Published online Feb 7, 2008. doi: 10.3748/wjg.14.701
Revised: September 30, 2007
Published online: February 7, 2008
AIM: To elucidate the influence of quasispecies on virological response and disease severity in patients with chronic hepatitis C.
METHODS: Forty seven patients with hepatitis C [32 with chronic active hepatitis (CAH), 9 with cirrhosis, and 6 with hepatocellular carcinoma (HCC)] were screened for the presence of quasispecies by single stranded conformational polymorphism (SSCP) analysis in the hypervariable region (HVR) and non-structural 5B (NS5B) viral genes of hepatitis C virus. The 41 patients excluding those with HCC were on therapy and followed up for a year with the determination of virological response and disease severity. Virus isolated from twenty three randomly selected patients (11 non-responders and 12 showing a sustained virological response) was sequenced for the assessment of mutations.
RESULTS: The occurrence of quasispecies was proportionately higher in patients with HCC and cirrhosis than in those with CAH, revealing a significant correlation between the molecular evolution of quasispecies and the severity of disease in patients with hepatitis C. The occurrence of complex quasispecies has a significant association (P < 0.05) with the non-responders, and leads to persistence of infection. Significant differences (P < 0.05) in viral load (log10 IU/mL) were observed among patients infected with complex quasispecies (CQS), those infected with simple quasispecies (SQS) and those with no quasispecies (NQS), after 12 wk (CQS-5.2 ± 2.3, SQS-3.2 ± 1.9, NQS-2.8 ± 2.4) and 24 wk (CQS-3.9 ± 2.2, SQS-3.0 ± 2.2, NQS-2.1 ± 2.3) in the HVR region. However, a statistically significant difference (P < 0.05) was observed between the viral loads of patients infected with CQS and those infected with NQS in NS5B viral gene after 24 wk (CQS-3.9 ± 2.2, SQS-3.0 ± 2.2, and NQS-2.1 ± 2.3) and 48 wk (CQS-3.1 ± 2.7, SQS-2.3 ± 2.4, NQS-2.0 ± 2.3) of therapy. Disease severity was significantly associated with viral load during therapy. The strains isolated from non-responders showed close pairing on phylogeny based on the NS5B gene, but dissimilar HVR regions. This revealed the possibility of the selection of resistant strains during the evolution of quasispecies in NS5B.
CONCLUSION: Viral quasispecies may be an important predictor of virological responses to combination therapy in patients with chronic hepatitis C. Complex quasispecies and resistant strains may lead to high viral loads during therapy, with a concerted effect on disease severity.
- Citation: Kumar D, Malik A, Asim M, Chakravarti A, Das RH, Kar P. Influence of quasispecies on virological responses and disease severity in patients with chronic hepatitis C. World J Gastroenterol 2008; 14(5): 701-708
- URL: https://www.wjgnet.com/1007-9327/full/v14/i5/701.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.701
Hepatitis C is a noncytopathic hepatotrophic virus having a single stranded, positive sense linear -9.5 kb RNA genome. Hepatitis C virus infection is the leading cause of acute, chronic, liver cirrhosis and hepatocellular carcinoma. Chronic infection due to hepatitis C virus (HCV) occurs in 170-200 million people worldwide[12]. In India, 15%-20% of all patients with chronic liver disease and 5%-10% of hepatocellular carcinoma patients are infected with hepatitis C[3]. The HCV continues to persist, unless the patients are put on interferon therapy. Treatment with interferon-based therapy and error-prone replication of the genome via RNA-dependent RNA polymerase (RDRP) accelerates the evolution of HCV variants during the course of infection[2]. The origination of highly heterogeneous population in hepatitis C infected patients closely related to HCV genome because of interferon therapy, immune pressure and replication strategy called quasispecies[4].
The hypervariable region 1 (HVR-1) is located at the 5'-end of the E2 envelope glycoprotein, and variations occur in this region in association with persistence of viral infection[5]. Significant mutations in HVR-1 arise due to the presence of a specific anti-E2 humoral immune response. It is emerging that genetic heterogeneity extends throughout the entire HCV genome, and that this allows HCV infection to persist. Anti-HCV-specific T cell responses efficiently clear viruses during the acute phase of infection[6]. HVR quasispecies may have important biological consequences for the persistence of hepatitis C viral infection and the severity of liver disease[57].
