Published online May 7, 2012. doi: 10.3748/wjg.v18.i17.2053
Revised: February 6, 2012
Accepted: February 16, 2012
Published online: May 7, 2012
AIM: To develop an affinity peptide that binds to gastric cancer used for the detection of early gastric cancer.
METHODS: A peptide screen was performed by biopanning the PhD-12 phage display library, clearing non-specific binders against tumor-adjacent normal appearing gastric mucosa and obtaining selective binding against freshly harvested gastric cancer tissues. Tumor-targeted binding of selected peptides was confirmed by bound phage counts, enzyme-linked immunosorbent assay, competitive inhibition, fluorescence microscopy and semi-quantitative analysis on immunohistochemistry using different types of cancer tissues.
RESULTS: Approximately 92.8% of the non-specific phage clones were subtracted from the original phage library after two rounds of biopanning against normal- appearing gastric mucosa. After the third round of positive screening, the peptide sequence AADNAKTKSFPV (AAD) appeared in 25% (12/48) of the analyzed phages. For the control peptide, these values were 6.8 ± 2.3, 5.1 ± 1.7, 3.5 ± 2.1, 4.6 ± 1.9 and 1.1 ± 0.5, respectively. The values for AAD peptide were statistically significant (P < 0.01) for gastric cancer as compared with other histological classifications and control peptide.
CONCLUSION: A novel peptide is discovered to have a specific binding activity to gastric cancer, and can be used to distinguish neoplastic from normal gastric mucosa, demonstrating the potential for early cancer detection on endoscopy.
- Citation: Zhang WJ, Sui YX, Budha A, Zheng JB, Sun XJ, Hou YC, Wang TD, Lu SY. Affinity peptide developed by phage display selection for targeting gastric cancer. World J Gastroenterol 2012; 18(17): 2053-2060
- URL: https://www.wjgnet.com/1007-9327/full/v18/i17/2053.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i17.2053
New methods for the early detection of gastric cancer (GC) are urgently needed. GC is the second most common cause of cancer-related mortality worldwide[1,2]. Early detection is of paramount importance to improve the 5-year survival rate of the patients. Periodic endoscopic surveillance is the only currently available means to diagnose early gastric cancer in high-risk populations who have pre-cancerous lesions such as atrophic gastritis and intestinal metaplasia. However, the current surveillance program and mode of endoscopic diagnosis are labor-intensive and economically unfeasible. White light endoscopy has limited effectiveness for early GC screening. Neoplastic lesions can be less than a millimeter in size which is difficult to localize within regions of pre-cancerous mucosa that usually are several square centimeters. Thus, a rigorous method is needed for selecting and validating molecular probes that bind specifically and highlight neoplastic lesions.
Molecular imaging is a technique that identifies and characterizes tumors and other lesions based on their protein expression pattern, rather than by their macroscopic morphology[3]. The molecular expression pattern of cells and tissues can be visualized with the help of disease-specific molecular probes such as antibodies, antibody fragments, peptides, radioactive probes and nanoparticles[4-6]. Such molecular probes enable the diagnosis of disease in situ and in real time. In a previous study, a heptapeptide was isolated from a phage library and conjugated with fluorescein for labeling of colonic dysplasia[7]. Although the molecular target of this sequence has not yet been identified, preferential binding of this targeting moiety to neoplastic cells in vivo with a high sensitivity and specificity was observed. In recent clinical studies, molecular imaging has been developed for guiding biopsy of high-grade dysplasia in Barrett’s esophagus using fluorescent-labeled peptides. An affinity peptide selected using phage display techniques was administered over a region of intestinal metaplasia in resected specimens of the distal esophagus. The wide-area stereoscopic images of increased fluorescence intensity could predict and localize high-grade dysplasia[8].
In this study, we screened a peptide that has highly specific binding activity to human GC tissues. When labeled with fluorescein isothiocyanate (FITC), the peptide has the potential for in vivo use to produce increased fluorescence intensity at the site of neoplastic mucosa. This method can be used as a more specific strategy for early detection of GC.
The human gastric cancer cell line BGC823 and Epstein-Barr virus-transformed human gastric epithelial cell line GES-1 were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were incubated at 37 °C in an atmosphere with 5% CO2.
Peptide screen was conducted in the patients (n = 3) with histologically validated intestinal-type gastric adenocarcinoma (Lauren’s classification). Paraffin-embedded human tissues from 36 cases of gastric cancer (21 intestinal and 15 diffuse) and 15 cases of adjacent normal appearing gastric mucosa, 12 cases of breast cancer, and 15 cases of colorectal cancer were used for validating the screened peptide. The study was approved by the Bioethics Committee of the First Affiliated Hospital of Xian Jiaotong University Medical College, and written informed consent was obtained from all the patients. For the peptide screen, fresh specimens of cancer and adjacent normal appearing gastric mucosa (5 cm away from the macroscopic margin of the tumor) were collected during subtotal gastrectomy. Half of the tissue was cut into 0.5 cm × 0.5 cm × 0.3 cm pieces immediately and washed with magnesium-free Dulbecco’s phosphate-buffered saline (PBS) for 2 min at 4 °C to be used for biopanning or immunofluorescence procedures[9]. The other half of the tissue was embedded in optimal cutting temperature freezing compound (Sakura Finetek United States, Torrance, CA) immediately. The tissue was cut into 6-μm sections, mounted onto Poly-D-Lysine-coated slides, and stored at -80 °C for the peptide binding assay. All the histopathological specimens were evaluated by two gastrointestinal pathologists who were blinded to each other according to the common procedural criteria for such studies and to the imaging results[10].
