Published online Apr 28, 2013. doi: 10.3748/wjg.v19.i16.2481
Revised: March 15, 2013
Accepted: March 21, 2013
Published online: April 28, 2013
AIM: To investigate the molecular mechanisms underlying the reversal effect of emodin on platinum resistance in hepatocellular carcinoma.
METHODS: After the addition of 10 μmol/L emodin to HepG2/oxaliplatin (OXA) cells, the inhibition rate (IR), 50% inhibitory concentration (IC50) and reversal index (IC50 in experimental group/IC50 in control group) were calculated. For HepG2, HepG2/OXA, HepG2/OXA/T, each cell line was divided into a control group, OXA group, OXA + fibroblast growth factor 7 (FGF7) group and OXA + emodin group, and the final concentrations of FGF7, emodin and OXA in each group were 5 ng/mL, 10 μg/mL and 10 μmol/L, respectively. Single-cell gel electrophoresis was conducted to detect DNA damage, and the fibroblast growth factor receptor 2 (FGFR2), phosphorylated extracellular signal-regulated kinase 1/2 (p-ERK1/2) and excision repair cross-complementing gene 1 (ERCC1) protein expression levels in each group were examined by Western blotting.
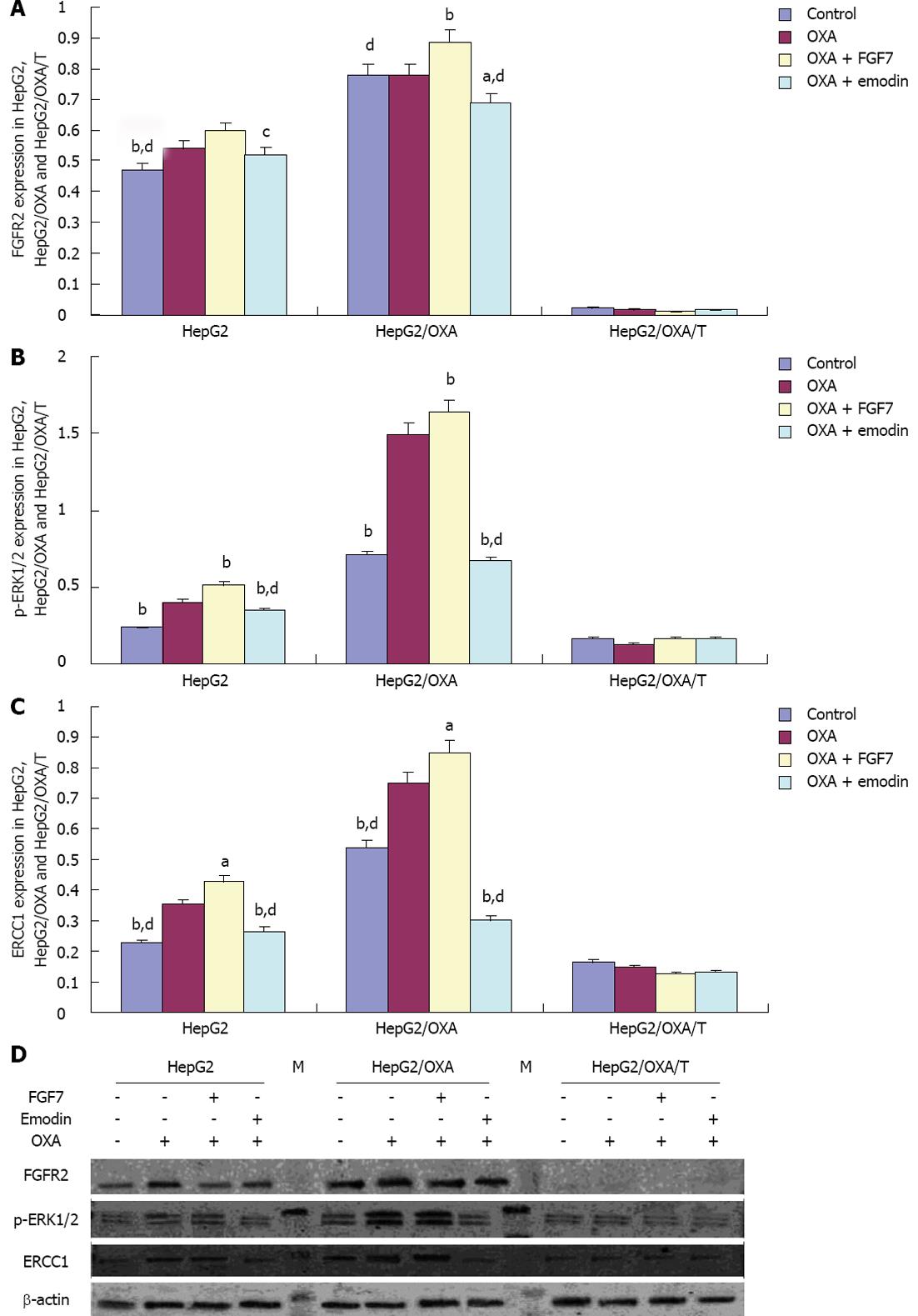
RESULTS: Compared with the IC50 of 120.78 μmol/L in HepG2/OXA cells, the IC50 decreased to 39.65 μmol/L after treatment with 10 μmol/L emodin; thus, the reversal index was 3.05. Compared with the control group, the tail length and Olive tail length in the OXA group, OXA + FGF7 group and OXA + emodin group were significantly increased, and the differences were statistically significant (P < 0.01). The tail length and Olive tail length were lower in the OXA + FGF7 group than in the OXA group, and this difference was also statistically significant. Compared with the OXA + FGF7 group, the tail extent, the Olive tail moment and the percentage of tail DNA were significantly increased in the OXA + emodin group, and these differences were statistically significant (P < 0.01). In comparison with its parental cell line HepG2, the HepG2/OXA cells demonstrated significantly increased FGFR2, p-ERK1/2 and ERCC1 expression levels, whereas the expression of all three molecules was significantly inhibited in HepG2/OXA/T cells, in which FGFR2 was silenced by FGFR2 shRNA. In the examined HepG2 cells, the FGFR2, p-ERK1/2 and ERCC1 expression levels demonstrated increasing trends in the OXA group and OXA + FGF7 group. Compared with the OXA group and OXA + FGF7 group, the FGFR2, p-ERK1/2, and ERCC1 expression levels were significantly lower in the OXA + emodin group, and these differences were statistically significant. In the HepG2/OXA/T cell line that was transfected with FGFR2 shRNA, the FGFR2, p-ERK1/2 and ERCC1 expression levels were significantly inhibited, but there were no significant differences in these expression levels among the OXA, OXA + FGF7 and OXA + emodin groups.
CONCLUSION: Emodin markedly reversed OXA resistance by enhancing OXA DNA damage in HepG2/OXA cells, and the molecular mechanism was related to the inhibitory effect on ERCC1 expression being mediated by the FGFR2/ERK1/2 signaling pathway.