HCV quasispecies populations result from the error rate (misincorporation frequencies averaging from about 10-4 to 10-5 per base) and the absence of proof-reading by RDRP. Most mutant particles cannot replicate, but the fittest infectious particles are selected on the basis of their replication capacities and selective pressure exerted by their interaction with host cell proteins and by the immune response[8]. Adaptive mutations in NS4B, NS5A or NS5B strongly enhance replication, but are incompatible to each other, whereas adaptive mutations in NS3 tend to be weak and cooperatively enhance replication when combined with strongly adaptive mutations[89]. The adaptive mutations in NS3 and NS5B map to surface residues distant from the enzyme active site, suggesting that these changes are likely to affect the interaction between non-structural protein and cellular factors[8].
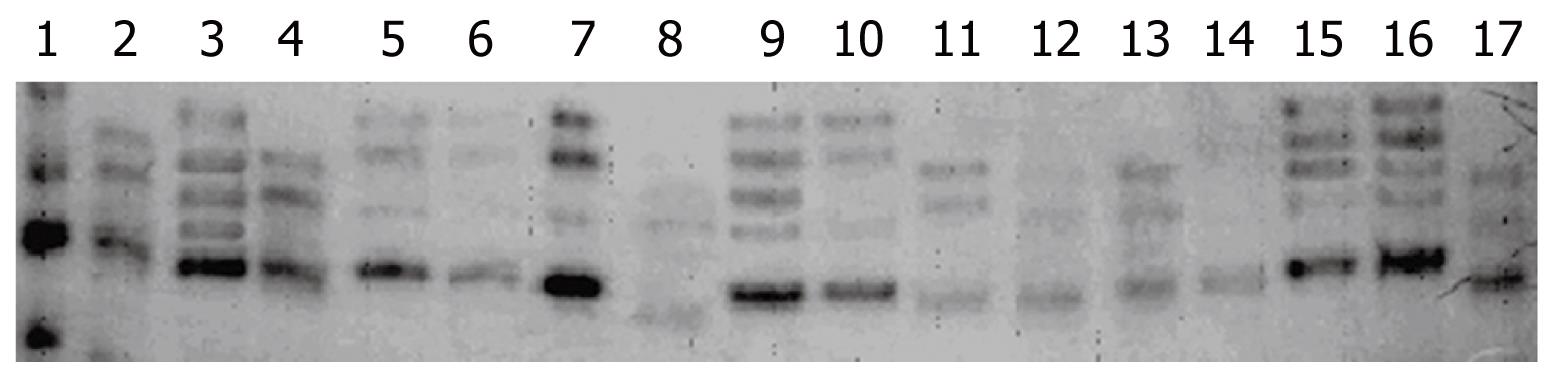
Hepatitis C viral quasispecies can be detected by cloning of reverse transcription - polymerase chain reaction products, but sequencing of 15-20 independent clones is expensive and laborious, and so is not practical for a large number of clinical samples. For large numbers of clinical samples, faster methodologies like single-strand conformational polymorphism (SSCP), heteroduplex analysis (HDA) and gel shift assay (GSA) have been developed based on the differential migration of amplified DNA in acrylamide gel electrophoresis. In the present study, we used the established and reliable method of SSCP to detect quasispecies[10–13]. The mobility of mutated DNA fragments is affected by particular changes in the single-strand DNA structure; therefore, every variant migrates in a different way.
In controlled trials on the effect of interferon and ribavirin combination therapy, the independent predictive influence of virological parameters on patients with sustained virological response (SVR) raised questions about the intrinsic properties of HCV strains, like genotype and quasispecies and their effect on sensitivity and resistance to combination therapy[14–16]. The influence of viral quasispecies with variations in the hypervariable region (HVR) and NS5A ISDR (interferon-sensitivity determining region) genes on the effectiveness of interferon has been explored world wide[1417–20]. These studies paid special attention to the importance of quasispecies as independent predictor of SVR. The cause and mechanism of HCV resistance to interferon needs to be well understood and several viral proteins may influence the therapy outcome. Gupta R et al reported HCV mutations in the 5’ untranslated region, ISDR and PePHD region, and their influence on therapy responses in a South-Indian population. However, the influence of quasispecies in the HVR and NS5B regions on the persistence of infection and disease severity in chronic hepatitis C patients needs to be explored.