Peptides were selected using the PhD-12TM phage display peptide library (New England BioLabs, Beverly, MA)[11-13]. This library has 1 × 1013 pfu/mL phages, with a diversity of 1.28 × 109 unique peptide sequences and about 70 copies of each sequence. For screening, non-specific binding phage was cleared from the library by panning against normal appearing gastric mucosa adjacent to the tumor. Tissue blocks were placed into 12-well cell culture plates and blocked by adding one mL of 1% bovine serum albumin (BSA) diluted in PBS for 30 min at 4 °C. Phage (1 × 1011 pfu) in one mL of blocking buffer was incubated with tissue at room temperature (RT) for 30 min with gentle agitation. The supernatant containing unbound phages was collected and added to another well for the second round of clearance. The resulting supernatant was incubated with the gastric cancer specimens for positive selection. After 30 min of biopanning at RT, the tissue specimens were transferred to 1.5 mL tubes and washed 10 times with PBST (PBS/0.1% Tween-20, v/v). The bound phages on the tissue surface were eluted with one mL of 0.2 mol glycine, pH 2.2, 0.1% BSA for 8 min and immediately neutralized with 150 μL of 1 mol Tris, pH 9.5. The eluted phage was amplified and tittered according to the manufacturer’s instructions. The resulting phage (1011 pfu) was used to perform another round of positive selection, as described above. In the last 2 rounds, elution was first performed for 2 min to remove the weakly bound phages, and new elution buffer was then added to obtain the stronger bound phage.
Phage clones (n = 48) obtained from the last round of biopanning were randomly selected and sequenced. Peptide sequences that appeared more than twice were selected as candidates for further analysis. These peptide sequences were analyzed by searching the UniProtKB/Swiss-Prot database for homology using the basic local assignment search tool (BLAST, National Center for Biotechnology Information, Bethesda, MD) with the option for short, nearly exact matches to identify potential human protein targets.
The protocol used for performing the cell enzyme-linked immunosorbent assay (C-ELISA) has been described previously[14]. BGC823 and GES-1 cells were allowed to reach an 80%-90% confluency in 96-well plates. The wells were blocked for 30 min at 37 °C with 200 μL BSA. Next, 2 × 107 pfu of candidate phages were incubated separately with each cell type in triplicate at RT for 30 min. The insertless wild-type phage (M13KE, New England Biolabs, Beverly, MA) was used as a control. Bound phages were detected using a horseradish peroxidase-conjugated polyclonal anti-M13 phage antibody (Pharmacia, United States). Tetramethylbenzidine working substrate solution (50 μL/well; Sigma, St Louis, MO) was added and incubated for 20 min at RT. The reaction was stopped by adding 4 mol H2SO4. Between each incubation step, the plates were washed three times with 300 μL TBST (0.5% Tween-20). Absorbance was measured at 490 nm using a microplate reader (Bio-Rad model 550, Hercules, CA). Untreated cells were used as controls. The absorbance (A) values between different groups were compared.
Specific binding of the candidate phages to gastric cancer was validated by incubating 2 × 1011 pfu of each phage (candidates and M13KE) with fresh gastric cancer or adjacent normal appearing gastric mucosa in wells in triplicate. The steps of incubation, two-step elution, and titration of phages were performed as described above. All of the eluted phages were tittered to determine the mean phage plaque numbers. The ratio of binding of each phage group to gastric cancer relative to that of M13KE was calculated. The level of binding of each phage clone to gastric cancer and normal appearing gastric mucosa was analyzed using the Student’s t test.
The candidate peptides were synthesized (Shanghai Biochem, Shanghai, China) using standard solid-phase fluorenylmethyloxycarbonyl chloride chemistry and purified to a minimum purity of 98% using high-performance liquid chromatography (HPLC). Analysis was performed by reverse phase HPLC and mass spectrometry[15]. FITC or biotin was conjugated to the C-terminus of the peptide via a flexible linker with the 5 amino acid sequence GGGSK (12-mer peptide-GGGSK-FITC or 12-mer peptide-GGGSK-biotin), the sequence of which is the same as that for the linker on the coat protein pIII of the M13 phage. For the control, the candidate peptide was scrambled to form a peptide sequence containing the same amino acids.
Preferential binding of the candidate peptide to gastric cancer was further validated by a competitive binding assay. The candidate peptides at concentrations of 0.5, 5, 50, 500 and 5000 μmol were incubated with fresh gastric cancer or adjacent normal appearing gastric mucosa in wells in triplicate. Each phage (2 × 1011 pfu; candidate or M13KE) was then added. Incubation, elution, and tittering of the binding phages were performed as described above. The ratio of binding of each phage clone to gastric cancer and normal appearing gastric mucosa was analyzed.
Peptide-based immunofluorescence analysis was performed to validate binding of the candidate peptide to human gastric cancer[16,17]. Frozen sections of human gastric cancer and adjacent normal appearing gastric mucosa tissues were blocked with PBS containing 3% BSA for 30 min at RT. Slides were then incubated with 100 μmol of the candidate peptide (peptide-FITC) for 30 min at 37 °C, rinsed 3 times with PBST and fixed in acetone at 4 °C for 90 s, counterstained with propidium iodide, and mounted using PBST. Fluorescent images of the sections were recorded at 400× magnification. A FITC-labeled scrambled peptide was used as a negative control.
The streptavidin-peroxidase-biotin immunohistochemical method was performed to detect candidate peptide binding on paraffin-embedded human tissues[18] from 36 cases of gastric cancer (21 intestinal and 15 diffuse) and 15 cases of adjacent normal appearing gastric mucosa, 12 cases of breast cancer, and 15 cases of colorectal cancer. In brief, paraffin-embedded specimens were cut into 4-μm sections and kept at 60 °C for 60 min. The sections were deparaffinized with xylene and rehydrated. Sections were submerged into ethylenediaminetetracetic acid antigenic retrieval buffer, microwaved for antigenic retrieval, and then cooled at RT for 20 min. The sections were pretreated with 3% hydrogen peroxide in methanol to quench the endogenous peroxidase activity, followed by incubation with normal serum to block non-specific binding. Then the sections were incubated with 100 μmol biotin-conjugated peptide for one hour at 37 °C. The unbound peptide was rinsed off with PBS. The tissue sections were incubated with the streptavidin-horseradish peroxidase complex (Zhongshan Biotechnology, Beijing, China), and stained with diaminobenzidene (DAB). Finally, the sections were counterstained with hematoxylin. A biotin-labeled scrambled peptide was used as a negative control.
