Published online Oct 21, 2009. doi: 10.3748/wjg.15.4896
Revised: August 4, 2009
Accepted: August 11, 2009
Published online: October 21, 2009
AIM: To reveal the characteristics of CD133+ cells in the liver.
METHODS: This study examined the histological characteristics of CD133+ cells in non-neoplastic and neoplastic liver tissues by immunostaining, and also analyzed the biological characteristics of CD133+ cells derived from human hepatocellular carcinoma (HCC) or cholangiocarcinoma cell lines.
RESULTS: Immunostaining revealed constant expression of CD133 in non-neoplastic and neoplastic biliary epithelium, and these cells had the immunophenotype CD133+/CK19+/HepPar-1-. A small number of CD133+/CK19-/HepPar-1+ cells were also identified in HCC and combined hepatocellular and cholangiocarcinoma. In addition, small ductal structures, resembling the canal of Hering, partly surrounded by hepatocytes were positive for CD133. CD133 expression was observed in three HCC (HuH7, PLC5 and HepG2) and two cholangiocarcinoma cell lines (HuCCT1 and CCKS1). Fluorescence-activated cell sorting (FACS) revealed that CD133+ and CD133- cells derived from HuH7 and HuCCT1 cells similarly produced CD133+ and CD133- cells during subculture. To examine the relationship between CD133+ cells and the side population (SP) phenotype, FACS was performed using Hoechst 33342 and a monoclonal antibody against CD133. The ratios of CD133+/CD133- cells were almost identical in the SP and non-SP in HuH7. In addition, four different cellular populations (SP/CD133+, SP/CD133-, non-SP/CD133+, and non-SP/CD133-) could similarly produce CD133+ and CD133- cells during subculture.
CONCLUSION: This study revealed that CD133 could be a biliary and progenitor cell marker in vivo. However, CD133 alone is not sufficient to detect tumor-initiating cells in cell lines.
- Citation: Yoshikawa S, Zen Y, Fujii T, Sato Y, Ohta T, Aoyagi Y, Nakanuma Y. Characterization of CD133+ parenchymal cells in the liver: Histology and culture. World J Gastroenterol 2009; 15(39): 4896-4906
- URL: https://www.wjgnet.com/1007-9327/full/v15/i39/4896.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.4896
| n | Age (yr) | Male/Female | Etiology | |
| Normal liver | 3 | 50 | 2/1 | |
| Chronic hepatitis/cirrhosis | 5 | 58 | 3/2 | HBV (3), HCV (2) |
| HCC | ||||
| Well-differentiated | 3 | 62 | 2/1 | HBV (1), HCV (2) |
| Moderately differentiated | 24 | 62 | 20/4 | HBV (11), HCV (7), alcohol (3), NASH (1), cryptogenic (1)1 |
| Poorly differentiated | 6 | 54 | 4/2 | HBV (3) |
| Cholangiocarcinoma | 6 | 60 | 3/3 | HCV (4) |
| Combined carcinoma | 5 | 59 | 4/1 | HBV (1), HCV (4) |
| F/R | Sequence | Temperature (°C) | Cycles | Size (bp) | |
| CD1331 | 1st F | GCCAGAAACTGTAATCTTAG | 48 | 35 | 275 |
| 1st R | TTACCTGGTGATTTGCCACA | ||||
| 2nd F | CCTGGGGCTGCTGTTTATTA | 55 | 35 | 153 | |
| 2nd R | ATCACCAACAGGGAGATTGC | ||||
| CK19 | F | TCCCGCGACTACAGCCACTACTACACGACC | 55 | 35 | 745 |
| R | CGCGACTTGATGTCCATGAGCCGCTGGTA | ||||
| CK7 | F | GGATGCTGCCTACATGAGC | 52 | 30 | 164 |
| R | CCAGGGAGCGACTGTTGT | ||||
| AFP | F | GGGAGCGGCTGACATTATTA | 50 | 30 | 231 |
| R | TCTTGCTTCATCGTTTGCAG | ||||
| Albumin | F | TGCTTGAATGTGCTGATGACAGGG | 50 | 30 | 161 |
| R | AAGGCAAGTCAGCAGGCATCTCATC | ||||
| β-actin | F | CAAGAGATGGCCACGGCTGCT | 55 | 30 | 334 |
| R | TCCTTCTGCATCCTGTCGGCA |
CD133 (also known as prominin-1 or AC133) was the first identified member of the prominin family of pentaspan membrane proteins[1-3]. In 1997, CD133 was reported as a marker of hematopoietic progenitor cells, using a novel monoclonal antibody that recognized the CD133 antigen[1-3]. Subsequently, it was reported that CD133 was also expressed in epithelial and non-epithelial progenitor cells in murine or human tissues including brain, kidney, prostate, pancreas, and skin[4-8]. The specific functions and ligands of CD133 have not been elucidated completely, although CD133 currently is recognized as a stem cell marker for normal and cancerous tissues in various organs[9-13].
Until now, there have been several reports regarding CD133 expression in hepatocellular carcinoma (HCC)[14-17]. Suetsugu et al[14] have examined CD133 expression in three cell lines of human HCC (HuH7, HepG2 and Hc). CD133 is expressed only on the surface of HuH7 cells. The CD133+ population of HuH7 cells is characterized by high proliferation activity and lower expression of mature hepatocellular markers. CD133+ cells can form tumors in SCID mice, whereas CD133- cells induce a very small number of tumors or none at all. It has been concluded that CD133 could be useable as a marker of cancer stem cells in human HCC[14]. Yin et al[15] and Ma et al[16] also have characterized CD133+ cells in HCC, and they have reached a conclusion similar to that of Suetsugu et al[14]. However, because these previous studies were mainly in vitro, the histological characteristics of hepatic CD133+ cells have not been fully examined so far. In particular, there are few data about CD133+ cells in non-neoplastic liver tissues and non-hepatocellular liver cancers.
In this study, CD133 expression in non-neoplastic and neoplastic liver tissues was examined. In vitro studies were also performed to examine the biological characteristics of CD133+ cells of HCC and cholangiocarcinoma cell lines. The goal of this study was to elucidate the histological and biological characteristics of CD133+ cells in non-neoplastic and neoplastic human livers.
Case selection: A total of 52 samples of liver tissues were obtained from the hepatobiliary disease file of the Division of Pathology, Kanazawa University Hospital in Japan between 2005 and 2009. This study consisted of three cases of normal liver, five cases of chronic viral hepatitis or liver cirrhosis, 33 cases of HCC, six cases of intrahepatic cholangiocarcinoma, and five cases of combined hepatocellular and cholangiocarcinoma (combined carcinoma). All cases used in this study were surgically resected cases. Normal liver tissues used in this study were background liver tissues of metastatic colon cancers. Age, sex and clinicopathological characteristics are shown in Table 1.