This study enrolled 47 patients with chronic liver disease [32 with chronic active hepatitis (CAH), 9 with cirrhosis and 6 with hepatocellular carcinoma (HCC)] verified by histological examination, Child-Pugh score, and FNAC (fine needle aspiration cytology) wherever applicable, who attended medical outpatient departments (OPDs) and wards of Lok Nayak Hospital, New Delhi, India between 2003 to 2005, on the basis of pre designed performa, with all relevant information about possible risk factors, symptoms, clinical examination, detailed biochemical investigation and serology. They had a mean age of 48.4 ± 14.5 years, a male:female ratio of 25:22 and an alanine aminotransferase (ALT) value (mean ± SD) of 79.6 ± 44.6 (26-296) IU/L. Out of 47 patients, 34 (24 with CAH, 6 with cirrhosis, and 4 with HCC) were infected with virus strain of genotype 3, 11 (7 with CAH, 2 with cirrhosis, and 2 with HCC) with virus of genotype 1 and 2 (1 with CAH and 1 with cirrhosis) with virus of mixed genotype (1 + 3). The most probable risk factor for transmission was transfusion of infected blood and contaminated blood products, in 63.8% (30/47) of the patients. Out of 47 patients, 41 (32 with CAH and 9 with cirrhosis) were on combination therapy, 30 (73.17%) attained sustained virological responses and 11 (26.83%) were non-responders. However, the 6 HCC patients were not on therapy. The 41 chronic hepatitis C (CH-C) patients (32 with chronic active hepatitis and 9 with cirrhosis) were on medication (interferon-α2b 3 MU + 1000 mg ribavarin daily), having ALT ≥ 60 IU/L and HAI score ≥ 3 at baseline. The severity of liver disease was determined by histopathological grading according to the modified Knodell Score for the CAH patients and by Child-Pugh score for the cirrhosis patients[21–23]. These 41 patients were put on 48 wk of therapy or follow up. The viral load was estimated after 0 d, 4, 8, 12, 24 and 48 wk of the therapy. Patients without any detectable HCV viremia (≤ log10 1 IU/mL) after stopping the therapy, and even after 6 mo of therapy withdrawal, were defined as responders, and, if these patients were found to show positivity 6 mo after withdrawal of therapy, they were defined as relapsers. However, the patients with detectable HCV viremia after the end of wk 48 of therapy were defined as non-responders. Liver biopsies of CAH patients and Child-Pugh scoring in cirrhosis patients were performed at baseline, and after 24 and 48 wk of therapy. The SSCP analysis of the HVR and NS5B regions was performed in all sera samples before the initiation of treatment and after 24 wk of therapy. Genotypes were also studied by RFLP for genotypic classification of these patients (data not shown). Patients with > 4 SSCP bands were considered to have complex quasispecies (CQS) of HCV. However, the patients with 3 or 4 bands in the SSCP gel were considered to have simple quasispecies (SQS) and those with 2 bands were considered to have no quasispecies (NQS). The patients were grouped based on the number of bands in the SSCP gel (Figure 1) to study the influence of quasispecies on virological responses and disease severity. RNA extracted from sera samples (11 non-responders and 12 responders) was amplified by RT-PCR using primers specific for the HVR and NS5B genes, and the products were sequenced (Macrogen Inc, Korea) on a commercial basis at three different time (0 d, 24 wk and 48 wk) intervals during therapy in non-responders and two times (0 d and last sample before the clearance of HCV infection) in responders. Sequencing results at baseline were submitted to the GenBank database and also used for phylogeny. A phylogenetic tree was drawn to analyze the distance matrix between the SVR and non-responder (NR) patients on the basis of sequences derived from the HVR and NS5B genes, using the Bio Edit program for sequence analysis.
Biochemical investigations, including routine and specific liver function tests, were carried out in all cases. Serological tests were performed using commercially available ELISA kits, according to the manufacturer’s instructions. Sera samples of all 47 patients were screened for HBsAg using a 3rd generation ELISA kit from Biokit, Spain; IgG HBc and HBeAg were screened using an ELISA kit from RADIM SpA, Italy; and anti-HCV antibody using a 3rd generation ELISA kit (General Biological Corporation, Taiwan). Exclusively anti-HCV-positive patients’ sera were used for the detection of HCV RNA.
