Published online Jan 28, 2014. doi: 10.3748/wjg.v20.i4.978
Revised: November 12, 2013
Accepted: December 12, 2013
Published online: January 28, 2014
Processing time: 120 Days and 3.4 Hours
Despite the recent advances in the therapeutic modalities, colorectal cancer (CRC) remains to be one of the most common causes of cancer-related death. CRC arises through accumulation of multiple genetic and epigenetic alterations that transform normal colonic epithelium into adenocarcinomas. Among crucial roles of epigenetic alterations, gene silencing by aberrant DNA methylation of promoter regions is one of the most important epigenetic mechanisms. Recent comprehensive methylation analyses on genome-wide scale revealed that sporadic CRC can be classified into distinct epigenotypes. Each epigenotype cooperates with specific genetic alterations, suggesting that they represent different molecular carcinogenic pathways. Precursor lesions of CRC, such as conventional and serrated adenomas, already show similar methylation accumulation to CRC, and can therefore be classified into those epigenotypes of CRC. In addition, specific DNA methylation already occurs in the normal colonic mucosa, which might be utilized for prediction of the personal CRC risk. DNA methylation is suggested to occur at an earlier stage than carcinoma formation, and may predict the molecular basis for future development of CRC. Here, we review DNA methylation and CRC classification, and discuss the possible clinical usefulness of DNA methylation as biomarkers for the diagnosis, prediction of the prognosis and the response to therapy of CRC.
Core tip: Colorectal cancer (CRC) is a heterogeneous disease which involves several distinct molecular carcinogenetic pathways. Recent comprehensive genome-wide analyses clarify detailed DNA methylation statuses of cancer-related genes in CRC. We and others have investigated the association between DNA methylation and genetic alterations, and performed classification of CRC/their precursors, including conventional adenomas, serrated adenomas, non-polypoid colorectal neoplasms and aberrant crypt foci. In addition, we also evaluated the usefulness of DNA methylation markers as surrogate biomarkers for diagnosis, prognosis and therapeutic application of CRC. Here, we review the DNA methylation status and classification of CRC to understand the roles of DNA methylation in colorectal carcinogenesis.
- Citation: Sakai E, Nakajima A, Kaneda A. Accumulation of aberrant DNA methylation during colorectal cancer development. World J Gastroenterol 2014; 20(4): 978-987
- URL: https://www.wjgnet.com/1007-9327/full/v20/i4/978.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i4.978
Colorectal cancer (CRC) arises through accumulation of multiple acquired genetic and epigenetic alterations that cause malignant transformation of normal colonic epithelium to adenocarcinoma[1,2]. These carcinogenetic processes were first described in the model of the adenoma-carcinoma sequence[3], and somatic mutations of tumor-suppressor genes (e.g., APC, p53 and DCC) and activating mutations in the KRAS oncogene are well-known genetic alterations involved in this model[3-5] (Table 1).
CRC can be biologically divided into those with microsatellite instability (MSI), characterized by DNA replication and repair defects, and those with chromosomal instability (CIN), characterized by aneuploidy, multiple chromosomal rearrangements and accumulation of somatic mutations in oncogenes[6]. These genomic instabilities have been reported to be closely associated with the molecular heterogeneity of CRC[7], which is a factor responsible for the significant variability in the prognosis and treatment response among patients with the same stage of CRC[8]. Since the distinct molecular subtypes of CRC are difficult to be accurately distinguished histologically or clinically, technologies that can detect significant molecular alterations in CRC on genome-wide scale had been expected to be developed. Recently, exome sequencing analyses revealed the involvement of many somatic mutations of genes, including SMAD4, FBXW7, TCF7L2, and FAM123B[9-11]. Although hundreds of mutations, on average, are found in genomes of CRC, only a small set of functionally important genes are proposed to be involved with cancer formation as driver genes in individual cancer[11]. Whereas key mutational changes are necessary for the initiation and progression of CRC, the number of genes silenced by epigenetic mechanisms is greater than the number of genetic mutations in CRC[11], suggesting a crucial role of epigenetic alterations.
Recently, we and other groups performed epigenotyping of CRC, by unsupervised hierarchical clustering method using comprehensive and quantitative methylation data (Table 2). These results demonstrated that CRC can be clearly clustered into three DNA methylation epigenotypes[12-14]. Interestingly, each of the epigenotypes showed a unique association with a variety of genetic mutations (i.e., BRAF, KRAS and TP53) and the genomic instability status of sporadic CRC, indicating that they develop through distinct carcinogenetic pathways. Moreover, the intermediate-methylation subtype with KRAS mutation showed a poorer prognosis than other subtypes, suggesting that the DNA methylation status could be used as prognostic markers. Subsequently, we conducted epigenotyping of conventional adenomas and serrated adenomas, and showed that these precursor lesions of CRC can also be classified into the three epigenotypes[15]. These findings suggested that epigenotype development occur at an earlier stage than carcinoma formation, and is already completed at the adenoma stage. In this review, we focus on the importance of DNA methylation and provide an overview of the classification of CRC and their precursors, and discuss the clinical applications of aberrant DNA methylation as biomarkers for the diagnosis, prediction of the prognosis and the response to therapy of CRC.