Core tip: In this study, our results indicated that emodin could significantly enhance the DNA damage caused by oxaliplatin (OXA) and induce OXA resistance reversal in HepG2/OXA cells. The molecular mechanism for this phenomenon is mediated by the inhibition of excision repair cross-complementing gene 1 expression by the fibroblast growth factor receptor 2/phosphorylated extracellular signal-regulated kinase 1/2 signaling pathway. The results for the reversal of platinum resistance by emodin and the emodin-based enhancement of the efficacy of platinum-based chemotherapy in hepatocellular carcinoma may provide an experimental basis for the further development and application of emodin in the reversal of platinum drug resistance in other types of malignant tumors.
- Citation: Chen G, Qiu H, Ke SD, Hu SM, Yu SY, Zou SQ. Emodin regulating excision repair cross-complementation group 1 through fibroblast growth factor receptor 2 signaling. World J Gastroenterol 2013; 19(16): 2481-2491
- URL: https://www.wjgnet.com/1007-9327/full/v19/i16/2481.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i16.2481
For up to 85%-95% of the cases of liver cirrhosis in patients suffering from hepatocellular carcinoma (HCC), the therapeutic effects of chemotherapy have not been proven to provide survival benefits[1]. For the patient and the oncologist, sorafenib, which is a unique targeting drug that generates proven survival benefits, does not produce many unexpectedly positive outcomes because of its limited cost-effectiveness[2]. In 2010, at the annual meeting of the American Society of Clinical Oncology, the results of an international multi-center randomized phase III clinical study provided the first evidence demonstrating that platinum-based chemotherapy produces survival benefits for patients with advanced HCC. This discovery has triggered widespread interest into the use of oxaliplatin-based chemotherapy for HCC[3]. Thus, the identification and characterization of a drug that strengthens platinum-based chemotherapy effects and protects normal liver cells could provide clinical benefits for the treatment of HCC in China.
Emodin (1,3,8-trihydroxy-6-methylanthraquinone) is a member of the family of anthraquinone alkaloids, which are the main active ingredients of rhubarb, Polygonum cuspidatum, Polygonum multiflorum, Amomum, lilies and other plants that are widely used in traditional Chinese medicine[4]. Modern studies have demonstrated that emodin produces anti-tumor biological effects on a variety of malignancies, including HCC[5]. In HCC, the primary anti-cancer mechanisms of emodin involve the induction of apoptosis and the inhibition of cell growth. Emodin can cause G2/M cell cycle arrest by regulating an assortment of cell cycle related genes, such as cyclin A, cyclin B, Chk2, CDK2 and p27, in diverse human hepatoma cell lines, including Huh7, Hep3B and HepG2[6]. Moreover, emodin can enhance the cytotoxicity of chemotherapy drugs, such as platinum-based compounds [cisplatin, carboplatin, and oxaliplatin (OXA)] in various types of malignant tumors, including liver cancer, gallbladder cancer[7,8], non-small cell lung cancer[9,10], and prostate cancer[11]. However, the mechanism underlying the synergistic effects of combinations of emodin and platinum-based drugs requires further elucidation.
The DNA damage that is caused by cisplatin, OXA and other platinum chemotherapy drugs is the root cause of the cytotoxicity of these compounds[12]. Nucleotide excision repair (NER) is the main pathway for repairing this damage, and excision repair cross-complementing gene 1 (ERCC1), which is the limiting enzyme in the NER pathway, plays an important role in this process[13,14]. Compared with normal liver tissue, fibrous tissue in the liver displays significantly increased levels of ERCC1 expression[15]. ERCC1 protein concentrations were significantly greater in liver cancers that were accompanied by hepatic fibrosis tissue than in liver cancers without hepatic fibrosis. In addition, high expression levels of ERCC1 are closely correlated with cisplatin resistance; thus, ERCC1 could be used as a predictor of sensitivity to platinum-based chemotherapy in cases of HCC[16]. The relationship between synergistic effects and the regulation of ERCC1 expression is worthy of further study in HCC treatments that combine emodin with platinum drugs.
Fibroblast growth factor receptor 2 (FGFR2), which is a member of a transmembrane tyrosine kinase receptor family (FGFRs), is an expression product of the bek oncogene that plays an important role in the differentiation of HCC, the clinical staging of tumors, the incidence of tumor thrombosis and the determination of alpha-fetoprotein levels[17]. As a molecular marker, FGFR2 can effectively predict the overall survival and progression-free survival of patients with HCC. Interestingly, ERCC1 is a downstream target gene of FGFR2[18], whereas emodin is a tyrosine kinase inhibitor[10,19]. Previously published research has indicated that emodin down-regulates ERCC1 expression in non-small cell lung cancer, and its effects may be relevant to the ERK1/2 signaling pathway[20]; however, the exact mechanism through which emodin produces these effects has not been well established. The ERK signaling pathway is one of the downstream components of the FGFR2 pathway[21,22]. Thus, we hypothesize that emodin reverses tumor drug resistance by increasing the DNA damage that is induced by platinum chemotherapy drugs, and we speculate that the molecular mechanism of this effect is related to the ERK1/2 pathway, which is mediated by FGFR2 signaling in hepatoma cells.
The primary aim of this study was to determine the molecular mechanisms underlying the reversal effect of emodin on platinum resistance in HCC.
Oxaliplatin and emodin were purchased from Sigma (St. Louis, MO, United States). Dulbecco’s modified Eagle’s medium/high glucose (DMEM/H) and fetal bovine serum were produced by Invitrogen (Carlsbad, CA, United States), trypsin and dimethyl sulfoxide was purchased from Guge Biology CO (Wuhan, China). FGF7, puromycin dihydrochloride (sc-108071), ERCC1 mouse anti-monoclonal antibody and p-ERK1/2 rabbit anti-monoclonal antibody (sc-16982-R) were purchased from Santa Cruz Biotechnology (United States). FGFR2 mouse anti-monoclonal antibody was produced (MAB684) by R-D Systems, Inc. (United States). Dylight™ 800-labeled anti-mouse immunoglobulin G (IgG) antibody and Dylight™ 800-labeled anti-rabbit IgG antibody were bought from Gaithersburg Biotechnology (MD, United States).
The human hepatoma cell line HepG2 was used in the present study. HepG2 cells were obtained from the China Center for Type Culture Collection. The HepG2 cells were cultured in DMEM/H supplemented with 10% (v/v) fetal bovine serum, 200 IU/mL penicillin (ICN Biomedical, Costa Mesa, CA, United States), 100 mg/mL streptomycin (ICN Biomedical) and 0.5 mmol/L sodium pyruvate (Cambrex, Walkersville, MD, United States). The cells were cultured at 37 °C in a humidified atmosphere of 5% CO2 in air. An OXA-resistant subline was established by discontinuously exposing parental HepG2 cells to high OXA concentration (25 μmol/L) medium over the course of one year until the resulting cells could grow exponentially in medium containing 1 μmol/L OXA. The HepG2/OXA cells were digested and subcultured three times prior to their use for this experiment.