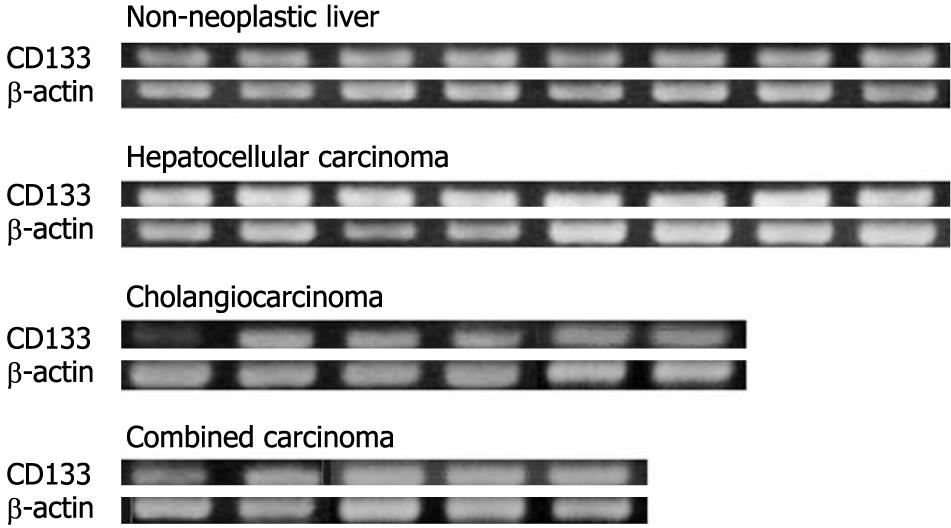
Expression of CD133 (mRNA level): Total RNA was extracted from the frozen section of all 47 cases using an RNeasy Mini kit (Qiagen, Valencia, CA, USA). Total RNA was dissolved in 50 μL of distilled water that contained 0.1% diethylpyrocarbonate, and quantitated using a spectrophotometer at OD260. Isolated RNA was used for the subsequent reverse transcriptase-polymerase chain reaction (RT-PCR). The expression of CD133 mRNA was examined by nested RT-PCR using two sets of primers. The oligonucleotide sequences, numbers of cycles, and annealing temperatures of these primers are shown in Table 2. After PCR, 5-μL aliquots of the products were subjected to 1.5% or 2.0% agarose gel electrophoresis and stained with ethidium bromide.
Immunostaining of CD133, cytokeratin 19 (CK19) and hepatocyte paraffin-1 (HepPar-1): Frozen sections of 52 samples of non-neoplastic and neoplastic liver tissues were used for immunostaining. Immunostaining for CD133, CK19 and HepPar-1 was performed using a mouse monoclonal antibody against human CD133 (clone AC133; Miltenyi Biotec, Auburn, CA, USA), a mouse monoclonal antibody against human CK19 (Dako Cytomation, Glostrup, Denmark), and a mouse monoclonal antibody against human HepPar-1 (Dako Cytomation).
Serial sections were used in each case to examine the co-localization of CD133, CK19 and HepPar-1 expression. Sliced frozen sections were fixed with acetone for 20 min. After blocking endogenous peroxidases, the sections were incubated in protein block solution (Dako Cytomation) for 20 min and incubated at 4°C with each primary antibody. These sections were incubated for 1 h at room temperature with goat anti-mouse immunoglobulins, which were conjugated to peroxidase-labeled polymer (Envision+; Dako Cytomation). 3,3'-Diaminobenzidine tetrahydrochloride was used as the chromogen, followed by light counterstaining with hematoxylin. Negative controls were evaluated by substituting the primary antibody with similarly diluted non-immunized mouse serum.
Cell culture: Three human HCC cell lines (HuH7, PLC5 and HepG2) and two human cholangiocarcinoma cell lines (CCKS1 and HuCCT1) were used in this study. HuH7, PLC5 and HepG2 were obtained from the Hearth Science Research Bank (Osaka, Japan). HuCCT-1 was obtained from the Cell Resource Center for Biochemical Research, Tohoku University, Sendai, Japan. CCKS1 was established in our laboratory[18]. HuH7 and PLC5 were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen Corp., Carlsbad, CA, USA), and HepG2 was maintained in minimum essential medium (Invitrogen Corp.) with 1% nonessential amino acids (Specialty Media, Phillipsburg, NJ, USA). CCKS1 and HuCCT1 were cultured in RPMI-1640 medium (Invitrogen Corp.) Each medium was supplemented with 10% fetal bovine serum (Invitrogen Corp.) and 1% antibiotic-antimycotic (Invitrogen Corp.).
Dual fluorescent immunostaining of CD133/CK19 and CD133/alpha-fetoprotein (AFP): Cell lines were cultured on Lab-Tek II chamber slides (Nalge Nunc International, Naperville, IL, USA) for fluorescent immunostaining. After culturing for 2 d, the specimens were fixed in 4% paraformaldehyde for 10 min at 4°C. After incubation in protein block solution (Dako Cytomation) for 10 min, the specimens were incubated with antibodies against CD133 and CK19, or antibodies against CD133 and AFP for 1 h at room temperature. The antibodies used were as follows: CD133, mouse monoclonal, clone AC133, Miltenyi Biotec; CK19, goat polyclonal, clone G-14, Santa Cruz Biotechnology (Santa Cruz, CA, USA); and AFP, a rabbit polyclonal, Dako Cytomation. The reaction product was visualized with fluorescent goat anti-mouse and anti-rabbit IgG antibodies (1:500, Molecular Probes Inc., Eugene, OR, USA). Specimens were counterstained with DAPI (Molecular Probes Inc.), and fluorescent signals were observed using a fluorescence microscope (Olympus, Tokyo, Japan).
Fluorescence-activated cell sorting (FACS) with reference to CD133 expression: HuH7 and HuCCT1 cells were used for FACS. Cultured cells were harvested after treatment with 0.25% of trypsin-EDTA solution (Sigma Chemical Co., St Louis, MO, USA) for 20 min, and washed three times in Hanks’ Balanced Salt Solution (Invitrogen Corp.). Cultured cells were stained live in a staining solution containing bovine serum albumin, insulin, and phycoerythrin (PE)-conjugated monoclonal antibody to CD133 (clone AC133; Miltenyi Biotec) for 30 min at 4°C. As negative controls, cultured cells were incubated similarly with non-immunized mouse immunoglobulin. Samples were analyzed and sorted by JSAN (Bay Bioscience, Kobe, Japan). Cell debris and cell aggregates were gated out electronically. For the positive population, only the top 5%-10% of the most brightly stained cells were selected. For the negative population, only the bottom 5%-10% of the most dimly stained cells were selected. Then, 1.0 × 105 cells were sorted from the positive or negative population at the most specific mode. Sorted cells were plated on culture dishes for subculture. After sorting, CD133+ and CD133- cells were cultured separately. After 4-wk culture, cultured cells were sorted again into CD133+ and CD133- cells using flow cytometry to evaluate how the CD133+ cell ratios were altered in each subpopulation. After subculturing for 3 or 4 wk, cultured cells were sorted again into CD133+ and CD133- cells to evaluate how the CD133+ or CD133- populations changed during subculture. The percentages of CD133+ cells were calculated in a total of 1000-5000 cells in each group.
RNA expression in culture cells: Total RNA was extracted from five types of cultured cells using an RNeasy Mini Kit (Qiagen). Total RNA was similarly extracted from CD133+ and CD133- cells. RT-PCR was performed for CD133, hepatocyte makers (AFP and albumin), biliary markers (CK19 and CK7 and β-actin. The oligonucleotide sequences, numbers of cycles and annealing temperatures of these primers are shown in Table 2. After PCR, 5-μL aliquots of the products were subjected to 1.5% or 2.0% agarose gel electrophoresis and stained with ethidium bromide.
Real-time RT-PCR: The alterations of CD133 expression levels were examined in non-sorted or sorted (CD133+ or CD133-) cultured cells time-dependently (days 0, 7, 14, 21 and 28) after the passage or sorting. Real-time analysis was performed using premade CD133 and β-actin-specific primers and probes with the ABI Prism 7700 sequence detection system (PE Applied Biosystems, Warrington, UK). RT-PCR was done with the TaqMan Universal PCR Master Mix (PE Applied Biosystems) using 2 μL cDNA in a 25-μL final reaction mixture. Cycling conditions were as follows: incubation at 50°C for 2 min, 10 min at 95°C, and 50 cycles of 15 s at 95°C and 1 min at 60°C. CD133 was normalized (ΔCt) to β-actin from the Ct value of CD133. Each experiment was performed in triplicate, and the mean adopted.