Semi-quantitative image analysis was performed as reported previously[19]. In brief, 3 images with typical features were selected from each slide. The quantitative labeling index was calculated as the ratio of brown membranous area stained by DAB to round blue areas stained by hematoxylin, for the assessment of tumor cell density in the selected image. The extractions of the brown vs blue signal were carried out based on an RGB color parameter. The blue areas larger than 0.005 mm2 were eliminated because of the nuclear staining in cells such as fibroblasts and lymphocytes but not in carcinoma cells. Images were analyzed using NIH Image J software.
Differences in the mean A value, number of eluted phages, and image intensity for all tissue classifications were compared using a one-way analysis of variance (ANOVA) or two-sided Student’s t test with unequal variance. Statistical significance was assessed at the level of P = 0.01. All results were presented as mean ± SD unless otherwise noted.
Approximately 92.8% of the non-specific phage clones were subtracted from the original phage library after two rounds of biopanning against normal appearing gastric mucosa. After the third round of positive screening, 50 phage clones that specifically bound to human gastric cancer were randomly selected from the enriched phage library. Phage clones were amplified and sequenced. The peptide sequence AADNAKTKSFPV (AAD) appeared in 25% (12/48) of the analyzed phages. Except for 2 phage clones which expressed the same peptide sequence IVWPTSPRALDA, the other 36 clones expressed unique amino acid sequences. These peptide sequences were analyzed by searching the UniProtKB/Swiss-Prot database using BLAST. Peptide AAD has identities = 10/14 (71%) with methyltransferase, which belongs to UbiE/COQ5 family.
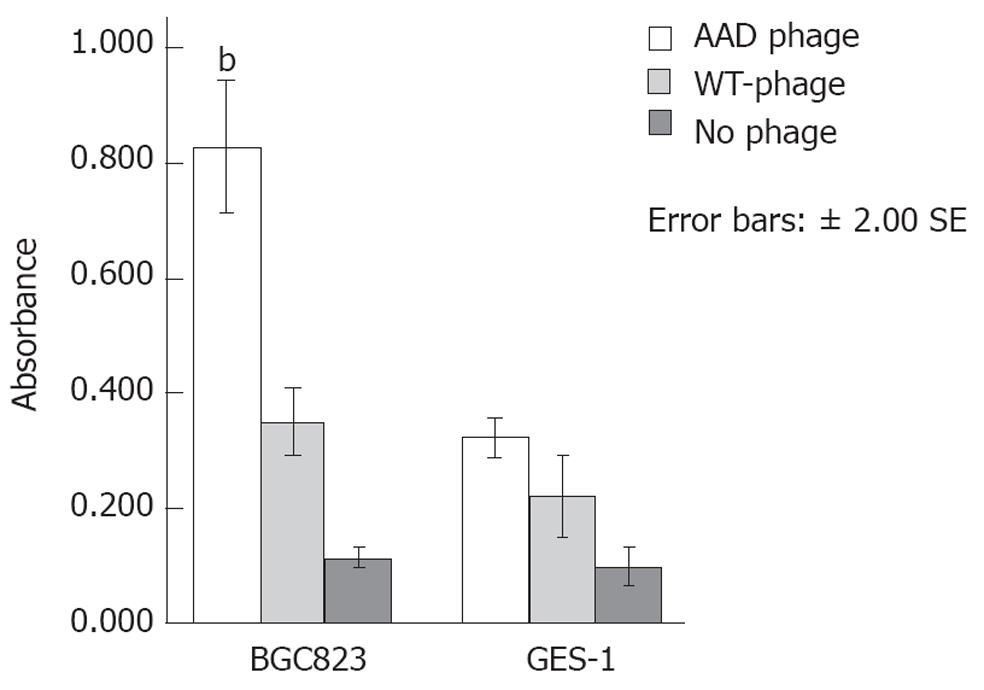
The C-ELISA demonstrated selective binding of the AAD phage to BGC823 cells. As shown in Figure 1, the A value for the AAD phage binding to BGC823 cells was 1.15 ± 0.09 compared to 0.61 ± 0.07 and 0.65 ± 0.05 for the wild type (WT)-phage (P < 0.01) and no phage (P < 0.01), respectively. The A for AAD phage binding to the GES-1 cells was 0.123 ± 0.035 compared to 0.189 ± 0.045 and 0.271 ± 0.035 for the WT-phage (P > 0.05) and for no phage (P > 0.05), respectively. These results suggest that the AAD phage binds specifically to the BGC823 (cancer) cells and not to the GES-1 (control) cells. WT-phage and no phage did not bind significantly to any of the cells.
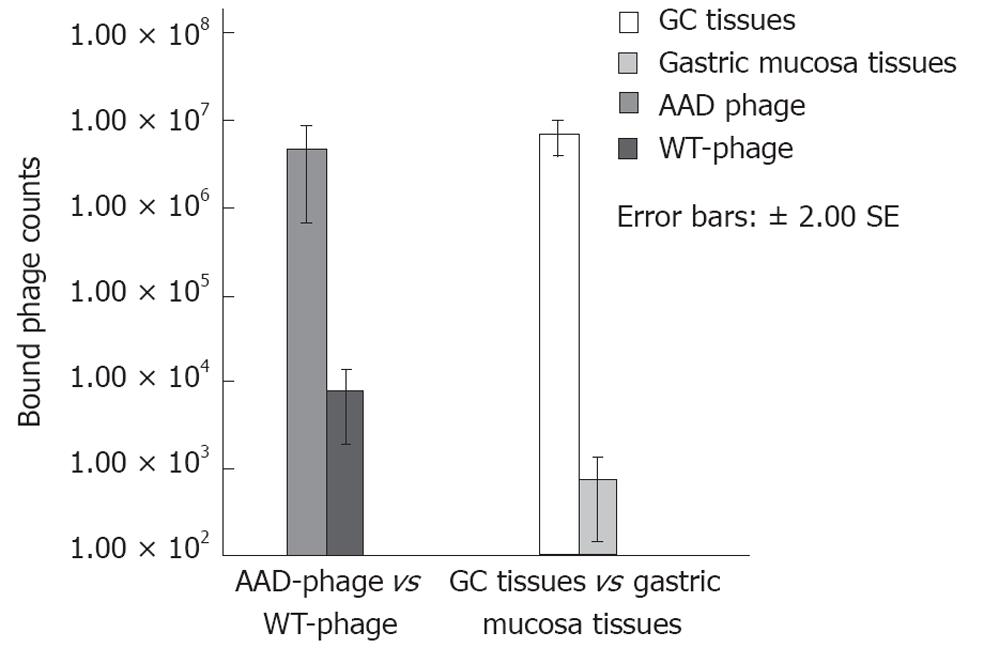
The AAD phage showed about 615 times greater binding to gastric cancer than did the WT-phage, with a total phage number of 4.8 × 106vs 7.9 × 103, as shown in Figure 2 (P < 0.01). Similarly, the binding of AAD phage was 591 times greater to gastric cancer than normal appearing gastric mucosa with a total phage number of 7.1 × 106vs 1.2 × 104, respectively (P < 0.01, Figure 2). These results suggest that AAD phage binds specifically to gastric cancer (target) and not to the adjacent normal appearing gastric mucosa (control).