Cells from the resistant cell line HepG2/OXA in logarithmic growth phase were grown in 96-well plates. In particular, 100 μL of cell suspension was inoculated into 8 mL of medium. Medium containing different concentrations of the chemotherapy drug OXA (3.125, 6.25, 12.5, 25, 50 or 100 μmol/L) was added after 12 h of culture. The experimental group (EG) also received a final concentration of 10 μmol/L emodin in medium. In addition, a control group (CG) was established. The medium was aspirated after 24 h of culture; subsequently, 110 μL of a mixture of DMEM/H and cell counting kit-8 (CCK-8) (at a ratio of 10 μL CCK-8:100 μL DMEM/H) was added, after which the samples were incubated for 2 h. A cell-free blank group (BG) was established. The optical density (OD) of each well was determined at 450 nm. The formula for calculating the inhibition rate (IR) at different concentrations was 1 - (ODEG - ODBG)/(ODCG - ODBG). Based on the IR at different concentrations of the anti-cancer drug, the concentration at which the inhibition rate was 50% (IC50) was calculated using the SPSS 13.0 software (SPSS Inc., Chicago, IL, United States). The reversal index was calculated using the formula (IC50EG/IC50CG). Five wells were established at different experimental concentrations, and each experiment was repeated three times.
Bek shRNA cell transfection was conducted in accordance with the protocol for the bek shRNA plasmid (h): sc-29218-S (Santa Cruz Biotechnology, Inc.). HepG2/OXA cells were cultured to 60%-80% adherence in 6-well cell culture plates by adding antibiotic-free fetal bovine serum growth medium. The shRNA plasmid DNA solution (Solution A) was added directly to the dilute shRNA plasmid transfection reagent (Solution B) using a pipette. The solution was mixed gently by pipetting up and down and was incubated for 30 min at room temperature. Subsequently, the cells were washed twice with 2 mL of shRNA transfection medium, after which the medium was aspirated. Immediately afterward, 200 μL shRNA plasmid DNA/shRNA plasmid transfection reagent was added. The cells were incubated for 6 h at 37 °C in a CO2 incubator. Following this incubation, 1 mL of normal growth medium containing 2 times the normal serum and antibiotics concentration was added to each well, and the cells were incubated for an additional 24 h under normal conditions. Forty-eight hours after the transfection, the medium was aspirated and replaced with fresh medium containing 5 μg/mL puromycin. Every 2 d afterward, the medium was aspirated and replaced with freshly prepared selective medium. Finally, an aliquot of the cells was washed once with phosphate buffer saline (PBS). The cell sample was lysed in 300 μL 1 × electrophoresis sample buffer by gently rocking the 6-well plate and was subjected to sodium dodecyl sulfate (SDS) gel electrophoresis to confirm the silencing of the FGFR2 gene. Thus, a Bek-silenced HepG2/OXA cell line (HepG2/OXA/T) was established.
HepG2, HepG2/OXA and HepG2/OXA/T cells were seeded in culture flasks. Each cell type was divided into four groups: a CG, an OXA group, an OXA + FGF7 group and an OXA + emodin group. A final concentration of 10 μmol/L OXA was added to the OXA group, the OXA + FGF7 group and the OXA + emodin group. and the final concentrations of FGF7, emodin and OXA in each group were 5 ng/mL, 10 μg/mL and 10 μmol/L, respectively. After being incubated for 24 h and digested with trypsin, the cells were divided into two portions, one for total protein preparation for Western blotting, and one for single-cell gel electrophoresis to assay the DNA damage.
After counting the cells and adjusting them to a concentration of 2000 cells/μL, the cells in each group were gently suspended into single-cell suspension. Subsequently, 110 μL of 0.5% normal melting point agarose (NMA) at 45 °C was poured onto Dakin slides, avoiding the production of air bubbles. The agarose solidified at room temperature. Subsequently, 5 μL PBS containing 10000 cells in each group was mixed well with 75 μL of 0.5% low melting point agarose (LMA). The upper cover-slip was carefully removed, the mixture was quickly added to the 0.5% NMA, the cells were spread evenly, and the slide was placed in a 4 °C refrigerator for 5 min until the agarose solidified. The cover-slip was removed, and 75 μL of 0.5% LMA was added, after which the slide was placed in the refrigerator at 4 °C again to solidify the agarose. The slide was slowly immersed in freshly prepared 4 °C pre-cooling cell lysate and then set in a 4 °C refrigerator for at least 1 h. The slide was then placed in a horizontal gel electrophoresis tank and incubated in the dark for 45 min, after which electrophoresis was performed for 30 min at 25 V at room temperature, and the height of the electrophoresis buffer liquid was adjusted to maintain a continuous 300 mA current. After the electrophoresis, each slide was dipped two times in a buffer in a darkroom for 5 min each. Following this treatment, 0.5 μL ethidium bromide dye was added to stain the DNA, the cover-slip was stamped, and the slide was observed and analyzed under a fluorescence microscope with a camera.
Total protein was collected from the different groups of the cultured HepG2, HepG2/OXA and HepG2/OXA/T cells. The protein concentration was measured by a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Jiangsu, China). Before electrophoresis, the protein was denatured in lithium dodecyl sulfate (LDS) sample buffer (106 mmol/L Tris-HCl, 141 mmol/L Tris base pH 8.5, 0.51 mmol/L ethylenediaminetetraacetic acid, 10% glycerol, 2% LDS, 0.22 mmol/L Serva blue G250, 0.175 mmol/L phenol red, and 0.1 mmol/L 2-mercaptoethanol) for 10 min at 95 °C. The total protein (20 μg per lane) was electrophoresed on a 8% SDS polyacrylamide gel electrophoresis gel and transferred onto a 0.45 μm nitrocellulose filter membrane (Roche, Indianapolis, IN, United States). The membranes were blocked with 5% (w/v) nonfat dry milk in PBS containing 0.05% Tween-20 (PBST) for 2 h at room temperature and incubated overnight at 4 °C with antibodies against FGFR2 (1:250), p-ERK1/2 (1:250) or ERCC1 (1:100) (Santa Cruz, CA, United States). Next, the membranes were incubated with a Dylight™ 800-labeled antibody for 1 h after being washed 4 times for 5 min in PBST. Finally, the immunoblot signals were scanned and analyzed using an Odyssey Infrared Imaging System (Li-Cor Biosciences, Nebraska, United States).
All of the digital results are displayed as the means ± SD. The quantitative ratios of different groups were compared using Student’s t-test with the SPSS 13.0 software. Probability values of P < 0.05 were regarded as statistically significant. All of the statistical tests were two-sided.