Cell proliferation assay of CD133+ and CD133-cells: CD133+ and CD133- cells were plated on a Lab-Tek II chamber slide (Nalge Nunc International), and cultured for 7 d before the cell proliferation assay. Cell proliferation was assayed using BrdU. Cultured cells were incubated on slides with BrdU solution (10 mmol/L) at 37°C for 30 min. After fixing with 70% ethanol (50 mmol/L glycine buffer solution, pH 2.0) for more than 20 min, the slides were incubated with anti-BrdU solution at 37°C for 30 min. After additional incubation with IgG fluorochrome solution for 30 min, positive signals were detected by a fluorescence microscope (Olympus).
Relationship between side population (SP) and CD133+ cells: SP is currently estimated as one of the most reliable stem cell phenotypes[19,20]. The relationship between SP and CD133+ cells was examined by FACS. After detaching and washing, the cultured cells were then incubated at 37°C for 90 min with 20 μg/mL Hoechst 33342 (Sigma Chemical Co.), PE-conjugated monoclonal antibody to CD133 (clone AC133; Miltenyi Biotec), bovine serum albumin, in the presence or absence of 100 μmol/L verapamil (Sigma Chemical Co.). After incubation, 1 μg/mL propidium iodide (Sigma Chemical Co.) was added and the cells were filtered through a 40-μm cell strainer (BD Biosciences, San Diego, CA, USA) to obtain single-cell suspensions. The relationship between SP and CD133 expression was analyzed by JSAN (Bay Bioscience). Hoechst 33342 was excited with a UV laser at 350 nm and fluorescence emission was measured with 405/BP30 (Hoechst blue) and 570/BP20 (Hoechst red) optical filters. Propidium iodide labeling was measured through a 630/BP30 filter for the discrimination of dead cells. Next, HuH7 cells were sorted into SP/CD133+, SP/CD133-, non-SP/CD133+, and non-SP/CD133-. After 4 wk subculturing, each population was analyzed again with regard to CD133 expression by FACS.
Differences between two groups were analyzed using the Mann-Whitney U test or χ2 test. Statistical analysis was performed using Statcel 2 software (OMS publishing, Tokorozawa, Japan). P < 0.05 was considered to be significant.
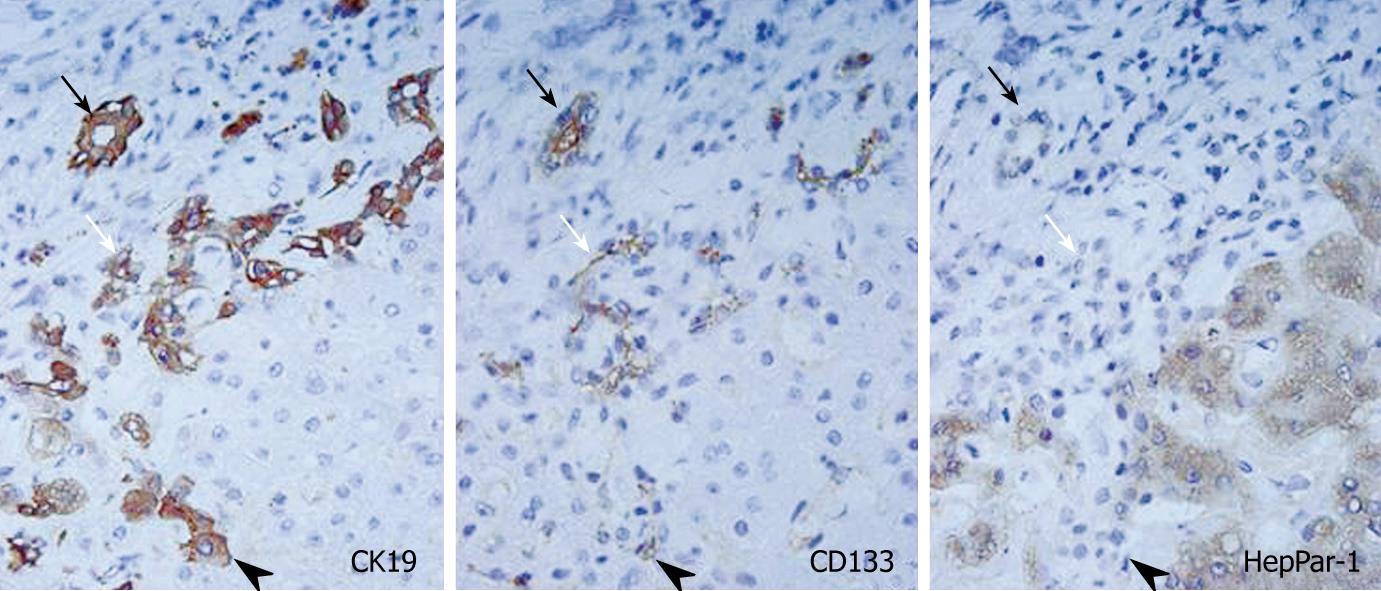
The expression of CD133 mRNA was identified in all non-neoplastic and neoplastic liver tissues examined in this study by nested RT-PCR (Figure 1). The results of immunostaining of CD133 are shown in Figures 2 and 3. In normal livers, CD133 was expressed constantly in biliary epithelium of intrahepatic large and small bile ducts. Mature hepatocytes were negative for CD133. In the livers of chronic hepatitis and liver cirrhosis patients, CD133 was expressed in bile ducts and proliferating bile ductules. In addition, small ductal structures, resembling the canal of Hering, partly surrounded by hepatocytes were also positive for CD133 (Figure 2). CD133 was expressed on cellular membrane with accentuation on the luminal side. Immunostaining of CK19 and HepPar-1 on serial sections revealed that CD133 and CK19 expressions were closely co-localized (Figure 2). In contrast, mature hepatocytes that expressed HepPar-1 were negative for CD133. CD133 expression was not evident in mesenchymal or inflammatory cells upon immunostaining.