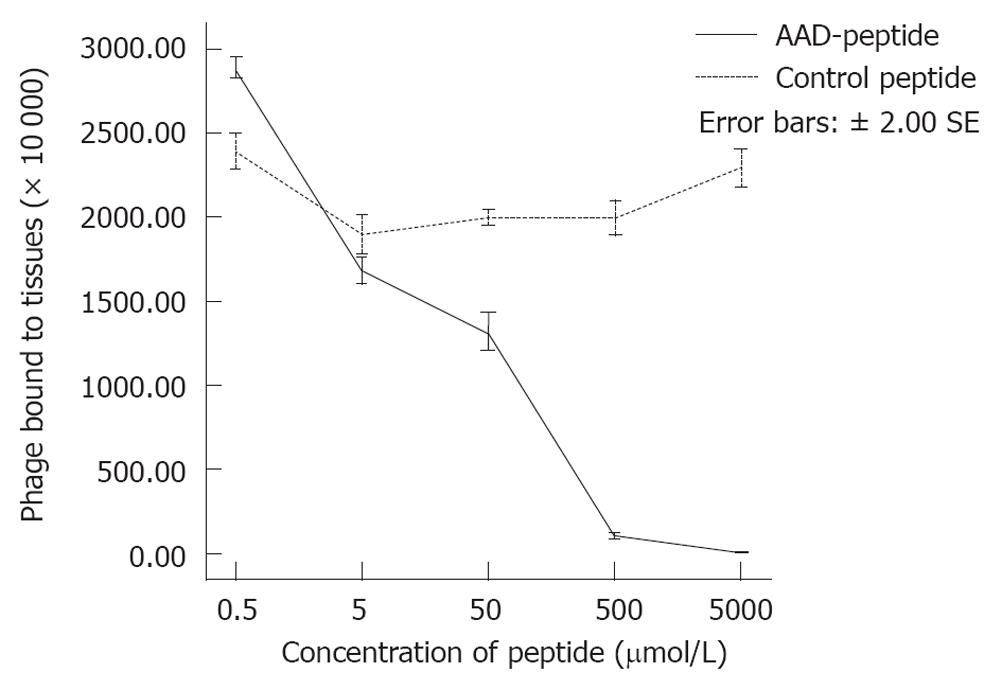
As shown in Figure 3, we observed that the addition of 0.5, 5, 50, 500 and 5000 μmol of the compound consisting of the AAD peptide with the GGGSK linker (AAD-GGGSK) resulted in a significant reduction in the number of bound phages, corresponding to values of 2900 × 104, 1680 × 104, 1320 × 104, 80 × 104 and 0 (P < 0.01), respectively. Moreover, we did not see any significant change in the number of bound phages with the addition of 0.5, 5, 50, 500 and 5000 μmol of the control peptide (PAKFKAANSDVT), which resulted in a total of (2300 ± 41) × 104 bound phage (P < 0.01) at 5000 μmol. These results suggest that the AAD peptide competes with the AAD phage for binding to gastric cancer, and that binding is determined by the specific sequence of the expressed peptide, rather than by the phage coat proteins.
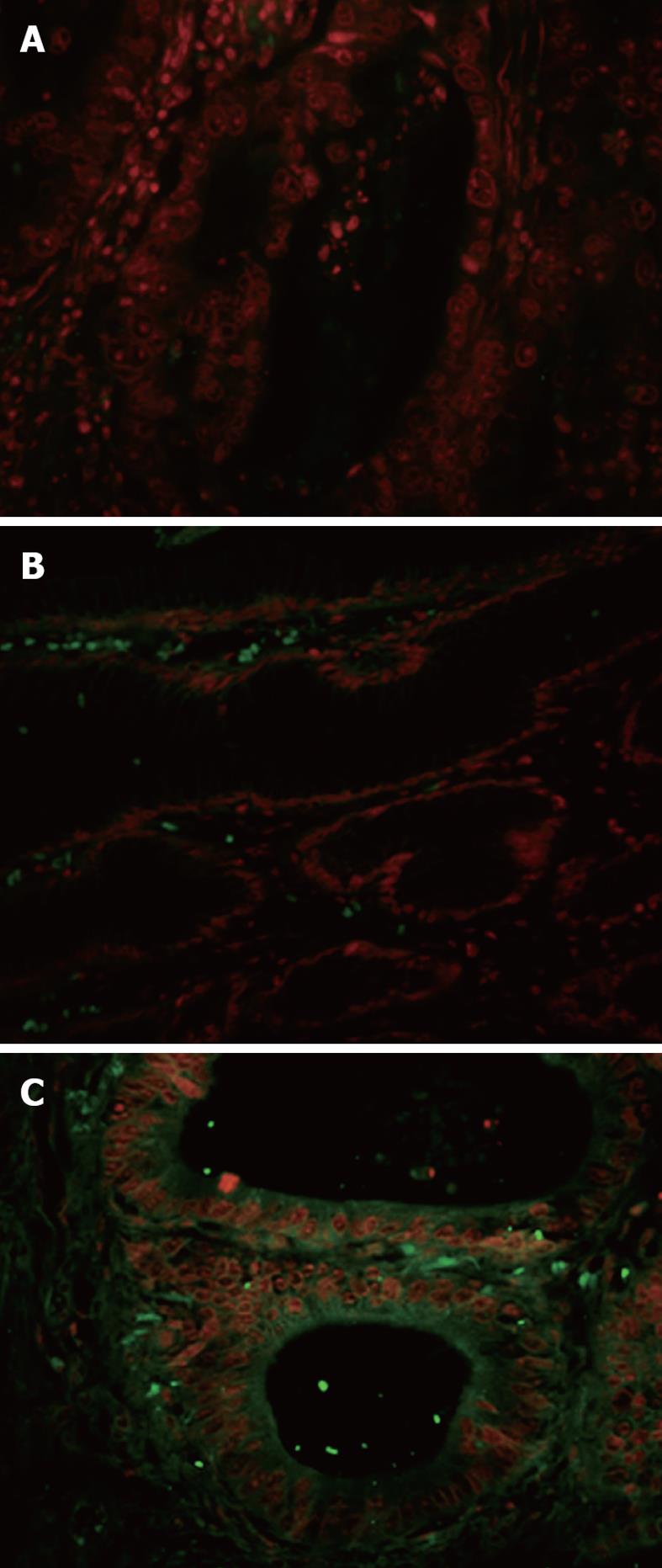
The peptide-based immunofluorescence assay was performed to confirm the selective binding of the AAD phage to fresh gastric cancer tissues. As shown in Figure 4, the fluorescence images displayed that the AAD peptide binds to both the tumor cell membrane and cytoplasm (C), but not to adjacent normal appearing gastric mucosa (B). Fluorescence was seen on the membrane and in the perinuclear cytoplasm of gastric cancer cells. The FITC-labeled scrambled control peptide, PAKFKAAN SDVT, did not bind to tumor tissues.