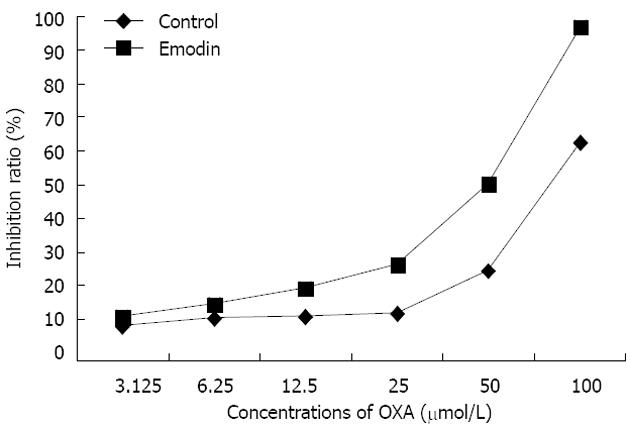
Compared with the CG, in which HepG2/OXA was treated with OXA alone, the inhibition ratio was significantly increased in the emodin group, in which HepG2/OXA cells were treated with a combination of OXA and emodin (Figure 1). Based on the inhibition ratios of different concentrations (μmol/L) of OXA, the IC50 of the OXA-resistant HepG2/OXA cells was 120.78 μmol/L; however, in the OXA-resistant HepG2/OXA cells that were treated with 10 μmol/L emodin, the IC50 was reduced to 39.65 μmol/L. This result indicated that the reversal index of 10 μmol/L emodin in HepG2/OXA cells was 3.05 (Table 1).
| OXA concentration (μmol/L) | 3.125 | 6.25 | 12.5 | 25 | 50 | 100 | IC50 (μmol/L) | Reversal index |
| IR in control (%) | 9.76 ± 1.18 | 11.94 ± 1.30 | 13.95 ± 1.11 | 14.58 ± 1.02 | 28.06 ± 2.01 | 63.95 ± 4.71 | 120.78 ± 9.68 | 3.05 |
| IR in emodin (%) | 12.35 ± 1.3a | 16.76 ± 1.3a | 21.69 ± 1.6a | 29.87 ± 1.55b | 48.14 ± 2.09b | 80.34 ± 3.00b | 39.65 ± 5.43 |
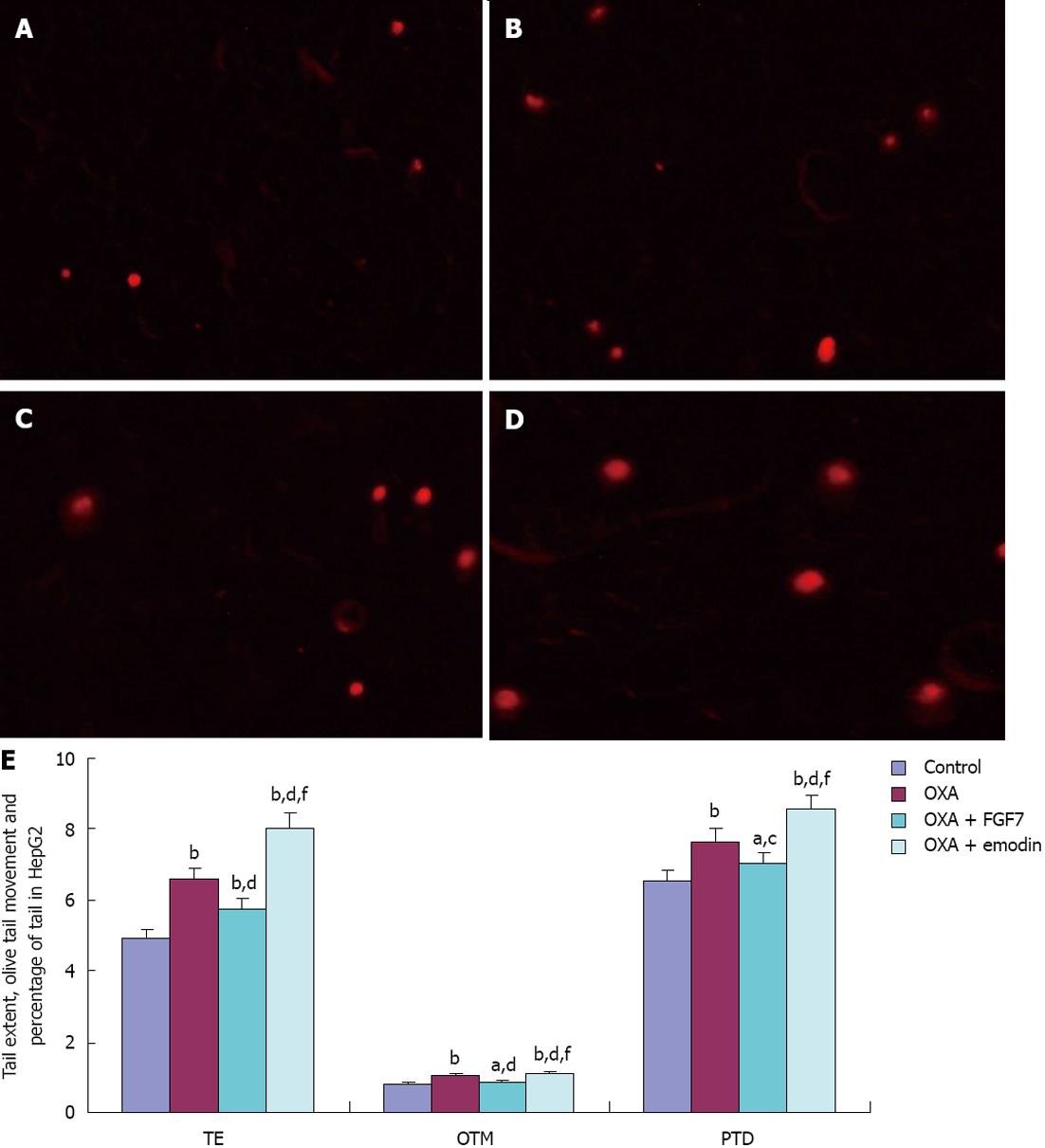
Compared with the CG of HepG2 cells, the tail extent (TE) and the Olive tail moment (OTM) were considerably increased in the OXA group, OXA + FGF7 group, and OXA + emodin group; these differences were statistically significant (P < 0.01). Compared with the OXA group, TE and OTM in the OXA + FGF7 group were considerably decreased, and these differences were statistically significant (P < 0.01). TE, OTM and the percentage of tail DNA (PTD) were significantly greater (P < 0.01) in the OXA + emodin group than in the OXA + FGF7 group. PTD in the CG, OXA group, and OXA + FGF7 group did not significantly differ (P > 0.05) (Figure 2).