| Ref. | Marker selection | Methylation analysis methods | Classification method | Methylation phenotypes |
| Toyota et al[20] | Genome-wide (MCA-RDA) | COBRA | Methlation frequency | CIMP+ |
| CIMP- | ||||
| Weisenberger et al[35] | MethyLight markers | MethyLight | Hierarchical clustering | CIMP+ |
| CIMP- | ||||
| Ogino et al[39] | Reported markers | MethyLight | Methylation frequency | CIMP-high |
| CIMP-low | ||||
| CIMP-0 | ||||
| Shen et al[13] | Reported markers | Pyrosequence | Hierarchical clustering | CIMP1 |
| COBRA | CIMP2 | |||
| MCA | CIMP-negative | |||
| MSP | ||||
| Yagi et al[14] | Genome-wide (MeDIP-chip) | MassARRAY | Hierarchical clustering | HME |
| IME | ||||
| LME | ||||
| Hinoue et al[12] | Genome-wide (Infinium 27k) | MethyLight | Hierarchical clustering | CIMP-H |
| CIMP-L | ||||
| Non-CIMP |
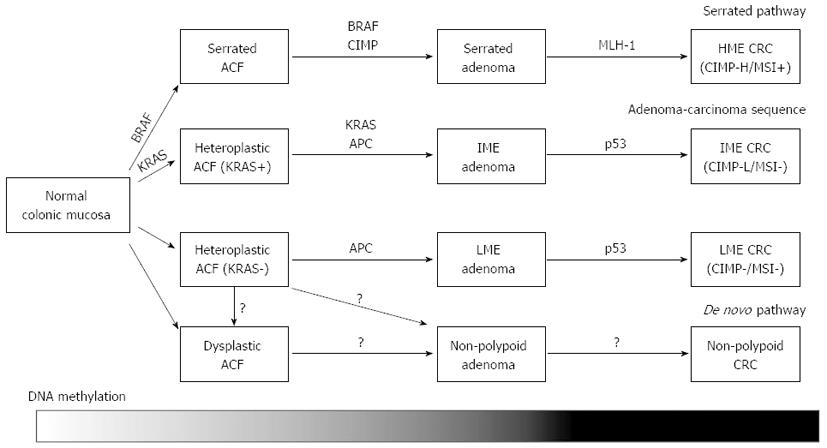
Two major types of epigenetic alterations closely linked to CRC are aberrant DNA methylation and covalent histone modifications[16]. Gene silencing by aberrant DNA methylation of its promoter region is one of the most important epigenetic mechanisms to inactivate the expression of tumor-suppressor genes. While the majority of CpG sites in the genome are known to be methylated in normal mammalian cells, unmethylated CpG sites are typically present in the genomic regions known as CpG islands. CpG islands are reported to overlap the promoter regions in 60%-70% of genes and tend to be protected from methylation; however they can be aberrantly methylated during the carcinogenetic process[17]. Investigation of genes using aberrant methylation as markers is useful to identify novel tumor-suppressor genes and methylation markers for cancer classification[18-23]. Therefore, several methods for genome-wide analysis have been developed since the 1990’s[24-28]. These epigenetic alterations have been noted to play crucial roles not only in cancer progression, but also in cancer initiation, since the alterations have been identified in the pre-cancerous “normal” tissues that could modify cancer risk[2,29-31]. Recently, genome-wide DNA methylation analysis tools have been developed to reveal the detailed epigenetic backgrounds of CRC[12-14]. Importantly, gene silencing resulting from aberrant DNA methylation cooperates with other genetic mechanisms to alter the key molecular pathways critical in colorectal carcinogenesis[29] (Figure 1).
In 1999, Toyota et al[21] reported that some CRCs show a significantly high frequency of aberrant DNA methylation in specific CpG islands, named CpG island methylator phenotype (CIMP). CIMP-positive CRC shows DNA hypermethylation at a specific subset of genomic loci[32,33] and is highly enriched for an activating mutation of BRAF[34,35]. Hypermethylation of CpG islands in gene promoter regions results in transcriptional repression. For example, CIMP-mediated gene silencing of the mismatch repair gene MLH1 by promoter hypermethylation is the molecular basis for MSI in sporadic microsatellite-unstable CRC, and most sporadic microsatellite-unstable CRC are therefore CIMP-positive[36]. CIMP-positive CRC inversely correlates with CRC with CIN[37,38], tends to occur in the proximal colon, and is commonly observed in women[32], suggesting that they appear to develop distinct carcinogenetic pathway from CIMP-negative CRC.
Since the first CIMP markers were identified by Toyota et al[20,21], many other CIMP markers have been described, e.g., MLH1, NEUROG1, SOCS1, RUNX3, IGF2 and CACNA1G[18-22,35]. Using quantitative real-time PCR, Ogino et al[39] selected five CIMP markers to distinguish high from low levels of CIMP-mediated gene promoter methylation, and found that CIMP-low CRC tends to be associated with male sex and KRAS mutations. CIMP-low appears to be independent of the MSI status, suggesting that CIMP-low might be a different subtype of CRC from CIMP-high and CIMP-0. However, no clear difference was observed between CIMP-low and CIMP-0, because these methylation markers were specific for CIMP-high and not ideal for identification of the CIMP-low subtype. Sensitive and specific markers for CIMP-low were needed to be determined.