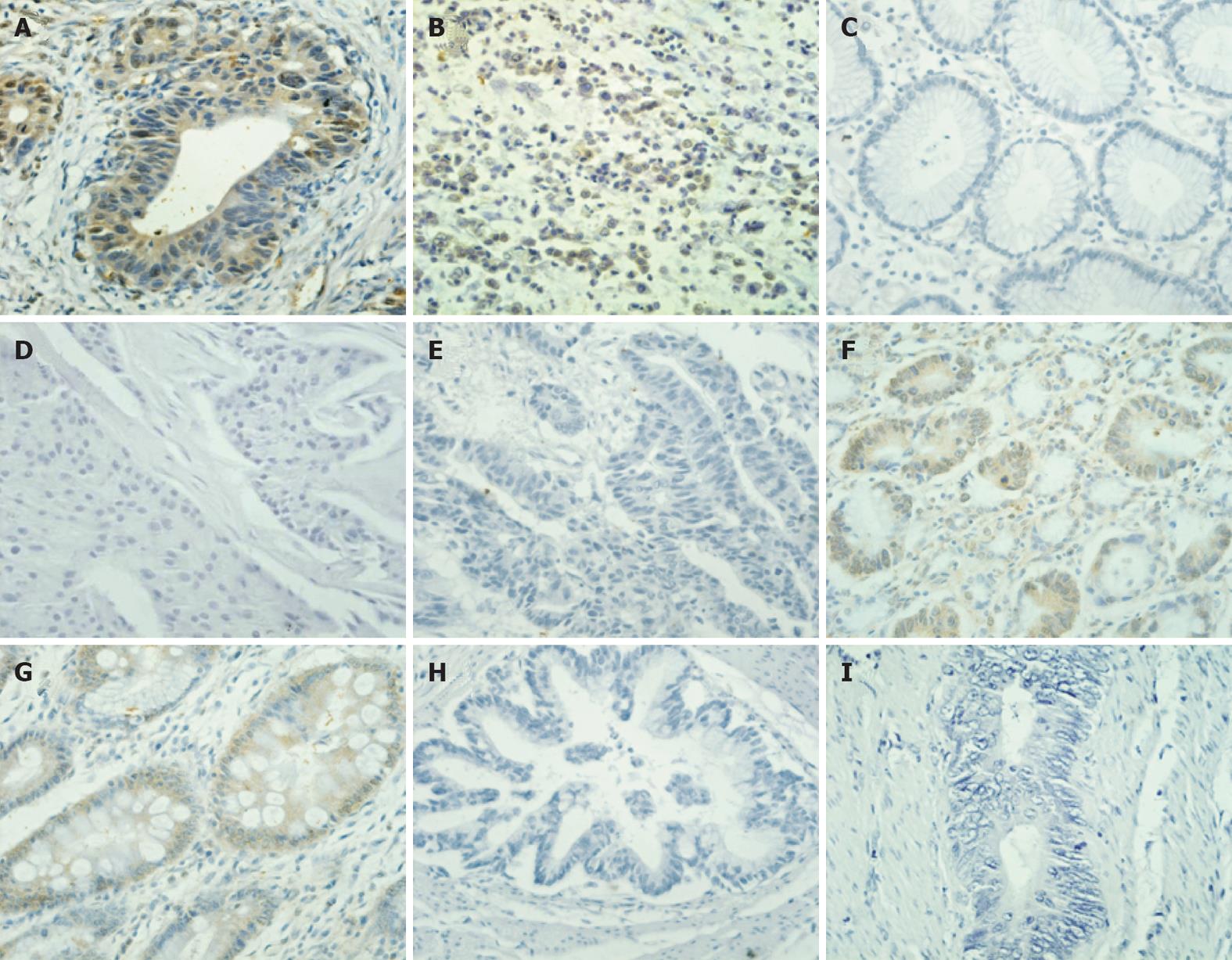
Tissue slides from multiple types of other human cancers were prepared to evaluate specific binding of biotin-labeled AAD peptide. From the results shown in Figure 5, biotin-AAD demonstrates specific binding to intestinal (Figure 5A) and diffuse (Figure 5B) gastric cancer. In contrast, no staining was observed in normal appearing gastric mucosa (Figure 5C), or breast cancer (Figure 5D) and colon cancer (Figure 5E). Weak binding of the AAD peptide to gastric mucosa dysplasia (Figure 5F) and intestinal metaplasia (Figure 5G) was also observed. The negative results were obtained when gastric cancer tissues were stained with biotin-conjugated scramble peptide (Figure 5H) and PBS (Figure 5I). In the positive slides, the area stained dark brown was located at the membrane and perinuclear cytoplasm, which is the same as FITC-conjugated AAD binding on fresh GC tissues, indicating the positive binding region of peptide AAD to GC cells.