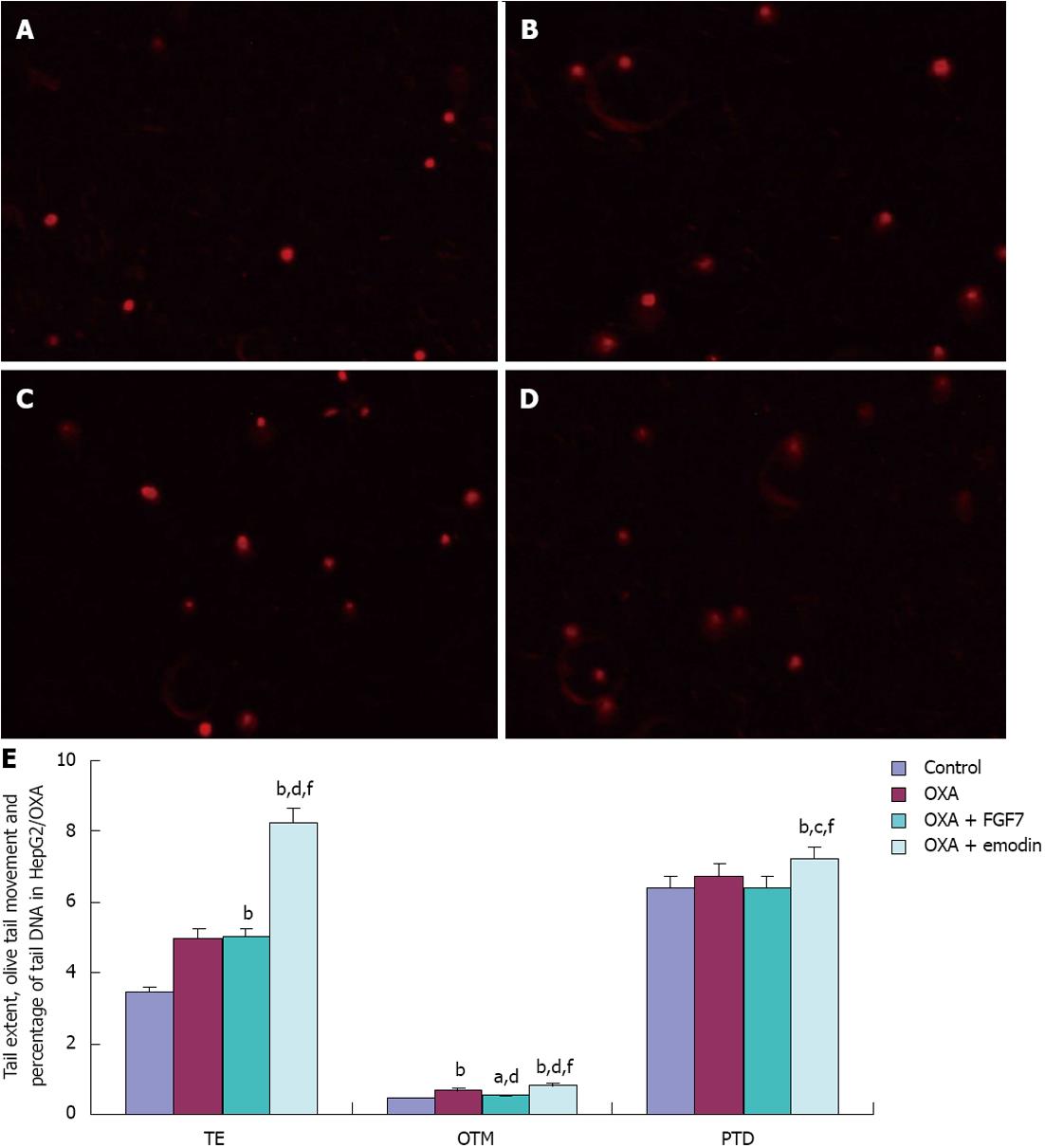
In HepG2/OXA cells, compared with the CG, TE, PTD and OTM in the OXA group, OXA + FGF7 group and OXA + emodin group were significantly increased; the differences were statistically significant (P < 0.01). Compared with the OXA group, OTM and PTD in the OXA + FGF7 group were reduced with statistically significant difference (P < 0.01). In the OXA + emodin group, TE, OTM and PTD were significantly increased; this difference was statistically significant (P < 0.01). Compared with the OXA + FGF7 group, TE, OTM and PTD in the OXA + emodin group were significantly increased (P < 0.01). These results are presented in Figure 3.
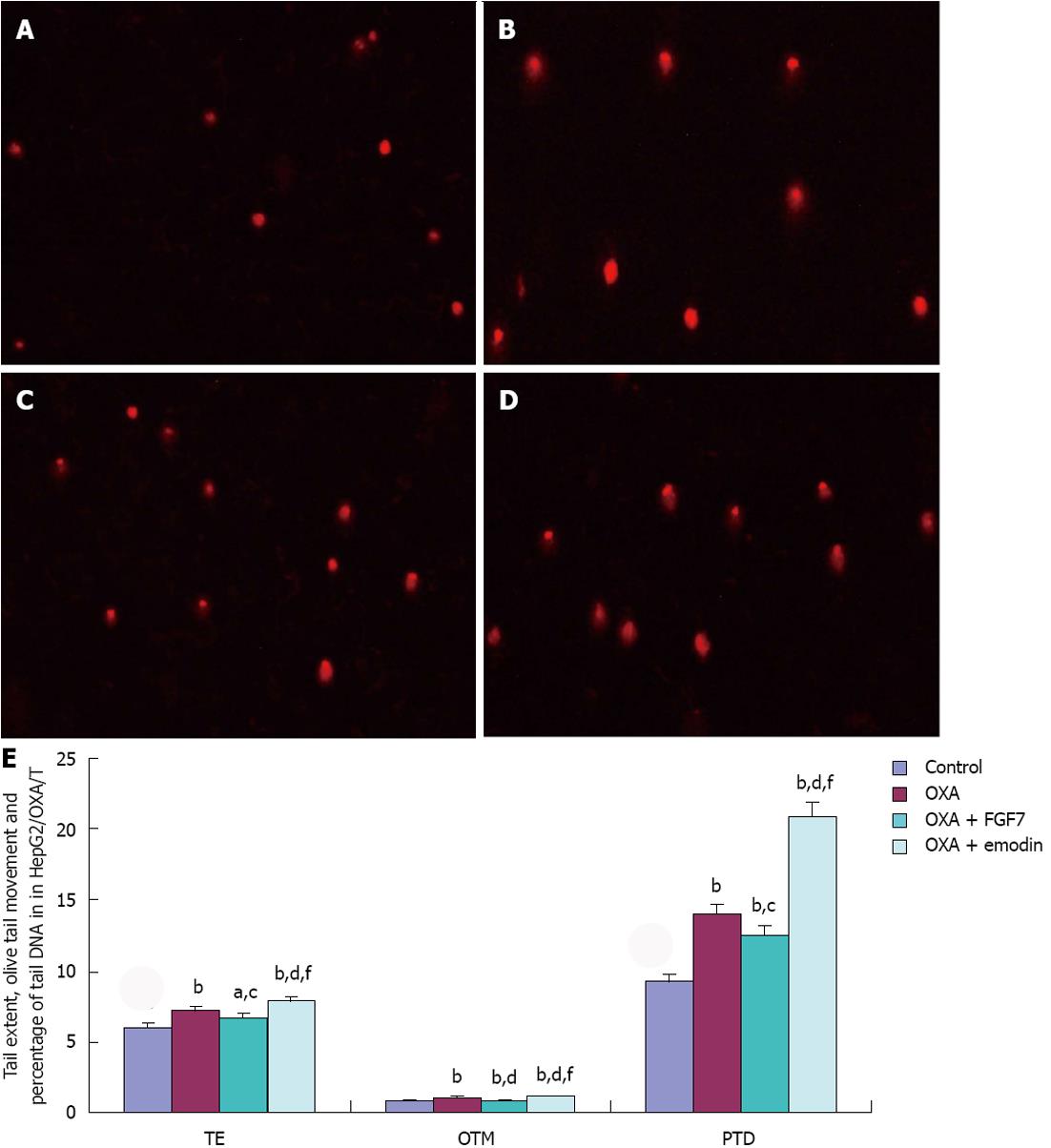
In HepG2/OXA/T cells, compared with the CG, TE, OTM and PTD in the OXA group, OXA + FGF7 group, and OXA + emodin group were significantly higher; the differences were statistically significant. Compared with the OXA group, OTM and PTD in the OXA + FGF7 group were reduced, and there was a significant difference; however, TE, OTM and PTD were significantly increased in the OXA + emodin group, with a statistically significant difference (P < 0.01). In comparison with the OXA + FGF7 group, TE, OTM and PTD were significantly increased in the OXA + emodin group, and the difference was statistically significant (P < 0.01), as shown in Figure 4.