According to the results of unsupervised two-way hierarchical clustering based on the quantitative DNA methylation data of 27 previously reported gene promoter and genetic alterations, including mutations of BRAF, KRAS, and p53, Shen et al[13] proposed that CRC can be classified into three subsets, CIMP1, CIMP2, and CIMP-negative. This report successfully showed the existence of three clusters of CRC with different molecular characteristics: (1) CIMP1 with MSI-high (80%), BRAF mutation (53%) and high-methylation; (2) CIMP2 with KRAS mutation (92%) and different methylation; and (3) CIMP-negative with p53 mutation (71%) and absence of these methylations. Integrated genetic and epigenetic analysis was found to be important, and genetic markers performed better than epigenetic markers in their classification of CRC[13].
To clarify whether CRC can be classified into more than two subsets using information on methylation accumulation alone, we performed comprehensive two-way unsupervised hierarchical clustering, using quantitative methylation data of genome-widely selected novel markers that were established through MeDIP-chip analysis. We demonstrated that CRC can be clearly classified into three distinct epigenotypes: high-, intermediate-, and low-methylation epigenotypes (HME, IME, and LME). HME was strongly correlated to the presence of the BRAF mutation (71%) and MSI-high (76%), and IME was strongly correlated to the presence of the KRAS mutation (63%)[14]. In our analysis, p53 mutation was absent in HME, but was detected in both IME and LME. It was noteworthy that the methylation markers were clustered into two groups: (1) Group-1 markers included most of the known CIMP markers and showed methylation specifically in HME CRC; and (2) Group-2 markers including novel methylation markers which showed methylation in both HME and IME. It was also noteworthy that patients with IME KRAS-mutation(+) CRC showed a significantly worse prognosis.
Subsequent to our report, Hinoue et al[12] performed DNA methylation profiling of CRC using Illumina Infinium DNA methylation beadarray, and reported that CRC can be classified into three distinct epigenotypes (CIMP-H, CIMP-L and Non-CIMP), consistent with previous reports[13,14]. Genetic and epigenetic features of CIMP-H/CIMP-L CRC are also in agreement with those observed in the CIMP1/CIMP2 CRC[13] and the HME/IME CRC[14]. According to the frequency of p53 mutation, they proposed that non-CIMP CRC could be classified into two distinct sub-groups; one with a significantly higher frequency of p53 mutations (65%) and frequent occurrence in the distal colon, and the other with absence of both cancer-specific DNA methylation and gene mutations, and more frequent occurrence in the rectum.
The majority of sporadic CRC is thought to develop from conventional adenomas through the adenoma-carcinoma sequence[3], whereas the serrated pathway has been considered as an alternative pathway distinct from the adenoma-carcinoma sequence. The serrated pathway is known to involve mutation of BRAF, MLH1 methylation and CIMP[34,35]. While serrated adenomas are commonly CIMP-high and carry the BRAF mutation[34,40-45], conventional adenomas rarely exhibit these genetic and epigenetic alterations[40]. In addition, the risk factors for CIMP-high serrated adenomas are reported to be similar to those of CRC with CIMP[40]. Serrated adenoma is therefore considered to be a precursor of CIMP-positive CRC. Although CIMP-high and BRAF mutation were frequently observed at the adenoma stage, the prevalence of MLH1 methylation was lower than that of CRC with CIMP[40-42,45]. Interestingly, MLH1 methylation was more frequently observed in proximal, large serrated adenomas[40], suggesting that MLH1 methylation is a late event in the serrated pathway, and heralds the transition from serrated adenoma to CRC, involving the mutator phenotype.
While the genetic and epigenetic features among conventional and serrated adenomas have been demonstrated to be widely different, existence of DNA methylation phenotypes within conventional adenomas and their correlation to genetic mutations were not fully investigated. We investigated whether conventional adenomas could be classified into epigenotypes, and our CRC classification markers successfully classified conventional adenomas into two distinct epigenotypes, IME and LME[15]. There were no remarkable differences in the morphological and pathological features among the two epigenotypes. While IME adenomas showed a significantly high frequency of the KRAS mutation (62%), LME adenomas did not show any genetic alterations, similar to the case of LME CRC. Interestingly, there was no difference in the methylation level between IME adenoma and IME cancer, suggesting that accumulation of aberrant DNA methylation is mostly completed at the adenoma stage. This indicated that additional aberration(s) other than DNA methylation are needed for adenomas to transform into CRC. The progression of adenomas to CRC is postulated to be associated with p53 abnormalities[3], and DNA methylation of some genes, e.g., MGMT, CXLC12, TIMP3, ID4 and IRF8, might also be involved in the development to CRC[46,47].