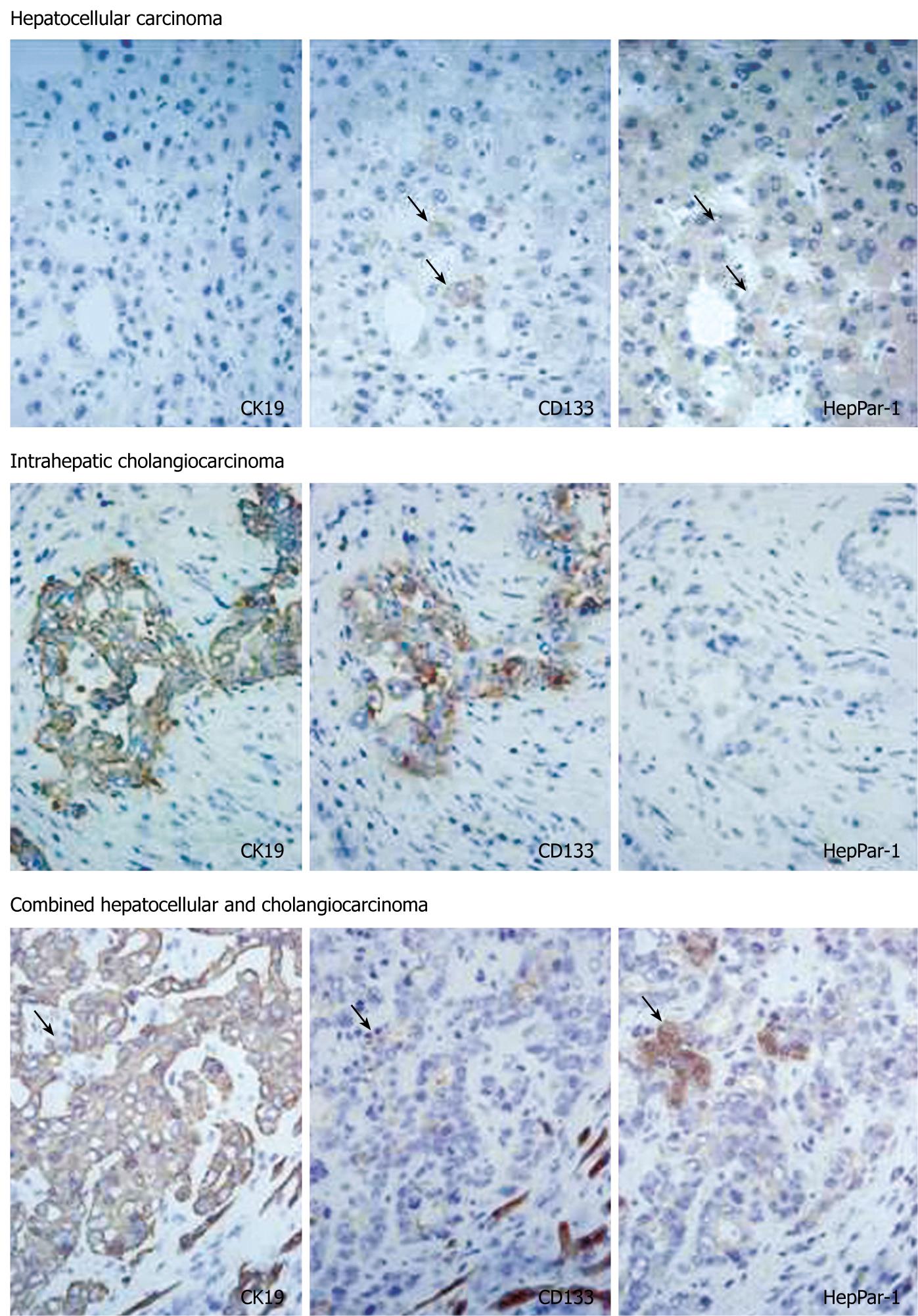
In HCC, eight of 33 cases (24%) had CD133+ cells. CD133+ cells were small in number and randomly distributed in these tumors. There were no morphological differences between CD133+ and CD133- cells. Serial sections stained with CK19 and HepPar-1 revealed that CD133+ cells in HCC were HepPar-1+ and CK19- (Figure 3). CD133+ cells were observed more often in less differentiated HCCs: 0/3 (0%) in well-differentiated, 4/24 (17%) in moderately differentiated, and 4/6 (67%) in poorly differentiated HCC cases. The expression of CD133 mRNA was detected in all HCC cases by nested RT-PCR, although CD133+ cells were identified in only 24% of cases by immunostaining. This discrepancy might have resulted from the small numbers of CD133+ cells in HCC.
In cholangiocarcinoma, CD133 was expressed diffusely in carcinoma cells in all cases examined (Figure 3). CD133 expression was mainly on cellular membranes. HepPar-1 expression was not observed in any cases of cholangiocarcinoma, and CD133+ cholangiocarcinoma cells were CK19+ and HepPar-1-. In combined carcinoma, all cases had CD133+ carcinoma cells. CD133 expression was observed mainly in adenocarcinoma components. Most CD133+ cells were CK19+ and HepPar-1-, although some CD133+ cells were CK19- and HepPar-1+ (Figure 3).
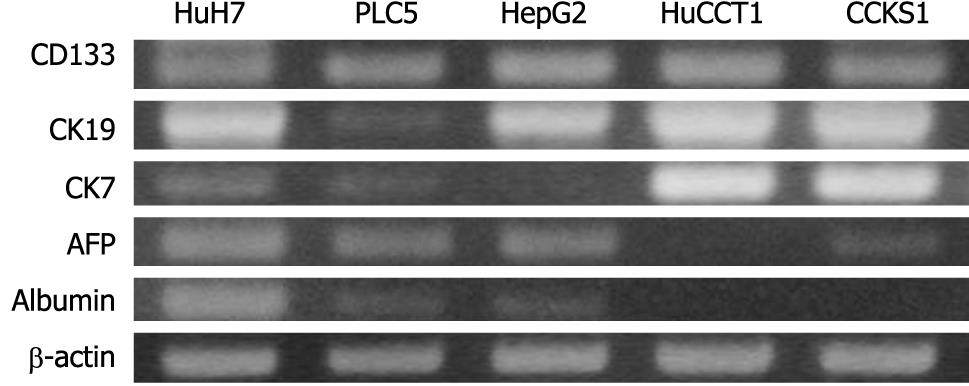
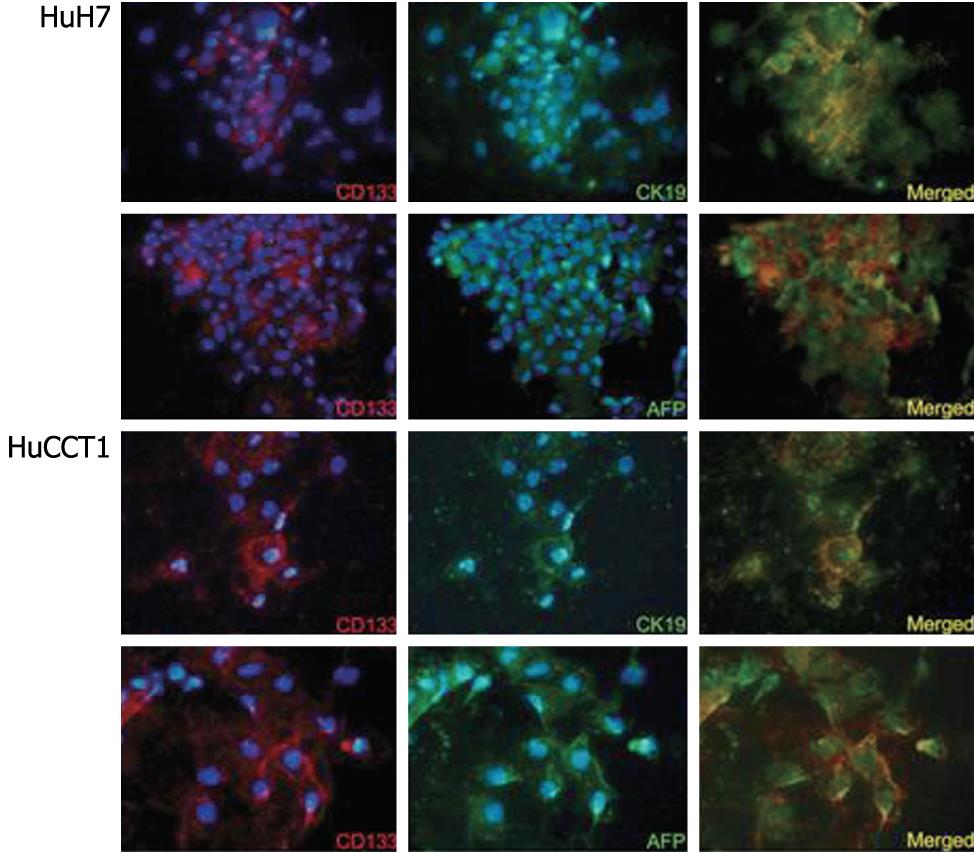
The expression of CD133 mRNA was identified in all cell lines by RT-PCR (Figure 4). Biliary markers (CK19 and CK7) were expressed strongly in CCKS1 and HuCCT1 cells, whereas hepatocellular markers (AFP and albumin) were expressed constantly in HuH7, PLC5 and HepG2 cells. In addition, a cholangiocarcinoma cell line, HuCCT1, also expressed AFP. Similarly, HCC cell lines also expressed CK19 or CK7. Next, the relationships between CD133 and CK19 or AFP expression levels were examined using HCC (HuH7) and cholangiocarcinoma (HuCCT1) cell lines, both of which expressed hepatocellular and biliary markers. Dual immunostaining of CD133/CK19 or CD133/AFP revealed that CD133+/CK19+ or CD133+/AFP+ cells were present in both HuH7 and HuCCT1 cells (Figure 5).