In comparison with the expression levels in the parental HepG2 cell line, the FGFR2, pERK1/2 and ERCC1 expression levels in the resistant cell line HepG2/OXA were significantly increased, whereas the pERK1/2 and ERCC1 expression levels in the shRNA-transfected cell line HepG2/OXA/T were significantly inhibited, as FGFR2 expression was silenced. Compared with the CG in HepG2 cells, FGFR2 expression was increased with statistical significance in the OXA + FGF7 treatment group; moreover, there was a significant difference between the OXA + emodin group and the OXA + FGF7 group. The same trend was observed in the HepG2 and HepG2/OXA cells with respect to the expression of pERK1/2 and ERCC1 among the different treatment groups. Compared with the pERK1/2 and ERCC1 expression in CG, the levels in the OXA group were significantly up-regulated. Compared with the CG, pERK1/2 and ERCC1 expression were increased in the OXA + FGF7 group and reduced in the OXA + emodin group. However, the expression of FGFR2, pERK1/2 and ERCC1 exhibited no significant differences among the treatment groups in the shRNA-transfected cell line HepG2/OXA/T (Figure 5).
Rhubarb and polygonum cuspidatum have been widely used in various heat syndromes to clear heat and detoxify in the body in accordance with the theories of traditional Chinese medicine. Emodin (1,3,8-trihydroxy-6-methylanthraquinone), the primary active ingredient in these traditional medicines, was identified by modern pharmacological studies as having a wide range of pharmacological effects, such as protecting the function of the liver and the kidney[23], producing anti-inflammatory effects[24] and regulating lipid metabolism[25,26]. In recent years, its anti-cancer function has been revealed in a variety of malignancies, including HCC[27]. In addition, combined with chemotherapy drugs such as platinum[7-11,16], emodin demonstrates a synergistic effect and reverses platinum resistance. In the human multidrug-resistant breast cancer cell line MCF-7/Adr, the reversal index of emodin was 2.86 and 1.79 in combination with doxorubicin and cisplatin, respectively[28]. Our study results are similar, yielding a value of 3.05 in resistance reversal effect for 10 μmol/L in HepG2/OXA cells.
DNA interstrand cross-linking or chain cross-linking caused by platinum drugs induces apoptosis in tumor cells[29]. Although the capacity to repair DNA damage by NER, which removes the chain of platinum drug-induced DNA adducts, is considered to be the main mechanism of tumor cell resistance to platinum, the synergistic effect and reversal of platinum resistance induced by emodin appears to function by enhancing the DNA damage[30]. This study showed that TE, OTM and PTD were significantly higher in HepG2 cells after OXA and OXA + emodin treatment; moreover, differences are also statistically significant between the OXA group and the OXA + emodin group. A similar trend was observed in the resistant cell line HepG2/OXA. Our results suggest that emodin promotes the DNA damage that is induced by OXA. As the rate-limiting enzyme of the NER process, ERCC1 and XPF form heterodimers that exhibit damage recognition and nucleic acid cutting activity at the 5’ end of the DNA in platinum-based chemotherapy. ERCC1 plays an important role in HCC, which was expected given that this gene is an effective predictor of the sensitivity of tumor cells to platinum-based chemotherapy[15,16]. This study observed that an increase in the DNA damage level occurred in the resistant cell line HepG2/OXA and their parental cell line HepG2 after being treated with emodin combined with OXA. Moreover, ERCC1 expression levels were significantly decreased. The study results suggested that emodin-mediated down-regulation of the expression of ERCC1 plays an important role in enhancing the DNA damage induced by OXA. Zhou et al[28] also found that ERCC1 protein expression decreases gradually in MCF-7/Adr cells in proportion to the duration of emodin treatment and that the effect of 20 μg/mL of emodin was greater than that of 10 μg/mL.
In the multidrug-resistant gastric cancer cell lines, our previous results indicated that ERCC1 was a target gene in the FGFR2 signaling pathway[18]. The same result was observed in HCC drug-resistant cell lines in this study. Accompanied with Bek shRNA silencing of FGFR2 gene expression, the expression of ERCC1 was significantly reduced in the drug-resistant cell line HepG2/OXA, whereas after FGF7 stimulation of the FGFR2 signaling pathway, ERCC1 expression was significantly increased. These results support the idea that ERCC1 is a downstream gene of the FGFR2 signaling pathway. FGFR2, a member of the transmembrane tyrosine kinase receptor family (FGFRs) and the expression product of the bek oncogene, plays an important role in the cell differentiation of stomach cancer[31,32] and HCC[17], which effectively predicts overall survival and progression-free survival as a molecular marker. The protein kinase C, Ras/Raf/MEK/ERK, janus kinase/signal transducer and activator of transcription, and PI3K signaling pathways are downstream cascades in the FGF-induced signaling pathways[33]. ERCC1 expression can be inhibited by the ERK inhibitor U0126, suggesting that ERCC1 is one of the target genes in the downstream of the ERK signaling pathway[8]. Our results suggested that ERCC1 expression was inhibited by bek gene silencing; additionally, ERCC1 expression was significantly increased by the positive stimulus of FGF7. In addition, p-ERK1/2, the key molecule in the Ras/Raf/MEK/ERK pathway, was increased or reduced in conjunction with FGF7 stimulus or bek gene silencing, suggesting that the Ras/Raf/MEK/ERK pathway may be an important pathway in the FGFR2 regulation of ERCC1 expression. The most interesting result of this study was that FGFR2 protein expression disappeared accompanied with bek gene being silenced, whereas ERCC1 and p-ERK1/2 expression were not completely inhibited, suggesting that other signaling pathways may be involved in the pathway by which FGFR2 regulates ERCC1 expression.