Non-polypoid colorectal neoplasms that do not exhibit a macroscopic protruding appearance have been documented not only in Japan[48-53], but also in western countries[54,55]. They are characterized by lateral extensions along the luminal wall with a low vertical axis, and such tumors with a diameter of > 10 mm are called laterally spreading tumors (LSTs)[48]. The incidence of genetic alterations such as KRAS, BRAF and p53[49,56-58] and MSI[59] were less common in these non-polypoid colorectal neoplasms than those in conventional adenomas. In addition, a high percentage of these lesions are reported to exhibit high-grade dysplasia and rapidly invade the submucosal layer despite their small sizes[50-52,60]. Therefore, non-polypoid colorectal neoplasms are hypothesized to develop through an alternative carcinogenetic pathway (i.e., de novo pathway) different from the adenoma-carcinoma sequence and the serrated pathway. LSTs are usually categorized into two subtypes based on their macroscopic morphology: the granular type and non-granular type[48]. Whereas the epigenetic features of LST have not yet been fully investigated, Hiraoka et al[61] reported frequent methylation of CIMP-markers and frequent KRAS mutation in the granular type, but not in the non-granular type. LST might be composed of several subtypes which exhibit distinct molecular pathway, and further investigations are needed to reveal the genetic and epigenetic features of nonpolypoid colorectal neoplasms and their association with colorectal carcinogenesis.
Aberrant crypt foci (ACF) are microscopic mucosal abnormalities, a subset of which postulated to be the earliest precursors of CRC[62]. ACF show increased expression of proliferative markers[63], and a significant correlation has been reported to exist between the presence of ACF and synchronous advanced neoplasia[62,64-71], suggesting a positive role of these lesions in colorectal carcinogenesis. Therefore, ACF have been recognized as a useful surrogate biomarker for CRC surveillance[72] and been used in recent chemoprevention trials[73-77]. Histopathologically, human ACF can be sub-classified into two categories: dysplastic and heteroplastic[62]. Dysplastic ACF resemble adenomas and sometimes lack mucin production[78], and are more common in familial adenomatous polyposis (FAP) patients than in sporadic CRC patients[79]. In contrast, heteroplastic ACF resemble hyperplastic polyps and lack dysplasia, and are highly identified in sporadic CRC patients.
Although all ACF from FAP patients carry the APC mutation[79], both the dysplastic and heteroplastic ACF from sporadic CRC patients frequently carry KRAS mutation, but not the APC mutation[79-81]. While BRAF mutation has rarely been identified in ACF[79,82], Rosenberg et al[83] reported that heteroplastic ACF with serrated pathology exclusively exhibit BRAF mutation. Although there was a report that CIMP-high was less frequently observed in ACF in sporadic CRC patients[80], the methylation status of ACF has not been well investigated. In our DNA methylation analysis in heteroplastic ACF, ACF showed frequent KRAS mutation, consist with previous reports. The levels of aberrant DNA methylation were significantly lower compared to adenomas[82], suggesting that DNA methylation accumulation might be requested during aberrant cell expansion in adenoma formation, but not in ACF formation.
Some of genes showing aberrant methylation in CRC, such as ESR1, IGF2 and TUSC3 are also methylated in histologically normal colonic epithelium. Aberrant DNA methylation of these genes is considered to increase in an age-dependent manner, and approximately half of them have also been shown to be involved in the pathogenesis of CRC[84-86].
The concept of “field cancerization” was proposed to explain the multiple primary lesions, local recurrence and increased susceptibility of normal tissue to malignant transformation[87]. The field changes occur at the molecular level, and these abnormalities of the normal colonic epithelium could be potential biomarkers for assessing the personal risk for future CRC development. Suzuki et al[88] reported that a higher incidence of hypermethylation and down-regulation of the SFRP genes, negative regulators of the WNT signaling pathway, were observed in the normal colonic mucosa from patients with CRC, than in that from patients without CRC. In addition, Kawakami et al[89] reported that higher methylation levels of age-related markers, such as ESR1 and MYOD, were observed in the normal colonic mucosa from patients with CIMP-positive CRC than in that from patients without CRC. It was reported, in contrast, that lower methylation levels of these markers were observed in the normal colonic mucosa from patients with CRC than from patients without CRC[90,91]. Recently, genome-wide DNA methylation analysis revealed that the gene methylation levels involved in the metabolic pathways of carbohydtates, lipids and amino acids were significantly different among normal colonic mucosa specimens obtained from patients with and without CRC[92]. While DNA methylation accumulation is expected to contribute to field cancerization in the colon, further studies are necessary to establish useful surrogate biomarkers for CRC surveillance.
Early CRC detection could contribute to a reduction of CRC-related mortality. However, strategies such as colonoscopy are invasive, whereas the less-invasive fecal blood test shows low sensitivity and specificity[93]. Identification of noninvasively testable, high-quality biomarkers for CRC is therefore necessary. Recent genome-wide analyses were conducted to identify candidate DNA methylation markers for early CRC detection, by comparing the DNA methylation levels between CRC and/or adenomas, and matched normal colonic mucosa[93-96]. For example, Mori et al[94] reported that the methylation status of VSX2 showed a high discriminative accuracy (83% sensitivity and 92% specificity). These potential biomarkers may allow reliable discrimination of CRC patients from tumor-free patients. Several clinical studies have been carried out to confirm the usefulness of stool and blood DNA-based methylation markers for early CRC detection[97,98]. In any application, classification marker genes are specifically methylated in some epigenotypes, therefore, genes that are commonly methylated in all CRCs, regardless of the epigenotype, would be useful markers for early CRC detection.
The MSI status has been proposed as a biomarker for determination of the prognosis and/or the effectiveness of FU chemotherapy in advanced CRC patients[99]. KRAS mutation in advanced CRC has been reported to be associated with a poor prognosis[100], and the usefulness of determining its presence for predicting a lack of response to EGFR-targeted therapy is well proven[101]. Our previous study revealed that IME CRC with KRAS mutation is associated with a poor prognosis. DNA methylation biomarkers for prediction of the therapeutic responses of CRC, however, have not been identified yet. Additional studies are needed to establish methylation biomarkers for application in clinical practice, e.g., for prediction of the prognosis and of the responses to therapy of CRC.
Recent comprehensive genome-wide methylation analyses revealed that sporadic CRC can be classified into three distinct epigenotypes. Each of these CRC epigenotypes cooperates with specific genetic alterations, suggesting that they develop through different molecular carcinogenetic pathways. Serrated adenomas are commonly CIMP-high and carry BRAF mutation, thus postulated to be precursor lesions of CIMP-positive, MSI-high proximal CRCs. MLH1 methylation has been suggested to be a late event in the serrated pathway, and heralds the transition from serrated adenoma to CIMP-positive CRC. Conventional adenomas can also be classified into two distinct epigenotypes. DNA methylation accumulation is mostly completed by the adenoma stage, and conventional adenomas are hypothesized to be precursors of CIMP1/IME/CIMP-low and CIMP0/LME/Non-CIMP CRCs. ACF showed significantly lower methylation levels than adenomas, suggesting that DNA methylation accumulation is a prerequisite for aberrant cell expansion in adenoma formation, but not in the formation of ACF.
DNA methylation may predict the molecular basis of CRC, and these markers might be present as useful surrogate markers for the diagnosis, prediction of the prognosis and the response to therapy of CRC. Some genes already showed aberrant methylation in apparently normal colonic mucosa, and their methylation may be related to field cancerization of CRC and predict cancer risk. Continued efforts to investigate the associations between molecular mechanisms of CRC and genetic/epigenetic alterations may allow us to understand colorectal carcinogenesis, and lead to the translation of these insights into clinical practice.
P- Reviewers: Lakatos PL, Milone M, Sipos F S- Editor: Gou SX L- Editor: A E- Editor: Ma S
| 1. | Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079-1099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 717] [Cited by in RCA: 712] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 2. | Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65:11236-11240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 3. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4462] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 4. | Jen J, Kim H, Piantadosi S, Liu ZF, Levitt RC, Sistonen P, Kinzler KW, Vogelstein B, Hamilton SR. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331:213-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 524] [Cited by in RCA: 493] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 6. | Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159-170. [PubMed] |
| 7. | Dunican DS, McWilliam P, Tighe O, Parle-McDermott A, Croke DT. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene. 2002;21:3253-3257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 105] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1973] [Cited by in RCA: 2078] [Article Influence: 159.8] [Reference Citation Analysis (2)] |
| 9. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6630] [Article Influence: 510.0] [Reference Citation Analysis (0)] |
| 10. | Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153-158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2512] [Cited by in RCA: 2295] [Article Influence: 127.5] [Reference Citation Analysis (0)] |
| 11. | Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2562] [Cited by in RCA: 2540] [Article Influence: 133.7] [Reference Citation Analysis (0)] |
| 12. | Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 484] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 13. | Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA. 2007;104:18654-18659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 417] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 14. | Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21-33. [PubMed] |
| 15. | Yagi K, Takahashi H, Akagi K, Matsusaka K, Seto Y, Aburatani H, Nakajima A, Kaneda A. Intermediate methylation epigenotype and its correlation to KRAS mutation in conventional colorectal adenoma. Am J Pathol. 2012;180:616-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3299] [Cited by in RCA: 3398] [Article Influence: 188.8] [Reference Citation Analysis (0)] |
| 17. | Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5074] [Cited by in RCA: 4862] [Article Influence: 211.4] [Reference Citation Analysis (0)] |
| 18. | Kaneda A, Kaminishi M, Yanagihara K, Sugimura T, Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 2002;62:6645-6650. [PubMed] |
| 19. | Kaneda A, Wakazono K, Tsukamoto T, Watanabe N, Yagi Y, Tatematsu M, Kaminishi M, Sugimura T, Ushijima T. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res. 2004;64:6410-6415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 138] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 20. | Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681-8686. [PubMed] |
| 21. | Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307-2312. [PubMed] |
| 22. | Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 359] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 23. | Ushijima T, Morimura K, Hosoya Y, Okonogi H, Tatematsu M, Sugimura T, Nagao M. Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc Natl Acad Sci USA. 1997;94:2284-2289. [PubMed] |
| 24. | Gu H, Bock C, Mikkelsen TS, Jäger N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133-136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 25. | Hayashi H, Nagae G, Tsutsumi S, Kaneshiro K, Kozaki T, Kaneda A, Sugisaki H, Aburatani H. High-resolution mapping of DNA methylation in human genome using oligonucleotide tiling array. Hum Genet. 2007;120:701-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 42] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 26. | Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1096] [Article Influence: 73.1] [Reference Citation Analysis (0)] |
| 27. | Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3346] [Cited by in RCA: 3358] [Article Influence: 209.9] [Reference Citation Analysis (0)] |
| 28. | Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1315] [Cited by in RCA: 1289] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 29. | Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1182] [Cited by in RCA: 1184] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 30. | Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1263] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 31. | Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976-1978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 232] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 32. | Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O’Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376-1387. [PubMed] |
| 33. | Whitehall VL, Wynter CV, Walsh MD, Simms LA, Purdie D, Pandeya N, Young J, Meltzer SJ, Leggett BA, Jass JR. Morphological and molecular heterogeneity within nonmicrosatellite instability-high colorectal cancer. Cancer Res. 2002;62:6011-6014. [PubMed] |
| 34. | Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 566] [Article Influence: 27.0] [Reference Citation Analysis (1)] |
| 35. | Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1423] [Cited by in RCA: 1491] [Article Influence: 78.5] [Reference Citation Analysis (0)] |
| 36. | Samowitz WS. The CpG island methylator phenotype in colorectal cancer. J Mol Diagn. 2007;9:281-283. [PubMed] |
| 37. | Cheng YW, Pincas H, Bacolod MD, Schemmann G, Giardina SF, Huang J, Barral S, Idrees K, Khan SA, Zeng Z. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005-6013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 38. | Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 211] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 39. | Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 253] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 40. | Burnett-Hartman AN, Newcomb PA, Potter JD, Passarelli MN, Phipps AI, Wurscher MA, Grady WM, Zhu LC, Upton MP, Makar KW. Genomic aberrations occurring in subsets of serrated colorectal lesions but not conventional adenomas. Cancer Res. 2013;73:2863-2872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 41. | Kim YH, Kakar S, Cun L, Deng G, Kim YS. Distinct CpG island methylation profiles and BRAF mutation status in serrated and adenomatous colorectal polyps. Int J Cancer. 2008;123:2587-2593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 42. | Maeda T, Suzuki K, Togashi K, Nokubi M, Saito M, Tsujinaka S, Kamiyama H, Konishi F. Sessile serrated adenoma shares similar genetic and epigenetic features with microsatellite unstable colon cancer in a location-dependent manner. Exp Ther Med. 2011;2:695-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 43. | O'Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, Amorosino M, Farraye FA. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491-1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 381] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 44. | Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, Goldblum JR, Guillem JG, Kahi CJ, Kalady MF. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315-129; quiz 1314, 1330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 825] [Cited by in RCA: 828] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 45. | Vaughn CP, Wilson AR, Samowitz WS. Quantitative evaluation of CpG island methylation in hyperplastic polyps. Mod Pathol. 2010;23:151-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 230] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 47. | Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113-7117. [PubMed] |
| 48. | Kudo S, Kashida H, Nakajima T, Tamura S, Nakajo K. Endoscopic diagnosis and treatment of early colorectal cancer. World J Surg. 1997;21:694-701. [PubMed] |
| 49. | Minamoto T, Sawaguchi K, Mai M, Yamashita N, Sugimura T, Esumi H. Infrequent K-ras activation in superficial-type (flat) colorectal adenomas and adenocarcinomas. Cancer Res. 1994;54:2841-2844. [PubMed] |
| 50. | Minamoto T, Sawaguchi K, Ohta T, Itoh T, Mai M. Superficial-type adenomas and adenocarcinomas of the colon and rectum: a comparative morphological study. Gastroenterology. 1994;106:1436-1443. [PubMed] |
| 51. | Tada S, Iida M, Matsumoto T, Yao T, Aoyagi K, Koga H, Tanoue Y, Fujishima M. Small flat cancer of the rectum: clinicopathologic and endoscopic features. Gastrointest Endosc. 1995;42:109-113. [PubMed] |
| 52. | Watari J, Saitoh Y, Obara T, Fujiya M, Maemoto A, Ayabe T, Ashida T, Yokota K, Orii Y, Kohgo Y. Natural history of colorectal nonpolypoid adenomas: a prospective colonoscopic study and relation with cell kinetics and K-ras mutations. Am J Gastroenterol. 2002;97:2109-2115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 53. | Muto T, Kamiya J, Sawada T, Konishi F, Sugihara K, Kubota Y, Adachi M, Agawa S, Saito Y, Morioka Y. Small “flat adenoma” of the large bowel with special reference to its clinicopathologic features. Dis Colon Rectum. 1985;28:847-851. [PubMed] |
