 19. | Liao B, Zhong BL, Li Z, Tian XY, Li Y, Li B. Macrophage migration inhibitory factor contributes angiogenesis by up-regulating IL-8 and correlates with poor prognosis of patients with primary nasopharyngeal carcinoma. J Surg Oncol. 2010;102:844-851. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Xia HH, Yang Y, Chu KM, Gu Q, Zhang YY, He H, Wong WM, Leung SY, Yuen ST, Yuen MF. Serum macrophage migration-inhibitory factor as a diagnostic and prognostic biomarker for gastric cancer. Cancer. 2009;115:5441-5449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Han I, Lee MR, Nam KW, Oh JH, Moon KC, Kim HS. Expression of macrophage migration inhibitory factor relates to survival in high-grade osteosarcoma. Clin Orthop Relat Res. 2008;466:2107-2113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Ren Y, Chan HM, Li Z, Lin C, Nicholls J, Chen CF, Lee PY, Lui V, Bacher M, Tam PK. Upregulation of macrophage migration inhibitory factor contributes to induced N-Myc expression by the activation of ERK signaling pathway and increased expression of interleukin-8 and VEGF in neuroblastoma. Oncogene. 2004;23:4146-4154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 72] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 23. | Nabizadeh Marvast M, Sima HR, Ghaffarzadehgan K, Taghizadeh Kermani A, Norouzi N. Clinicopathological significance of macrophage migration inhibitory factor and its relation with p53 in gastric cancer. J Gastrointest Cancer. 2011;42:5-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | Czauderna F, Fechtner M, Dames S, Aygün H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705-2716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 447] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 25. | Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, Karpilow J, Marshall WS, Khvorova A. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12:988-993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 243] [Cited by in F6Publishing: 231] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 26. | Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 649] [Cited by in F6Publishing: 637] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 27. | Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757-1766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 502] [Cited by in F6Publishing: 548] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 28. | Dessein AF, Stechly L, Jonckheere N, Dumont P, Monté D, Leteurtre E, Truant S, Pruvot FR, Figeac M, Hebbar M. Autocrine induction of invasive and metastatic phenotypes by the MIF-CXCR4 axis in drug-resistant human colon cancer cells. Cancer Res. 2010;70:4644-4654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 29. | Munaut C, Boniver J, Foidart JM, Deprez M. Macrophage migration inhibitory factor (MIF) expression in human glioblastomas correlates with vascular endothelial growth factor (VEGF) expression. Neuropathol Appl Neurobiol. 2002;28:452-460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Shimizu T, Abe R, Nakamura H, Ohkawara A, Suzuki M, Nishihira J. High expression of macrophage migration inhibitory factor in human melanoma cells and its role in tumor cell growth and angiogenesis. Biochem Biophys Res Commun. 1999;264:751-758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 157] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 31. | Li Z, Ren Y, Wu QC, Lin SX, Liang YJ, Liang HZ. Macrophage migration inhibitory factor enhances neoplastic cell invasion by inducing the expression of matrix metalloproteinase 9 and interleukin-8 in nasopharyngeal carcinoma cell lines. Chin Med J (Engl). 2004;117:107-114. [PubMed] [Cited in This Article: ] |
| 32. | Meyer-Siegler K, Hudson PB. Enhanced expression of macrophage migration inhibitory factor in prostatic adenocarcinoma metastases. Urology. 1996;48:448-452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 99] [Article Influence: 3.5] [Reference Citation Analysis (0)] |