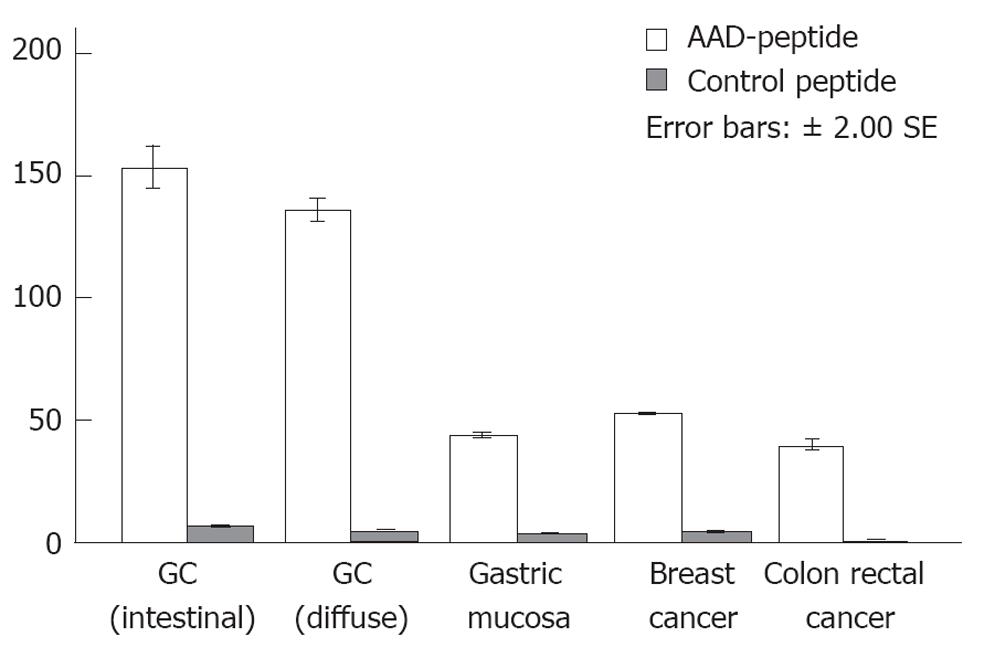
Semi-quantitative image analysis was then performed. For the AAD peptide, the values in 37 specimens of gastric cancer (21 intestinal and 15 diffuse), 15 specimens of normal appearing adjacent gastric mucosa, 12 specimens of breast cancer, and 15 specimens of colon rectal cancer were 150.0 ± 11.0, 135.5 ± 13.2, 43.5 ± 3.4, 52.3 ± 6.4 and 39.6 ± 5.0, respectively (Figure 6). For the control peptide, these values were 6.8 ± 2.3, 5.1 ± 1.7, 3.5 ± 2.1, 4.6 ± 1.9, and 1.1 ± 0.5, respectively. A one-way ANOVA showed an F-value of 1149.2 (P < 0.01), and the pair-wise t test yielded a t value of 15.3 (P < 0.01), demonstrating that the result for the AAD peptide is statistically significant for gastric cancer as compared with other histological classifications and control peptides.
Other investigators have used phage display technology to select peptides that target specific organs, tumors, and proteins without prior knowledge of the target’s molecular structure[20-22]. These libraries often contain more than 10 billion unique sequences, which enable peptide selection with highly specific binding properties. Peptides specific for endothelial markers in dysplasia have been identified in mice[23-26]. Biopanning using freshly harvested human tissues has successfully isolated peptides that specifically bind to polarized luminal surfaces of dysplastic colonocytes[27,28]. In this study, we selected the 12-mer peptide AAD using the PhD-12 library. This peptide exhibited specific binding to human gastric cancer cells in culture and tissues.
The biopanning protocol used in this study was different from that used by most other investigators. The original phage library was first panned against freshly harvested normal-appearing mucosa adjacent to cancer to clear non-specific phages. After the clearing of normal mucosa binding phage from the original phage library, the likelihood of obtaining gastric cancer-specific peptides in the following tumor-targeted screen increased. We removed 92.8% of the phage clones from the original library after two rounds of subtractive biopanning. To avoid biasing the library, we did not amplify the remaining phage pool between each round. The peptide sequence AAD appeared in more than 20% (12/50) of the analyzed phages after the third round of positive screening. This peptide was found to have no more than a 50% amino acid residue homology to the reported protein sequence. Phage expressing this peptide demonstrated preferential binding to cultured gastric cancer cells and fresh gastric cancer mucosa, and was validated by ELISA and bound phage counts. The binding was inhibited by the addition of competing peptide AAD, thus supporting cell surface binding. Moreover, when conjugated with FITC or biotin, the peptide AAD can be used as an in vitro peptide probe to distinguish tumor-adjacent mucosa from gastric cancer.
There is a great clinical need to improve the cancer screening and surveillance methods for diseases such as Barrett’s esophagus, gastric intestinal metaplasia, flat and depressed sporadic colonic adenomas, and bladder carcinoma in situ. In nuclear medicine, imaging with radioactively labeled probes is routinely used. In contrast, fluorescent-labeled probes in gastrointestinal endoscopy are still being developed. Tumor-specific molecular probes have been used to improve the lesion contrast during gastrointestinal endoscopy to guide tissue biopsies[29]. Most digestive tract neoplasia arises from the epithelial layer, which is compatible with topical administration of the probe. Thus, molecular imaging has a particular advantage in the diagnosis or treatment of disorders of the gastrointestinal and other hollow organs, compared with lesions from solid tumors. Antibodies against epitopes that are over-expressed in gastrointestinal cancers, such as vascular endothelial growth factor (VEGF) or epidermal growth factor receptor (EGFR), have been fluorescently labeled and used for in vivo imaging[30,31]. These antibodies have highly selective binding affinities to their target structures, with an optimized signal-to-background ratio. With the disclosure of the biologic relevance of their targets, therapeutic antibodies were developed such as cetuximab and panitumumab against EGFR, and bevacizumab against VEGF.