FACS was performed using two cell lines (HuH7 and HuCCT1). The flow cytometry analysis with regard to CD133 expression is shown in Figure 6. The percentages of CD133+ cells from flow cytometry were 65.0% in HuH7 and 48.5% in HuCCT1 cells. After cell sorting, CD133+ and CD133- cells derived from HuH7 or HuCCT1 cells were cultured separately for 4 wk. After 4 wk subculturing, both CD133+ and CD133- subpopulations returned to almost the pre-sorting cellular population that comprised both CD133+ and CD133- cells (Figure 6). These results suggested that CD133- HuH7 and HuCCT1 cells generated CD133+ and CD133- progenies during subculture.
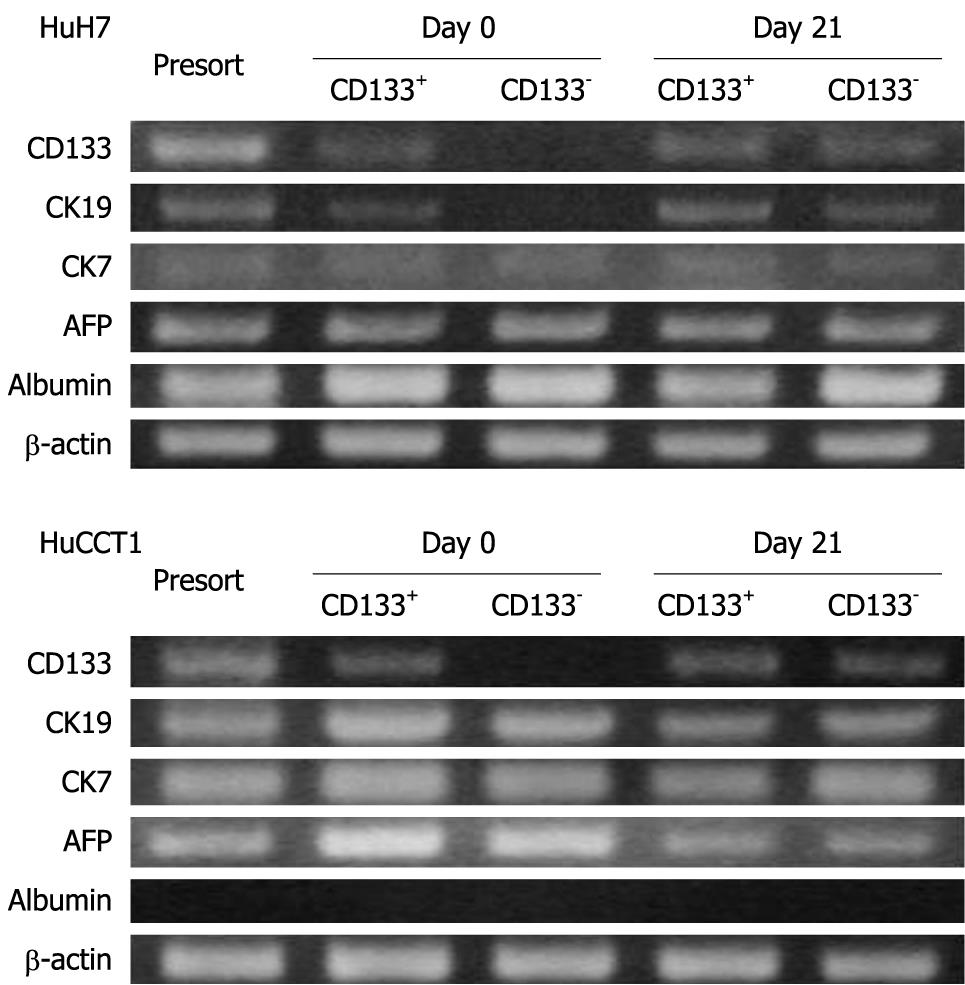
The expression patterns of CD133, CK19, CK7, AFP and albumin were examined in HuH7 and HuCCT1 cells before sorting, just after sorting (day 0), and after 3 wk subculture (day 21). At day 0, the expression of CD133 mRNA was observed in only the CD133+ population in both HuH7 and HuCCT1 cells. However, CD133+ and CD133- populations expressed the CD133 mRNA at similar levels at day 21 (Figure 7). These results suggested that CD133- cells began to express CD133 or produce CD133+ progeny during subculture. Acquisition of CD133 expression in CD133- cells was consistent with the results of FACS.
On day 0, CK19 was expressed only in CD133+ HuH7 cells. However, CK19 expression was also identified in CD133- cells at day 21. At day 21, CD133+ and CD133- HuH7 or HuCCT1 cells showed similar expression patterns for mRNA, except for the slightly more intense expression of CK19 and albumin in CD133+ and CD133-HuH7 cells, respectively (Figure 7).