| 54. | Soetikno R, Friedland S, Kaltenbach T, Chayama K, Tanaka S. Nonpolypoid (flat and depressed) colorectal neoplasms. Gastroenterology. 2006;130:566-576; quiz 588-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 80] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 55. | Testoni PA, Notaristefano C, Vailati C, Di Leo M, Viale E. High-definition colonoscopy with i-Scan: better diagnosis for small polyps and flat adenomas. World J Gastroenterol. 2012;18:5231-5239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 56. | Morita T, Tomita N, Ohue M, Sekimoto M, Yamamoto H, Ohnishi T, Tada M, Ikenaga M, Miyake Y, Sakita I. Molecular analysis of diminutive, flat, depressed colorectal lesions: are they precursors of polypoid adenoma or early stage carcinoma? Gastrointest Endosc. 2002;56:663-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 57. | Saitoh Y, Waxman I, West AB, Popnikolov NK, Gatalica Z, Watari J, Obara T, Kohgo Y, Pasricha PJ. Prevalence and distinctive biologic features of flat colorectal adenomas in a North American population. Gastroenterology. 2001;120:1657-1665. [PubMed] |
| 58. | Yashiro M, Carethers JM, Laghi L, Saito K, Slezak P, Jaramillo E, Rubio C, Koizumi K, Hirakawa K, Boland CR. Genetic pathways in the evolution of morphologically distinct colorectal neoplasms. Cancer Res. 2001;61:2676-2683. [PubMed] |
| 59. | Kinney TP, Merel N, Hart J, Joseph L, Waxman I. Microsatellite analysis of sporadic flat and depressed lesions of the colon. Dig Dis Sci. 2005;50:327-330. [PubMed] |
| 60. | Rembacken BJ, Fujii T, Cairns A, Dixon MF, Yoshida S, Chalmers DM, Axon AT. Flat and depressed colonic neoplasms: a prospective study of 1000 colonoscopies in the UK. Lancet. 2000;355:1211-1214. [PubMed] |
| 61. | Hiraoka S, Kato J, Tatsukawa M, Harada K, Fujita H, Morikawa T, Shiraha H, Shiratori Y. Laterally spreading type of colorectal adenoma exhibits a unique methylation phenotype and K-ras mutations. Gastroenterology. 2006;131:379-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 62. | Takayama T, Katsuki S, Takahashi Y, Ohi M, Nojiri S, Sakamaki S, Kato J, Kogawa K, Miyake H, Niitsu Y. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N Engl J Med. 1998;339:1277-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 397] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 63. | Shpitz B, Bomstein Y, Mekori Y, Cohen R, Kaufman Z, Neufeld D, Galkin M, Bernheim J. Aberrant crypt foci in human colons: distribution and histomorphologic characteristics. Hum Pathol. 1998;29:469-475. [PubMed] |
| 64. | Adler DG, Gostout CJ, Sorbi D, Burgart LJ, Wang L, Harmsen WS. Endoscopic identification and quantification of aberrant crypt foci in the human colon. Gastrointest Endosc. 2002;56:657-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 65. | Hurlstone DP, Karajeh M, Sanders DS, Drew SK, Cross SS. Rectal aberrant crypt foci identified using high-magnification-chromoscopic colonoscopy: biomarkers for flat and depressed neoplasia. Am J Gastroenterol. 2005;100:1283-1289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 66. | Kim J, Ng J, Arozulllah A, Ewing R, Llor X, Carroll RE, Benya RV. Aberrant crypt focus size predicts distal polyp histopathology. Cancer Epidemiol Biomarkers Prev. 2008;17:1155-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 67. | Moxon D, Raza M, Kenney R, Ewing R, Arozullah A, Mason JB, Carroll RE. Relationship of aging and tobacco use with the development of aberrant crypt foci in a predominantly African-American population. Clin Gastroenterol Hepatol. 2005;3:271-278. [PubMed] |
| 68. | Rudolph RE, Dominitz JA, Lampe JW, Levy L, Qu P, Li SS, Lampe PD, Bronner MP, Potter JD. Risk factors for colorectal cancer in relation to number and size of aberrant crypt foci in humans. Cancer Epidemiol Biomarkers Prev. 2005;14:605-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 69. | Sakai E, Takahashi H, Kato S, Uchiyama T, Hosono K, Endo H, Maeda S, Yoneda M, Taguri M, Nakajima A. Investigation of the prevalence and number of aberrant crypt foci associated with human colorectal neoplasm. Cancer Epidemiol Biomarkers Prev. 2011;20:1918-1924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 70. | Seike K, Koda K, Oda K, Kosugi C, Shimizu K, Nishimura M, Shioiri M, Takano S, Ishikura H, Miyazaki M. Assessment of rectal aberrant crypt foci by standard chromoscopy and its predictive value for colonic advanced neoplasms. Am J Gastroenterol. 2006;101:1362-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 71. | Stevens RG, Swede H, Heinen CD, Jablonski M, Grupka M, Ross B, Parente M, Tirnauer JS, Giardina C, Rajan TV. Aberrant crypt foci in patients with a positive family history of sporadic colorectal cancer. Cancer Lett. 2007;248:262-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 72. | Uchiyama T, Takahashi H, Endo H, Kato S, Sakai E, Hosono K, Yoneda M, Inamori M, Hippo Y, Nakagama H. Number of aberrant crypt foci in the rectum is a useful surrogate marker of colorectal adenoma recurrence. Dig Endosc. 2012;24:353-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 73. | Cho NL, Redston M, Zauber AG, Carothers AM, Hornick J, Wilton A, Sontag S, Nishioka N, Giardiello FM, Saltzman JR. Aberrant crypt foci in the adenoma prevention with celecoxib trial. Cancer Prev Res (Phila). 2008;1:21-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 74. | Higurashi T, Hosono K, Endo H, Takahashi H, Iida H, Uchiyama T, Ezuka A, Uchiyama S, Yamada E, Ohkubo H. Eicosapentaenoic acid (EPA) efficacy for colorectal aberrant crypt foci (ACF): a double-blind randomized controlled trial. BMC Cancer. 2012;12:413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 75. | Higurashi T, Takahashi H, Endo H, Hosono K, Yamada E, Ohkubo H, Sakai E, Uchiyama T, Hata Y, Fujisawa N. Metformin efficacy and safety for colorectal polyps: a double-blind randomized controlled trial. BMC Cancer. 