Peptides have several advantages over antibodies as disease-specific probes for molecular imaging. Peptides consist of only a few amino acids, and have much smaller structures with lower molecular weight. Therefore, peptides have better tissue penetration, shorter plasma half-life, and less associated immunogenicity[32-34]. In this study, peptide AAD showed a weak binding affinity to gastric dysplasia, but a significantly higher binding affinity to gastric cancer. A possible reason is that the targets are expressed at a lower level in pre-cancerous lesions as compared with cancer cells. As a molecular probe, peptide AAD may be used for grading dysplastic tissue and diagnosis of cancerous mucosa in early stage gastric cancer. The difference in binding affinity was not as significant between peptide AAD and the control peptide as reported in some published studies[35,36]. This may indicate a lower sensitivity as a tumor-specific probe and influence its future use. However, because of their pharmacokinetic advantages for in vivo imaging, tumor-targeting peptides do not necessarily have the highest binding affinity. Multiple excitation and detection wavelengths, in conjunction with multiple labels, may further enhance the applicability of this strategy. More than one tumor-specific peptide, each with a different target and fluorescent-label, could be mixed to increase the sensitivity, which may be used as a promising strategy for in vivo detection. Even with the limitations of current approaches, molecular imaging has the potential to greatly affect future imaging in gastroenterology. Future efforts should focus on the validation of peptides binding to malignantly-transformed mucosa in vivo.
Great progress has been made in molecular imaging in recent years, and technological and scientific advancement in endoscope compatible instruments have provided new imaging tools to improve the detection of early neoplastic lesions. Fluorescence endoscopes and confocal microendoscopes have been developed with a high sensitivity[37-39]. Once integrated with novel screening and surveillance methods, molecular endoscopy will prove effective real-time localization of dysplasia or neoplastic mucosa. Molecular probes that bind to suspected mucosal lesions may guide the doctor to perform a targeted biopsy. In vivo molecular imaging of live tissues may be less sensitive to bias from sampling error and tissue processing artifact than conventional histopathology, thus increasing the efficiency of endoscopic screening and surveillance. The peptide AAD identified in this study has the potential to guide tissue biopsy and improve the detection of pre-cancerous lesions in gastric mucosa.
Periodic endoscopy in high risk populations is most helpful in improving the early detection of gastric cancer (GC). However, the current endoscopic surveillance program for GC is labor-intensive and ineffective. Molecular probes are being developed to increase image contrast from early cancer during endoscopy to guide biopsy in some pioneered reports.
Molecular imaging is a technique that identifies and characterizes tumors and other lesions based on their protein expression pattern, rather than by their macroscopic morphology. The molecular expression pattern of cells and tissues can be visualized with the help of disease-specific molecular probes such as antibodies, antibody fragments, peptides, activatable probes and nanoparticles. Such molecular probes enable the diagnosis of disease in situ and in real time.
In this study the authors discovered a novel peptide that has specific binding activity to GC and can be used to distinguish neoplastic from normal gastric mucosa.
The peptide AADNAKTKSFPV identified in this study has the potential to guide tissue biopsy and improve the detection of pre-cancerous lesions in gastric mucosa.
The topic of the study is interesting and the authors tried to tackle very relevant and clinical important issues, the early detection of GC. The authors identified a peptide that seems to bind to GC tissue like an antibody.
Peer reviewers: Thomas Wex, Clinic of Gastroenterology and Hepatology, Otto-von-Guericke University, Leipziger Str. 44, 39120 Magdeburg, Germany; Takaaki Arigami, Department of Surgical Oncology and Digestive, Kagoshima University Gradu-ate School of Medical and Dental Sciences, 8-35-1 Sakuragaoka, Kagoshima 890-8520, Japan; Hikaru Nagahara, Professor, Gastroenterology, Aoyama Hospital, Tokyo Women’s Medical University, 2-7-13 Kitaaoyama Minato-ku, Tokyo 107-0061, Japan
S- Editor Gou SX L- Editor Ma JY E- Editor Li JY
| 1. | Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E. Gastric cancer. Crit Rev Oncol Hematol. 2009;71:127-164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 282] [Cited by in F6Publishing: 318] [Article Influence: 21.2] [Reference Citation Analysis (1)] |
| 2. | Clark CJ, Thirlby RC, Picozzi V, Schembre DB, Cummings FP, Lin E. Current problems in surgery: gastric cancer. Curr Probl Surg. 2006;43:566-670. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Kuipers EJ, Haringsma J. Diagnostic and therapeutic endoscopy. J Surg Oncol. 2005;92:203-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Tung CH. Fluorescent peptide probes for in vivo diagnostic imaging. Biopolymers. 2004;76:391-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 131] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 5. | Kumar S, Richards-Kortum R. Optical molecular imaging agents for cancer diagnostics and therapeutics. Nanomedicine (Lond). 2006;1:23-30. [PubMed] [Cited in This Article: ] |
| 6. | Klohs J, Wunder A, Licha K. Near-infrared fluorescent probes for imaging vascular pathophysiology. Basic Res Cardiol. 2008;103:144-151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 7. | Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008;14:454-458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 334] [Cited by in F6Publishing: 315] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 8. | Li M, Anastassiades CP, Joshi B, Komarck CM, Piraka C, Elmunzer BJ, Turgeon DK, Johnson TD, Appelman H, Beer DG. Affinity peptide for targeted detection of dysplasia in Barrett's esophagus. Gastroenterology. 2010;139:1472-1480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 9. | Chang CC, Hsieh YY, Wang YK, Hsu KH, Tsai HD, Tsai FJ, Lin CS. Identification of novel peptides specifically binding to endometriosis by screening phage-displaying peptide libraries. Fertil Steril. 2009;92:1850-1855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 10. | Shukla GS, Krag DN. Phage display selection for cell-specific ligands: development of a screening procedure suitable for small tumor specimens. J Drug Target. 