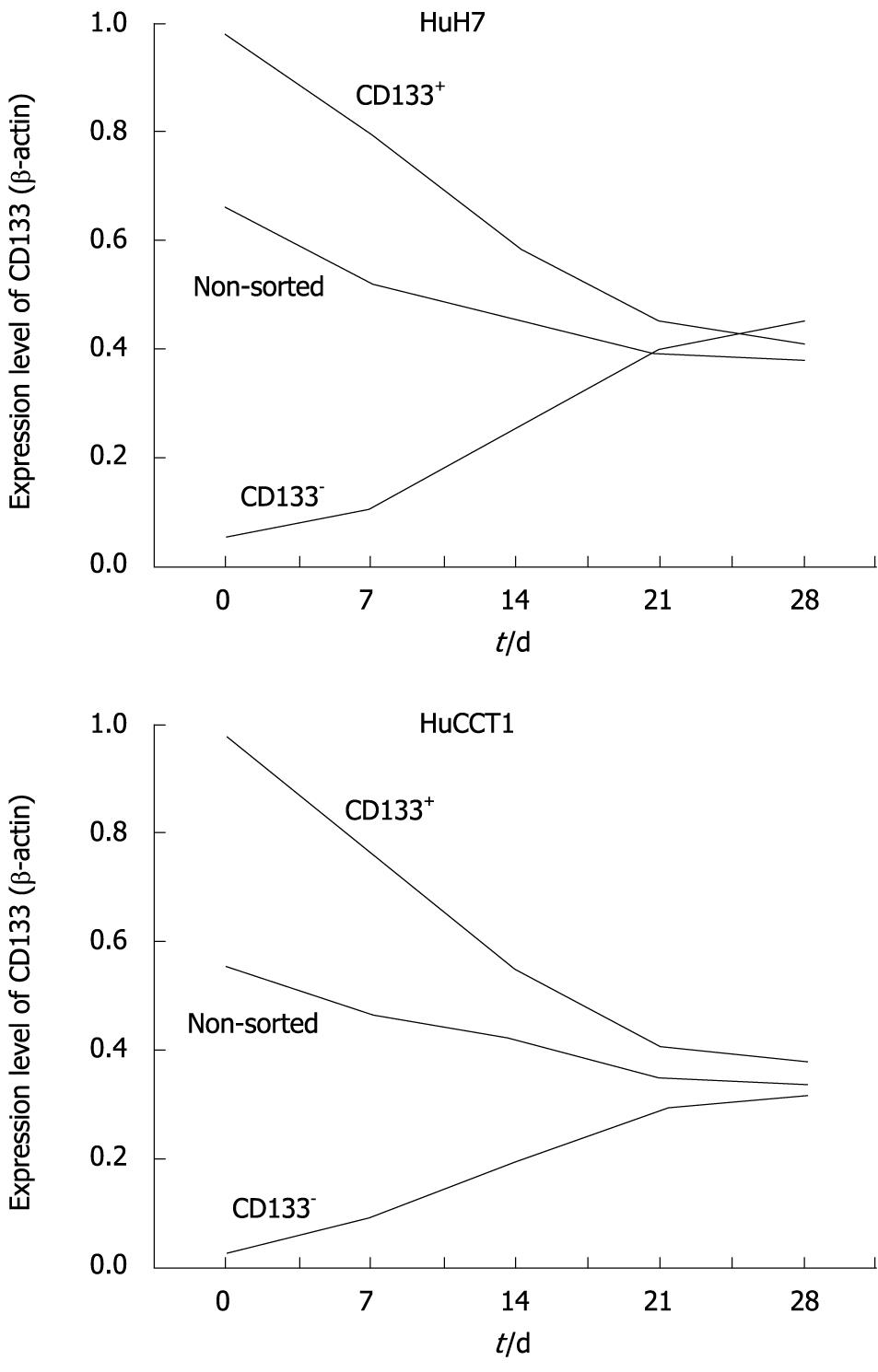
Alterations of CD133 expression levels in HuH7 and HuCCT1 cells were examined by real-time quantitative RT-PCR. HuH7 and HuCCT1 cells showed similar alteration patterns. As shown in Figure 8, CD133 expression levels in non-sorted HuH7 and HuCCT1 cells gradually decreased after passage. CD133 expression levels in CD133+ populations were high just after the sorting (day 0) in both cell lines. These expression levels decreased time-dependently. In contrast, CD133 expression levels in CD133- cells were very low at day 0, and time-dependently increased. CD133 expression in CD133+ and CD133- cells reached a similar level around day 21 or 28. In addition, their expression level was also similar to the level of CD133 expression in non-sorted cells at day 21 (Figure 8).
The proliferation of CD133+ and CD133- cells were examined using BrdU after 7 d subculture. The percentages of BrdU-labeled cells were as follows: CD133+ HuH7 cells, 22%; CD133- HuH7 cells, 24%; CD133+ HuCCT1 cells, 39%; and CD133-HuCCT1 cells, 42%. No significant differences were observed in proliferation of CD133+ and CD133- HuH7 and HuCCT1 cells.
Previous studies have reported that CD133+ HCC cells have a greater colony-forming efficiency, higher proliferative activity, and greater ability to form tumors in vivo[14-16]. It has been suggested that CD133- cells are not capable of producing CD133+ cells. However, CD133+ and CD133- HuH7 cells returned to almost identical cell populations after 4 wk subculture in this study. To resolve this discrepancy, the relationship between SP and CD133+ cells in HuH7 cells was examined because SP is one of the most reliable stem cell markers currently available.
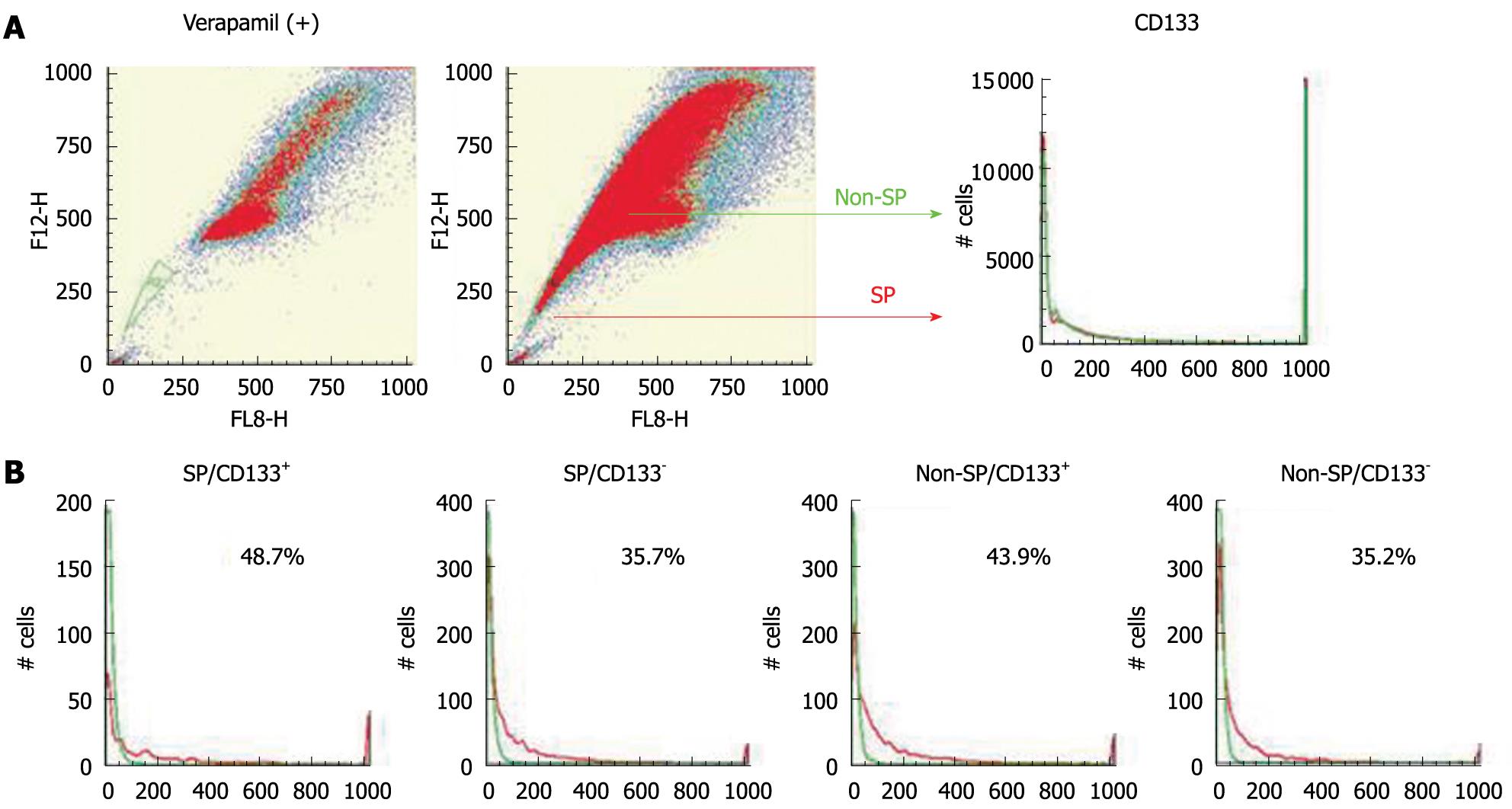
As in the previous study[21], an SP fraction was identified in HuH7 cells. The percentages of CD133+ cells in SP and non-SP fractions were examined using Hoechst 33342 and a PE-conjugated antibody against CD133. The ratios of CD133+ cells were almost the same in both the SP and non-SP fractions (Figure 9). HuH7 cells were sorted into four populations: SP/CD133+, SP/CD133-, non-SP/CD133+, and non-SP/CD133-. Each population was cultured separately for 4 wk and was analyzed again with regard to CD133 expression by FACS. CD133+ and CD133- cells were produced at similar levels in the four populations (Figure 9). These results suggested no relationship between SP phenotype and CD133 expression.
This study involved the histological characterization of CD133+ cells in the liver and the biological characteristics of CD133+ cells derived from human HCC and cholangiocarcinoma cell lines. The results obtained can be summarized as follows. (1) CD133 was expressed constantly in the biliary epithelium in non-neoplastic liver tissues. Most of the CD133+ cells were CK19+ and HepPar-1- in non-neoplastic liver tissues. (2) In HCC, the expression of CD133 mRNA was observed in all cases by nested RT-PCR, whereas CD133+ cells were identified in only 24% of cases by immunostaining. CD133+ cells were small in number in all the cases of HCC examined. (3) In cholangiocarcinoma, CD133 was expressed diffusely in most carcinoma cells. (4) In combined carcinoma, most of the CD133+ cells were CK19+ and HepPar-1-, although some CD133+ cells were CK19- and HepPar-1+. (5) In human HCC and cholangiocarcinoma cell lines, CD133+ cells co-expressed CK19 and AFP. (6) CD133+ or CD133- cells derived from HuH7 and HuCCT1 cell lines similarly produced CD133+ and CD133- progeny during subculturing. (7) There was no relationship between CD133+ cells and SP phenotype.
In the histological examination, CD133 expression was related closely to CK19 expression. CK19 has been used as not only a biliary marker, but also as a progenitor cell marker. CK19 is expressed usually in the bile ducts, bile ductules, and the canal of Hering[22-24]. Small ductal structures partly surrounded by hepatocytes (the canal of Hering) are currently estimated as hepatic stem/progenitor cells, and these structures are also positive for CD133[25,26]. Before starting this study, it was speculated that CD133 was expressed only in hepatic progenitor cells. However, this study revealed that CD133 is not only a progenitor cell marker, but can also be used as a novel biliary marker.
Some might argue about the discrepancy between the results with nested RT-PCR and immunohistochemistry for HCC. Nested RT-PCR could detect CD133 expression in all HCC cases; whereas, its expression was observed in only 24% of cases by immunostaining. We speculate that this difference might have been caused by the low expression level of CD133 in HCC. Indeed, non-nested conventional PCR showed CD133 expression in less than half of HCC cases in the preliminary study. That is, it might be difficult to detect CD133 expression in HCC by immunostaining because of the low expression level or the lower number of positive cells.
Other investigators have examined CD133 expression in liver tissues. Yin et al[15] have reported that CD133 expression is observed in a small subset of hepatocytes, biliary epithelium, and epithelial clusters in the portal tracts in cirrhotic livers, but CD133 expression is not seen in normal liver[15]. In addition, Ma et al[16] have also reported that CD133+ cells are almost absent in non-neoplastic liver tissues. The discrepancy between the previous and current studies might have been cause by the method of immunostaining. Both previous studies used a goat polyclonal antibody and paraffin-embedded specimens, but preliminary trials in the current study could not detect any positive signals for CD133 in paraffin-embedded specimens using any antibodies for CD133; therefore, frozen sections were used instead. In addition, CD133, which is usually expressed on the cellular membrane, was detected in the cytoplasm in previous studies. More recently, Shmelkov et al[27] Have examined CD133 in various organs using a unique transgenic mouse model, in which endogenous promoters for CD133 drove the expression of the reporter gene lacZ. CD133 was expressed widely in differentiated ductal structures in various organs including bile ducts in the liver[27]. These previous results are consistent with the results of the current study.
CD133 expression was also related closely to CK19 expression in neoplastic liver tissues. In cholangiocarcinoma and combined carcinoma, CK19+ cells constantly co-expressed CD133. CD133+ cells comprised 48.5% of cells in the HuCCT1 cell line, and there were no differences between CD133+ and CD133- cells in terms of proliferation or mRNA expression. We speculate that CD133 expression in cholangiocarcinoma reflects the biliary phenotype and not the progenitor phenotype. In contrast, some CD133+ cells in HCC and combined carcinoma were CK19- and HepPar-1+. CD133+/CK19-/HepPar-1+ cells could not be identified in non-neoplastic livers, although this suggests that CD133+ cells are pluripotent and can differentiate into CK19+/HepPar-1- and CK19-/HepPar-1+ cells. In particular, it is interesting that CD133+/CK19-/HepPar-1+ cells are observed in combined carcinoma because the involvement of hepatic progenitor cells is suggested in tumorigenesis of combined carcinoma[28-30].
Until now, some investigators have examined the characterization of CD133+ cells in HCC, and have suggested that CD133+ HCC cells are characterized by higher proliferative activity, expression of “stemness” genes, the ability to self-renew, and greater ability to form tumors in vivo[14-16]. They have concluded that CD133+ cells are tumorigenic cancer cells, and located at a higher rank in the cancer-cell hierarchy. However, in the current study, CD133+ HuH7 cells were not different from CD133- cells in terms of proliferation. In addition, CD133- cells could generate both CD133+ and CD133- cells in subcultures, which did not support the existence of a cancer-cell hierarchy with respect to CD133 expression. As noted in previous studies, CD133+ cells comprised about half of the carcinoma cells in the HuH7 cell line (65.0% in the current study, 46.7% or 65.0% in previous studies[14,16]). The percentage of CD133+ cells seems too high to suggest that CD133 cells are tumor-initiating cells. Indeed, CD133- cells were able to give rise to CD133+ cells in the subculture system in the previous study[16]. Moreover, a more recent study has revealed that CD133 expression is not restricted to stem cells, and CD133+ and CD133- cells derived from colon cancer are capable of initiating tumors in immunodeficient mice[27].
To resolve the uncertainties regarding CD133 expression and tumor-initiating cells, the relationship between CD133+ cells and SP phenotype was examined in our study. SP is a minor population with extreme tumorigenic potential, and it is supposed that tumor-initiating cells exist in SP cells. If CD133+ cells are tumor-initiating cells, CD133+ cells should be related closely to the SP phenotype, and CD133- cells should not exist in the SP fraction. However, there was no difference in the CD133+/CD133- cellular population between SP and non-SP fractions. In addition, four cell populations (SP/CD133+, SP/CD133-, non-SP/CD133+, and non-SP/CD133-) could similarly produce CD133+ and CD133- cells during subculture. It is speculated that CD133 expression might reflect the progenitor phenotype in HCC; however, CD133 alone is not sufficient to detect tumor-initiating cells.
It seems important to know that CD133 is one of the progenitor cell markers in the liver, but this is not specific. We have to determine the conditions to identify a pure hepatic progenitor or stem cell population, using multiple surface markers including CD133. From the biliary aspect, CD133 could become a useful marker, because this is a surface antigen. We can use this molecule for sorting or purification of biliary epithelium.
In conclusion, this study revealed that CD133 can be a biliary and progenitor cell marker in liver tissues. However, CD133 alone is not sufficient to detect tumor-initiating cells in cultured cells.
CD133 is recognized as a stem cell marker for normal and cancerous tissues in various organs. The histological characteristics of hepatic CD133+ cells have not been examined fully, especially in non-neoplastic liver tissues and non-hepatocellular liver cancers.
Previous studies have shown that CD133 can be used as a maker of cancer stem cells in human hepatocellular carcinoma (HCC). The current study elucidated the histological and biological characteristics of CD133+ cells in non-neoplastic and neoplastic human livers.
Immunohistochemical analysis showed that CD133 was expressed constantly in the non-neoplastic biliary epithelium. In cholangiocarcinoma, CD133 was expressed diffusely in most carcinoma cells, whereas CD133+ cells were identified in only a small number of cases of HCC. In combined carcinoma, most of the CD133+ cells were CK19+. In human HCC and cholangiocarcinoma cell lines, CD133+ cells co-expressed CK19 or alpha-fetoprotein. CD133+ or CD133- cells derived from human HCC and cholangiocarcinoma cell lines similarly could produce CD133+ and CD133- progeny during subculturing, and there was no relationship between CD133+ cells and the side population (SP) phenotype.
This study demonstrated that CD133 could be a biliary and progenitor cell marker in vivo. However, CD133 alone is not sufficient to detect tumor-initiating cells in cell lines. Theses results may provide insights into understanding the pathogenesis of various hepatobiliary diseases.
CD133 (also known as prominin-1 or AC133) is a marker of hematopoietic progenitor cells. It has also been reported that CD133 is expressed in epithelial and non-epithelial progenitors in various tissues, in which the specific functions and ligands of CD133 have not been fully elucidated. SP is a minor population with extreme tumorigenic potential, and it is supposed that tumor-initiating cells exist in SP cells.
The article presents interesting and novel data about CD133 expression in non-neoplastic and neoplastic liver tissues, and examines some biological characteristics of CD133+ cells in HCC and cholangiocarcinoma cell lines. Although the authors presented some controversial data about the presence of CD133 expression in HCC, the experiments were designed appropriately, the methodology was precise, and the discussion supports the results.
Peer reviewer: Maria Concepción Gutiérrez-Ruiz, PhD, Departamento de Ciencias de la Salud, Universidad Autónoma Metropolitana-Iztapalapa, DCBS, Av San Rafael Atlixco 186, Colonia Vicentina, México, DF 09340, México
S- Editor Li LF L- Editor Kerr C E- Editor Ma WH
| 1. | Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, Bray RA, Waller EK, Buck DW. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90:5013-5021. [Cited in This Article: ] |
| 2. | Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002-5012. [Cited in This Article: ] |
| 3. | Shmelkov SV, St Clair R, Lyden D, Rafii S. AC133/CD133/Prominin-1. Int J Biochem Cell Biol. 2005;37:715-719. [Cited in This Article: ] |
| 4. | Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97:14720-14725. [Cited in This Article: ] |
| 5. | Sagrinati C, Netti GS, Mazzinghi B, Lazzeri E, Liotta F, Frosali F, Ronconi E, Meini C, Gacci M, Squecco R. Isolation and characterization of multipotent progenitor cells from the Bowman's capsule of adult human kidneys. J Am Soc Nephrol. 2006;17:2443-2456. [Cited in This Article: ] |
| 6. | Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539-3545. [Cited in This Article: ] |
| 7. | Oshima Y, Suzuki A, Kawashimo K, Ishikawa M, Ohkohchi N, Taniguchi H. Isolation of mouse pancreatic ductal progenitor cells expressing CD133 and c-Met by flow cytometric cell sorting. Gastroenterology. 2007;132:720-732. [Cited in This Article: ] |
| 8. | Ito Y, Hamazaki TS, Ohnuma K, Tamaki K, Asashima M, Okochi H. Isolation of murine hair-inducing cells using the cell surface marker prominin-1/CD133. J Invest Dermatol. 2007;127:1052-1060. [Cited in This Article: ] |
| 9. | Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946-10951. [Cited in This Article: ] |
| 10. | O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-110. [Cited in This Article: ] |
| 11. | Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111-115. [Cited in This Article: ] |
| 12. | Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821-5828. [Cited in This Article: ] |
| 13. | Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396-401. [Cited in This Article: ] |
| 14. | Suetsugu A, Nagaki M, Aoki H, Motohashi T, Kunisada T, Moriwaki H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun. 2006;351:820-824. [Cited in This Article: ] |
| 15. | Yin S, Li J, Hu C, Chen X, Yao M, Yan M, Jiang G, Ge C, Xie H, Wan D. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer. 2007;120:1444-1450. [Cited in This Article: ] |
| 16. | Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, Zheng BJ, Guan XY. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542-2556. [Cited in This Article: ] |
| 17. | Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749-1758. [Cited in This Article: ] |
| 18. | Sugawara H, Yasoshima M, Katayanagi K, Kono N, Watanabe Y, Harada K, Nakanuma Y. Relationship between interleukin-6 and proliferation and differentiation in cholangiocarcinoma. Histopathology. 1998;33:145-153. [Cited in This Article: ] |
| 19. | Goodell MA, Rosenzweig M, Kim H, Marks DF, DeMaria M, Paradis G, Grupp SA, Sieff CA, Mulligan RC, Johnson RP. Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat Med. 1997;3:1337-1345. [Cited in This Article: ] |
| 20. | Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H, Taniguchi H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240-251. [Cited in This Article: ] |
| 21. | Fujii T, Zen Y, Harada K, Niwa H, Masuda S, Kaizaki Y, Watanabe K, Kawashima A, Nakanuma Y. Participation of liver cancer stem/progenitor cells in tumorigenesis of scirrhous hepatocellular carcinoma--human and cell culture study. Hum Pathol. 2008;39:1185-1196. [Cited in This Article: ] |
| 22. | Roskams TA, Theise ND, Balabaud C, Bhagat G, Bhathal PS, Bioulac-Sage P, Brunt EM, Crawford JM, Crosby HA, Desmet V. Nomenclature of the finer branches of the biliary tree: canals, ductules, and ductular reactions in human livers. Hepatology. 2004;39:1739-1745. [Cited in This Article: ] |
| 23. | Haque S, Haruna Y, Saito K, Nalesnik MA, Atillasoy E, Thung SN, Gerber MA. Identification of bipotential progenitor cells in human liver regeneration. Lab Invest. 1996;75:699-705. [Cited in This Article: ] |
| 24. | Zen Y, Fujii T, Yoshikawa S, Takamura H, Tani T, Ohta T, Nakanuma Y. Histological and culture studies with respect to ABCG2 expression support the existence of a cancer cell hierarchy in human hepatocellular carcinoma. Am J Pathol. 2007;170:1750-1762. [Cited in This Article: ] |
| 25. | Theise ND, Saxena R, Portmann BC, Thung SN, Yee H, Chiriboga L, Kumar A, Crawford JM. The canals of Hering and hepatic stem cells in humans. Hepatology. 1999;30:1425-1433. [Cited in This Article: ] |
| 26. | Saxena R, Theise N. Canals of Hering: recent insights and current knowledge. Semin Liver Dis. 2004;24:43-48. [Cited in This Article: ] |
| 27. | Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111-2120. [Cited in This Article: ] |
| 28. | Kim H, Park C, Han KH, Choi J, Kim YB, Kim JK, Park YN. Primary liver carcinoma of intermediate (hepatocyte-cholangiocyte) phenotype. J Hepatol. 2004;40:298-304. [Cited in This Article: ] |
| 29. | Zhang F, Chen XP, Zhang W, Dong HH, Xiang S, Zhang WG, Zhang BX. Combined hepatocellular cholangiocarcinoma originating from hepatic progenitor cells: immunohistochemical and double-fluorescence immunostaining evidence. Histopathology. 2008;52:224-232. [Cited in This Article: ] |
| 30. | Tickoo SK, Zee SY, Obiekwe S, Xiao H, Koea J, Robiou C, Blumgart LH, Jarnagin W, Ladanyi M, Klimstra DS. Combined hepatocellular-cholangiocarcinoma: a histopathologic, immunohistochemical, and in situ hybridization study. Am J Surg Pathol. 2002;26:989-997. [Cited in This Article: ] |