By contrast to FGF7 stimulation of the FGFR2 signal pathway in the resistant hepatic cancer cell line HepG2/OXA and the parental cell line HepG2, the p-ERK1/2 phosphorylation level was significantly inhibited by emodin treatment; meanwhile ERCC1 expression levels were significantly decreased. These data were consistent with the results of Ko et al[9], who observed that emodin could significantly enhance the cytotoxicity of platinum drugs in lung cancer cell lines, and its mechanism is closely related to the inhibition of ERCC1 expression, and the downward effect of ERCC1 expression was achieved through the inactivation of the ERK1/2 pathway. Emodin, a tyrosine kinase inhibitor[19,34] and FGF7, a positive stimulator, had no effect on the expression of ERCC1 and p-ERK1/2 if FGFR2 expression was inhibited by shRNA silencing, which suggested that the emodin regulation of ERCC1 expression by the ERK1/2 pathway was closely related to the inhibition of FGFR2 tyrosine kinase activity.
In summary, the results of this study indicated that emodin could significantly enhance the DNA damage that was caused by OXA and induce OXA resistance reversal in HepG2/OXA cells. The molecular mechanism for this phenomenon is mediated by the inhibition of ERCC1 expression by the FGFR2/ERK1/2 signaling pathway.
As the effect of platinum-based chemotherapy on advanced hepatocellular carcinoma (HCC) was re-proved in 2010, the drugs which could strengthen chemotherapy effects and protect normal liver cells were the hot research area. Emodin, as the main active ingredient in many Chinese herbs, interested us for its low toxicity and synergistic effect combined with platinum in HCC cells. Excision repair cross-complementing gene 1 (ERCC1), which was the limiting enzyme in the nucleotide excision repair pathway, plays an important role in the process of platinum drug resistance. The relationship between synergistic effects and the regulation of ERCC1 expression is worthy of further study in HCC treatments that combine emodin with platinum drugs.
Fibroblast growth factor receptor 2 (FGFR2), as a transmembrane tyrosine kinase, plays an important role in the differentiation of HCC, the clinical staging of tumors, the incidence of tumor thrombosis and the determination of alpha-fetoprotein levels. Interestingly, ERCC1 is a downstream target gene of FGFR2, whereas emodin is a tyrosine kinase inhibitor. Current research has indicated that emodin down-regulates ERCC1 expression in non-small cell lung cancer, and its effects may be relevant to the extracellular signal-regulated kinase 1/2 (ERK1/2) signaling pathway, which is one of the downstream components of the FGFR2 pathway. However, the exact mechanism through which emodin produces these effects has not been well established.
In this study, the resistance reversal effect for 10 μmol/L emodin was 3.05 in HepG2/oxaliplatin (OXA) cells. Meanwhile, the tail length, the olive tail length and the percentage of tail DNA were significantly higher after treatment combined with emodin, which suggest that emodin promotes the DNA damage induced by OXA. Accompanied with bek shRNA silencing and fibroblast growth factor 7 (FGF7) stimulation of FGFR2 gene expression, the expression of phosphorylated extracellular signal-regulated kinase 1/2 (p-ERK1/2) and ERCC1 was significantly reduced and increased in the drug-resistant cell line HepG2/OXA, which supports the idea that ERCC1 is a downstream gene of the FGFR2/p-ERK1/2 signaling pathway. Furthermore, ERCC1 expression levels in HepG2/OXA and HepG2 cells were significantly decreased after emodin treatment with significant inhibition of the p-ERK1/2 phosphorylation level. However, if FGFR2 expression was inhibited by shRNA silencing, the inhibition effect of emodin and the stimulation effect of FGF7 on the expression of ERCC1 and p-ERK1/2 disappeared, which suggested that emodin regulation of ERCC1 expression by the ERK1/2 pathway was closely related to the inhibition of FGFR2 tyrosine kinase activity.
The results of emodin enhancing the DNA damage caused by OXA and its molecular mechanism associated with the inhibition of ERCC1 expression by the FGFR2/ERK1/2 signaling pathway may provide an experimental basis for the further development and application of emodin in the reversal of platinum drug resistance in HCC and other types of malignant tumors.
Platinum drug resistance: Platinum-based compounds, including cisplatin, carboplatin and OXA, are widely used in a number of carcinomas, and compose a mainstay of chemotherapeutic treatment. The cytotoxicity of platinum is attributed to apoptosis induced by DNA damage through the formation of platinum crosslinks on DNA. Cancer cells have the capacity to decrease the platinum concentration and repair DNA damage, which is associated with platinum drug resistance. The capacity to remove the chain of platinum drug-induced DNA adducts, is considered to be the main mechanism of tumor cell resistance to platinum.
This is a good descriptive study in which authors proved emodin could significantly enhance the DNA damage caused by OXA and induce OXA resistance reversal in HepG2/OXA cells, and investigated the molecular mechanism for this effect from the point of the inhibition of ERCC1 expression mediated by the FGFR2/ERK1/2 signaling pathway. The results are interesting and suggest that emodin is a potential therapeutic substance that could be used in reversing platinum drug resistance in the HCC and other types of malignant tumors.
P- Reviewer Abdel-Hamid NM S- Editor Huang XZ L- Editor O’Neill M E- Editor Xiong L
| 1. | El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752-1763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 768] [Cited by in F6Publishing: 786] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 2. | Connock M, Round J, Bayliss S, Tubeuf S, Greenheld W, Moore D. Sorafenib for the treatment of advanced hepatocellular carcinoma. Health Technol Assess. 2010;14 Suppl 1:17-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Qin S, Bai Y, Ye S, Fan J, Lim H, Cho JY, Thongprasert S, Chao Y, Rau K, Sun Y. Phase III study of oxaliplatin plus 5-fluorouracil/leucovorin (FOLFOX4) versus doxorubicin as palliative systemic chemotherapy in advanced HCC in Asian patients. J Clin Oncol. 2010;28:4008. [Cited in This Article: ] |
| 4. | Srinivas G, Babykutty S, Sathiadevan PP, Srinivas P. Molecular mechanism of emodin action: transition from laxative ingredient to an antitumor agent. Med Res Rev. 2007;27:591-608. [PubMed] [Cited in This Article: ] |
| 5. | Shieh DE, Chen YY, Yen MH, Chiang LC, Lin CC. Emodin-induced apoptosis through p53-dependent pathway in human hepatoma cells. Life Sci. 2004;74:2279-2290. [PubMed] [Cited in This Article: ] |