2005;13:7-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 11. | Van Nieuwenhove LC, Rogé S, Balharbi F, Dieltjens T, Laurent T, Guisez Y, Büscher P, Lejon V. Identification of peptide mimotopes of Trypanosoma brucei gambiense variant surface glycoproteins. PLoS Negl Trop Dis. 2011;5:e1189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386-390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1590] [Cited by in F6Publishing: 1506] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 13. | Zang L, Shi L, Guo J, Pan Q, Wu W, Pan X, Wang J. Screening and identification of a peptide specifically targeted to NCI-H1299 from a phage display peptide library. Cancer Lett. 2009;281:64-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 14. | Du B, Qian M, Zhou Z, Wang P, Wang L, Zhang X, Wu M, Zhang P, Mei B. In vitro panning of a targeting peptide to hepatocarcinoma from a phage display peptide library. Biochem Biophys Res Commun. 2006;342:956-962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 15. | Radcliff G, Waite R, LeFevre J, Poulik MD, Callewaert DM. Quantification of effector/target conjugation involving natural killer (NK) or lymphokine activated killer (LAK) cells by two-color flow cytometry. J Immunol Methods. 1991;139:281-292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Chen Y, Huang K, Li X, Lin X, Zhu Z, Wu Y. Generation of a stable anti-human CD44v6 scFv and analysis of its cancer-targeting ability in vitro. Cancer Immunol Immunother. 2010;59:933-942. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Garcia-Hernandez Mde L, Gray A, Hubby B, Kast WM. In vivo effects of vaccination with six-transmembrane epithelial antigen of the prostate: a candidate antigen for treating prostate cancer. Cancer Res. 2007;67:1344-1351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 85] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Kelly KA, Bardeesy N, Anbazhagan R, Gurumurthy S, Berger J, Alencar H, Depinho RA, Mahmood U, Weissleder R. Targeted nanoparticles for imaging incipient pancreatic ductal adenocarcinoma. PLoS Med. 2008;5:e85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 171] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 19. | Hatanaka Y, Hashizume K, Kamihara Y, Itoh H, Tsuda H, Osamura RY, Tani Y. Quantitative immunohistochemical evaluation of HER2/neu expression with HercepTestTM in breast carcinoma by image analysis. Pathol Int. 2001;51:33-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 20. | Stefan N, Martin-Killias P, Wyss-Stoeckle S, Honegger A, Zangemeister-Wittke U, Plückthun A. DARPins recognizing the tumor-associated antigen EpCAM selected by phage and ribosome display and engineered for multivalency. J Mol Biol. 2011;413:826-843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 99] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 21. | Heemstra HE, van Weely S, Büller HA, Leufkens HG, de Vrueh RL. Translation of rare disease research into orphan drug development: disease matters. Drug Discov Today. 2009;14:1166-1173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Rivinoja A, Laakkonen P. Identification of homing peptides using the in vivo phage display technology. Methods Mol Biol. 2011;683:401-415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Ludtke JJ, Sololoff AV, Wong SC, Zhang G, Wolff JA. In vivo selection and validation of liver-specific ligands using a new T7 phage peptide display system. Drug Deliv. 2007;14:357-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Kolonin MG, Sun J, Do KA, Vidal CI, Ji Y, Baggerly KA, Pasqualini R, Arap W. Synchronous selection of homing peptides for multiple tissues by in vivo phage display. FASEB J. 2006;20:979-981. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 106] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 25. | Joyce JA, Laakkonen P, Bernasconi M, Bergers G, Ruoslahti E, Hanahan D. Stage-specific vascular markers revealed by phage display in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2003;4:393-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 192] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 26. | Fedorova A, Zobel K, Gill HS, Ogasawara A, Flores JE, Tinianow JN, Vanderbilt AN, Wu P, Meng YG, Williams SP. The development of peptide-based tools for the analysis of angiogenesis. Chem Biol. 2011;18:839-845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 27. | McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487-513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 541] [Cited by in F6Publishing: 553] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 28. | Fiske WH, Threadgill D, Coffey RJ. ERBBs in the gastrointestinal tract: recent progress and new perspectives. Exp Cell Res. 2009;315:583-601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Polglase AL, McLaren WJ, Delaney PM. Pentax confocal endomicroscope: a novel imaging device for in vivo histology of the upper and lower gastrointestinal tract. Expert Rev Med Devices. 2006;3:549-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Goetz M, Ziebart A, Foersch S, Vieth M, Waldner MJ, Delaney P, Galle PR, Neurath MF, Kiesslich R. In vivo molecular imaging of colorectal cancer with confocal endomicroscopy by targeting epidermal growth factor receptor. Gastroenterology. 2010;138:435-446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 171] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 31. | Barrett T, Koyama Y, Hama Y, Ravizzini G, Shin IS, Jang BS, Paik CH, Urano Y, Choyke PL, Kobayashi H. In vivo diagnosis of epidermal growth factor receptor expression using molecular imaging with a cocktail of optically labeled monoclonal antibodies. Clin Cancer Res. 2007;13:6639-6648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 93] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 32. | Brasnjevic I, Steinbusch HW, Schmitz C, Martinez-Martinez P. Delivery of peptide and protein drugs over the blood-brain barrier. Prog Neurobiol. 2009;87:212-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 177] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 33. | Pan W, Kastin AJ. Why study transport of peptides and proteins at the neurovascular interface. Brain Res Brain Res Rev. 2004;46:32-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 34. | Patel MM, Goyal BR, Bhadada SV, Bhatt JS, Amin AF. Getting into the brain: approaches to enhance brain drug delivery. CNS Drugs. 2009;23:35-58. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat Med. 2002;8:751-755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 368] [Cited by in F6Publishing: 348] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 36. | Jäger S, Jahnke A, Wilmes T, Adebahr S, Vögtle FN, Delima-Hahn E, Pfeifer D, Berg T, Lübbert M, Trepel M. Leukemia-targeting ligands isolated from phage-display peptide libraries. Leukemia. 2007;21:411-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 37. | Wong Kee Song LM, Wilson BC. Endoscopic detection of early upper GI cancers. Best Pract Res Clin Gastroenterol. 2005;19:833-856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 38. | Wong Kee Song LM. Optical spectroscopy for the detection of dysplasia in Barrett's esophagus. Clin Gastroenterol Hepatol. 2005;3:S2-S7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 39. | Shahid MW, Wallace MB. Endoscopic imaging for the detection of esophageal dysplasia and carcinoma. Gastrointest Endosc Clin N Am. 2010;20:11-24, v. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |