Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Nov 28, 2013; 19(44): 8000-8010
Published online Nov 28, 2013. doi: 10.3748/wjg.v19.i44.8000
Published online Nov 28, 2013. doi: 10.3748/wjg.v19.i44.8000
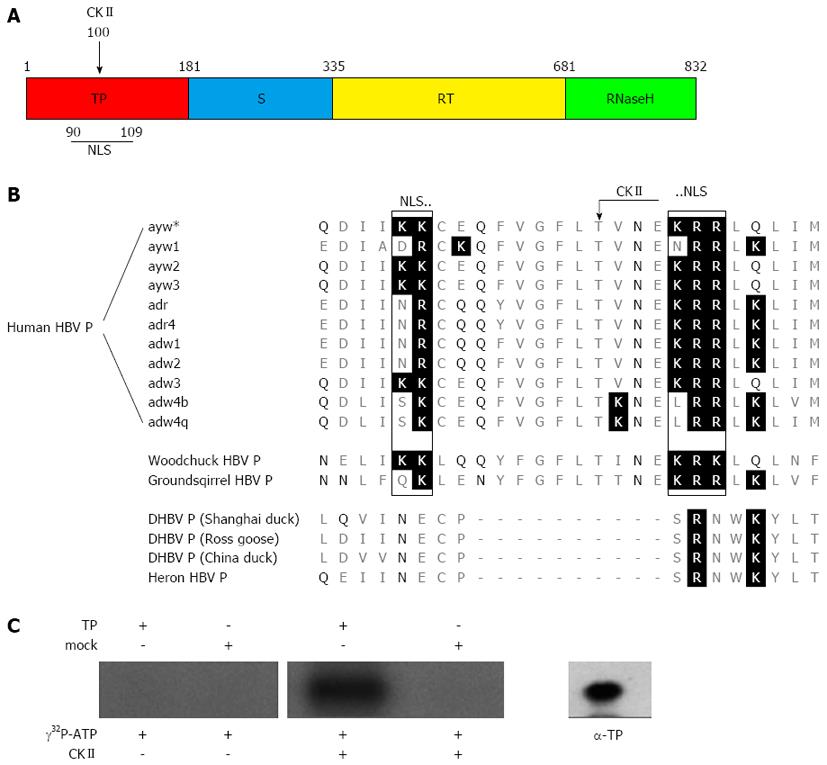
Figure 1 Sequence alignment of the hepatitis B virus polymerase from various virus subtypes and species.
A: Scheme of the hepatitis B virus (HBV) polymerase showing the different domains. The terminal protein domain (TP) is shown in red, the spacer domain (S) in blue, the reverse transcriptase domain (RT) in yellow and the RNase H domain in green. The numbers designate the amino acids referred to HBV genotype D. The positions of the casein kinase II (CKII) phosphorylation site and of the bipartite nuclear localization signal (NLS) identified in this study (see Figure 1B) are indicated; B: This figure shows amino acid alignment of Q86-M111 referred to the sequence of subtype ayw*, which was used in this study. Basic amino acids are highlighted by a black background and polar (δ+) amino acids are highlighted by black letters. A conserved protein kinase CKII recognition site (arrow) was identified in orthohepadnaviruses at Thr100 (protein kinase CKII: T/S-X-X-E/D; X = any amino acid). A bipartite nuclear localization signal was identified in orthohepadnaviruses, which is flanking the CKII recognition site by its two basic amino acid clusters (rectangles). All identified putative motifs were not found in the aligned P proteins of the compared avihepadnaviruses; C: Purified TP domain and mock purified proteins (empty vector products) were incubated with [γ-32P] ATP and recombinant protein kinase CKII. To control auto-phosphorylation TP domain was incubated with [γ-32P] ATP in absence of the kinase. The protein specificity was verified by Western blotting using a TP specific antibody (α-TP) on a separate lane. All experiments were performed in triplicate. One representative is shown.
- Citation: Lupberger J, Schaedler S, Peiran A, Hildt E. Identification and characterization of a novel bipartite nuclear localization signal in the hepatitis B virus polymerase. World J Gastroenterol 2013; 19(44): 8000-8010
- URL: https://www.wjgnet.com/1007-9327/full/v19/i44/8000.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i44.8000