| 6. | Hsu CM, Hsu YA, Tsai Y, Shieh FK, Huang SH, Wan L, Tsai FJ. Emodin inhibits the growth of hepatoma cells: finding the common anti-cancer pathway using Huh7, Hep3B, and HepG2 cells. Biochem Biophys Res Commun. 2010;392:473-478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 7. | Wang W, Sun Y, Li X, Li H, Chen Y, Tian Y, Yi J, Wang J. Emodin potentiates the anticancer effect of cisplatin on gallbladder cancer cells through the generation of reactive oxygen species and the inhibition of survivin expression. Oncol Rep. 2011;26:1143-1148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Wang W, Sun YP, Huang XZ, He M, Chen YY, Shi GY, Li H, Yi J, Wang J. Emodin enhances sensitivity of gallbladder cancer cells to platinum drugs via glutathion depletion and MRP1 downregulation. Biochem Pharmacol. 2010;79:1134-1140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 9. | Ko JC, Su YJ, Lin ST, Jhan JY, Ciou SC, Cheng CM, Chiu YF, Kuo YH, Tsai MS, Lin YW. Emodin enhances cisplatin-induced cytotoxicity via down-regulation of ERCC1 and inactivation of ERK1/2. Lung Cancer. 2010;69:155-164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Zhang L, Hung MC. Sensitization of HER-2/neu-overexpressing non-small cell lung cancer cells to chemotherapeutic drugs by tyrosine kinase inhibitor emodin. Oncogene. 1996;12:571-576. [PubMed] [Cited in This Article: ] |
| 11. | Huang XZ, Wang J, Huang C, Chen YY, Shi GY, Hu QS, Yi J. Emodin enhances cytotoxicity of chemotherapeutic drugs in prostate cancer cells: the mechanisms involve ROS-mediated suppression of multidrug resistance and hypoxia inducible factor-1. Cancer Biol Ther. 2008;7:468-475. [PubMed] [Cited in This Article: ] |
| 12. | Dip R, Camenisch U, Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Repair (Amst). 2004;3:1409-1423. [PubMed] [Cited in This Article: ] |
| 13. | Park CJ, Choi BS. The protein shuffle. Sequential interactions among components of the human nucleotide excision repair pathway. FEBS J. 2006;273:1600-1608. [PubMed] [Cited in This Article: ] |
| 14. | Altaha R, Liang X, Yu JJ, Reed E. Excision repair cross complementing-group 1: gene expression and platinum resistance. Int J Mol Med. 2004;14:959-970. [PubMed] [Cited in This Article: ] |
| 15. | Fautrel A, Andrieux L, Musso O, Boudjema K, Guillouzo A, Langouët S. Overexpression of the two nucleotide excision repair genes ERCC1 and XPC in human hepatocellular carcinoma. J Hepatol. 2005;43:288-293. [PubMed] [Cited in This Article: ] |
| 16. | Ueda S, Shirabe K, Morita K, Umeda K, Kayashima H, Uchiyama H, Soejima Y, Taketomi A, Maehara Y. Evaluation of ERCC1 expression for cisplatin sensitivity in human hepatocellular carcinoma. Ann Surg Oncol. 2011;18:1204-1211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Harimoto N, Taguchi K, Shirabe K, Adachi E, Sakaguchi Y, Toh Y, Okamura T, Kayashima H, Taketomi A, Maehara Y. The significance of fibroblast growth factor receptor 2 expression in differentiation of hepatocellular carcinoma. Oncology. 2010;78:361-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 18. | Qiu H, Yashiro M, Zhang X, Miwa A, Hirakawa K. A FGFR2 inhibitor, Ki23057, enhances the chemosensitivity of drug-resistant gastric cancer cells. Cancer Lett. 2011;307:47-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Jayasuriya H, Koonchanok NM, Geahlen RL, McLaughlin JL, Chang CJ. Emodin, a protein tyrosine kinase inhibitor from Polygonum cuspidatum. J Nat Prod. 1992;55:696-698. [PubMed] [Cited in This Article: ] |
| 20. | Ko JC, Su YJ, Lin ST, Jhan JY, Ciou SC, Cheng CM, Lin YW. Suppression of ERCC1 and Rad51 expression through ERK1/2 inactivation is essential in emodin-mediated cytotoxicity in human non-small cell lung cancer cells. Biochem Pharmacol. 2010;79:655-664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Yang H, Xia Y, Lu SQ, Soong TW, Feng ZW. Basic fibroblast growth factor-induced neuronal differentiation of mouse bone marrow stromal cells requires FGFR-1, MAPK/ERK, and transcription factor AP-1. J Biol Chem. 2008;283:5287-5295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 22. | Lunn JS, Fishwick KJ, Halley PA, Storey KG. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev Biol. 2007;302:536-552. [PubMed] [Cited in This Article: ] |
| 23. | Bhadauria M. Dose-dependent hepatoprotective effect of emodin against acetaminophen-induced acute damage in rats. Exp Toxicol Pathol. 2010;62:627-635. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 24. | Alisi A, Pastore A, Ceccarelli S, Panera N, Gnani D, Bruscalupi G, Massimi M, Tozzi G, Piemonte F, Nobili V. Emodin prevents intrahepatic fat accumulation, inflammation and redox status imbalance during diet-induced hepatosteatosis in rats. Int J Mol Sci. 2012;13:2276-2289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Meng G, Liu Y, Lou C, Yang H. Emodin suppresses lipopolysaccharide-induced pro-inflammatory responses and NF-κB activation by disrupting lipid rafts in CD14-negative endothelial cells. Br J Pharmacol. 2010;161:1628-1644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 26. | Feng Y, Huang SL, Dou W, Zhang S, Chen JH, Shen Y, Shen JH, Leng Y. Emodin, a natural product, selectively inhibits 11beta-hydroxysteroid dehydrogenase type 1 and ameliorates metabolic disorder in diet-induced obese mice. Br J Pharmacol. 2010;161:113-126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 27. | Huang Q, Lu G, Shen HM, Chung MC, Ong CN. Anti-cancer properties of anthraquinones from rhubarb. Med Res Rev. 2007;27:609-630. [PubMed] [Cited in This Article: ] |
| 28. | Zhou J, Fu JM, Shi J, Xie JS. Emodin reversing multidrug resistance in breast cancer cells and its influence on expression of ERCC1. Zhonghua Zhongliu Fangzhi Zazhi. 2010;17:27-29. [Cited in This Article: ] |
| 29. | Brulikova L, Hlavac J, Hradil P. DNA interstrand cross-linking agents and their chemotherapeutic potential. Curr Med Chem. 2012;19:364-385. [PubMed] [Cited in This Article: ] |
| 30. | Chang LC, Sheu HM, Huang YS, Tsai TR, Kuo KW. A novel function of emodin: enhancement of the nucleotide excision repair of UV- and cisplatin-induced DNA damage in human cells. Biochem Pharmacol. 1999;58:49-57. [PubMed] [Cited in This Article: ] |
| 31. | Matsunobu T, Ishiwata T, Yoshino M, Watanabe M, Kudo M, Matsumoto K, Tokunaga A, Tajiri T, Naito Z. Expression of keratinocyte growth factor receptor correlates with expansive growth and early stage of gastric cancer. Int J Oncol. 2006;28:307-314. [PubMed] [Cited in This Article: ] |
| 32. | Spencer-Dene B, Sala FG, Bellusci S, Gschmeissner S, Stamp G, Dickson C. Stomach development is dependent on fibroblast growth factor 10/fibroblast growth factor receptor 2b-mediated signaling. Gastroenterology. 2006;130:1233-1244. [PubMed] [Cited in This Article: ] |
| 33. | Katoh M, Katoh M. FGF signaling network in the gastrointestinal tract (review). Int J Oncol. 2006;29:163-168. [PubMed] [Cited in This Article: ] |
| 34. | Zhang L, Chang CJ, Bacus SS, Hung MC. Suppressed transformation and induced differentiation of HER-2/neu-overexpressing breast cancer cells by emodin. Cancer Res. 1995;55:3890-3896. [PubMed] [Cited in This Article: ] |