2012;12:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 76. | Hosono K, Endo H, Takahashi H, Sugiyama M, Sakai E, Uchiyama T, Suzuki K, Iida H, Sakamoto Y, Yoneda K. Metformin suppresses colorectal aberrant crypt foci in a short-term clinical trial. Cancer Prev Res (Phila). 2010;3:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 242] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 77. | Takayama T, Nagashima H, Maeda M, Nojiri S, Hirayama M, Nakano Y, Takahashi Y, Sato Y, Sekikawa H, Mori M. Randomized double-blind trial of sulindac and etodolac to eradicate aberrant crypt foci and to prevent sporadic colorectal polyps. Clin Cancer Res. 2011;17:3803-3811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 78. | Sakai E, Morioka T, Yamada E, Ohkubo H, Higurashi T, Hosono K, Endo H, Takahashi H, Takamatsu R, Cui C. Identification of preneoplastic lesions as mucin-depleted foci in patients with sporadic colorectal cancer. Cancer Sci. 2012;103:144-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 79. | Takayama T, Ohi M, Hayashi T, Miyanishi K, Nobuoka A, Nakajima T, Satoh T, Takimoto R, Kato J, Sakamaki S. Analysis of K-ras, APC, and beta-catenin in aberrant crypt foci in sporadic adenoma, cancer, and familial adenomatous polyposis. Gastroenterology. 2001;121:599-611. [PubMed] |
| 80. | Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823-1830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 172] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 81. | Otori K, Konishi M, Sugiyama K, Hasebe T, Shimoda T, Kikuchi-Yanoshita R, Mukai K, Fukushima S, Miyaki M, Esumi H. Infrequent somatic mutation of the adenomatous polyposis coli gene in aberrant crypt foci of human colon tissue. Cancer. 1998;83:896-900. [PubMed] |
| 82. | Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 520] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 83. | Rosenberg DW, Yang S, Pleau DC, Greenspan EJ, Stevens RG, Rajan TV, Heinen CD, Levine J, Zhou Y, O’Brien MJ. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer Res. 2007;67:3551-3554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 137] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 84. | Ahuja N, Issa JP. Aging, methylation and cancer. Histol Histopathol. 2000;15:835-842. [PubMed] |
| 85. | Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 864] [Cited by in RCA: 830] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 86. | Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Semin Cancer Biol. 1999;9:349-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 188] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 87. | Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963-968. [PubMed] |
| 88. | Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 837] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 89. | Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Grieu F, Watanabe G, Iacopetta B. DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer. 2006;94:593-598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 90. | Belshaw NJ, Elliott GO, Foxall RJ, Dainty JR, Pal N, Coupe A, Garg D, Bradburn DM, Mathers JC, Johnson IT. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. Br J Cancer. 2008;99:136-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 91. | Hiraoka S, Kato J, Horii J, Saito S, Harada K, Fujita H, Kuriyama M, Takemoto K, Uraoka T, Yamamoto K. Methylation status of normal background mucosa is correlated with occurrence and development of neoplasia in the distal colon. Hum Pathol. 2010;41:38-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 92. | Silviera ML, Smith BP, Powell J, Sapienza C. Epigenetic differences in normal colon mucosa of cancer patients suggest altered dietary metabolic pathways. Cancer Prev Res (Phila). 2012;5:374-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 93. | Hewitson P, Glasziou P, Watson E, Towler B, Irwig L. Cochrane systematic review of colorectal cancer screening using the fecal occult blood test (hemoccult): an update. Am J Gastroenterol. 2008;103:1541-1549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 676] [Cited by in RCA: 718] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 94. | Mori Y, Olaru AV, Cheng Y, Agarwal R, Yang J, Luvsanjav D, Yu W, Selaru FM, Hutfless S, Lazarev M. Novel candidate colorectal cancer biomarkers identified by methylation microarray-based scanning. Endocr Relat Cancer. 2011;18:465-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 95. | Simmer F, Brinkman AB, Assenov Y, Matarese F, Kaan A, Sabatino L, Villanueva A, Huertas D, Esteller M, Lengauer T. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics. 2012;7:1355-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 96. | Yi JM, Dhir M, Guzzetta AA, Iacobuzio-Donahue CA, Heo K, Yang KM, Suzuki H, Toyota M, Kim HM, Ahuja N. DNA methylation biomarker candidates for early detection of colon cancer. Tumour Biol. 2012;33:363-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 97. | Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat LF. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248-56; quiz e25-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (1)] |
| 98. | Tóth K, Sipos F, Kalmár A, Patai AV, Wichmann B, Stoehr R, Golcher H, Schellerer V, Tulassay Z, Molnár B. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One. 2012;7:e46000. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 151] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 99. | Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1344] [Article Influence: 67.2] [Reference Citation Analysis (1)] |
| 100. | Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103-2114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 766] [Cited by in RCA: 763] [Article Influence: 54.5] [Reference Citation Analysis (2)] |
| 101. | Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091-2096. [PubMed] |