Total RNA was isolated using Tri Reagent (Sigma-Aldrich) according to the manufacturer’s instructions and amplified by reverse transcription-PCR. For reverse transcription, 5 &mgr;L of extracted template RNA was added to 20 &mgr;L of reaction master mix containing reaction buffer, dNTPs, RNAsin (10 U), M-MuLV reverse transcriptase (40 units) and outer antisense primer, and incubated at 42°C for one hour for cDNA synthesis. First-round PCR amplification was carried out using outer sense and antisense primers, 2 &mgr;L of cDNA template, reaction buffer containing magnesium, dNTPs, and Taq DNA polymerase (1 unit), in a 25-&mgr;L reaction, by denaturing at 94°C for 5 min and 35 cycles with 60 s each of denaturation at 94°C, annealing at 55°C and extension at 72°C, in a DNA thermal cycler (M.J. Research, USA). After the last cycle, the extension was prolonged for 8 min at 72°C. Second-round PCR was performed using 2 &mgr;L of the first PCR product as template, which was amplified with the second PCR mix containing inner primers (Table 1). The reagent composition and temperature profile were the same as those for the first PCR. Both positive and negative controls were run in parallel with the test samples. The expected PCR products for HVR and NS5B were 403 and 293 bp, respectively; these were detected in ethidium bromide-stained 30 g/L Nusieve agarose gels in a UV Transilluminator.
| Region/Nucleotide position | Primer set | Forward/Reverse | Sequence (5’-3’) | Amplicon size (bp) | |
| HVR | Outer | Forward | GTGGTTTGGATGTGTGTGGATGAAC | ||
| 2040-2443 | Reverse | TATTGCACGTCCACGATGTTCTG | 403 | ||
| Inner | Forward | GGCTCCCCCTTGCAACATCT | |||
| Reverse | GTGGAGGTGTATGAGCCCTGTTG | ||||
| NS5B | Outer | Forward | TGGGGATCCCGTATGATACCCGCTGCTTTGA | ||
| 8301-8593 | Reverse | GGCGGAAT (T/A) CCTGGTCATAGCTCCGTGAA | 293 | ||
| Inner | Forward | CTCAAC (C/T) GTCACTGAACAGGACAT | |||
| Reverse | CCACGAC (T/C) AGATC (A/G) TCTCCG |
Briefly, single-strand conformational polymorphism (SSCP) was performed by radiolabelling the PCR products for an additional 10 cycles where dCTP was replaced with (α-p32) dCTP (specific activity, 4000 CI/nmol) in the PCR mix. One &mgr;L of radiolabelled PCR product was diluted with 9 volumes (10 times) of denaturing solution (950 g/L formamide, 20 mmol/L EDTA pH 8.0, 0.5 g/L xylene cyanol and 0.5 g/L bromophenol blue), heat-denatured for 5 min at 95°C, and chilled on ice for 3 min; 3 &mgr;L of this diluted radiolabel product was subjected to non-denaturing electrophoresis in a 60 g/L polyacrylamide gel containing 100 mL/L glycerol. Gels were run in 1 × TBE for 16 h at 250 V in a DNA sequencing apparatus, at 17 ± 1°C, dried on a gel drier, and exposed to X-ray film with an intensifying screen at -20°C. Electrophoretic mobility shifts of single-strand DNA bands were analyzed following development of the X-ray film.
Total RNA was used for real time quantification. The RealArt HCV PCR Reagents constitute a ready-to use system for PCR amplification and the detection of HCV RNA on the Light Cycler instrument (Rotor Gene 3000, Corbett Research Inc.). The RealArt HCV PCR master mixture contains reagents and enzymes for the amplification of a 240-bp region of the HCV genome with the parallel detection (A.Ch 1-FAM) and quantification of the amplified PCR products in comparison to five standard samples diluted 10-fold, which were used to generate the standard curve.
Statistical analysis was performed using the Student’s t-test, ANOVA, Chi-square, and Fischer’s exact test, wherever applicable, using the software EPISTAT and SPSS. Statistical analysis was performed by comparisons of two groups descriptively. Patients were classified as being infected with complex quasispecies (CQS), simple quasispecies (SQS) and no quasispecies (NQS) on the basis of their SSCP pattern, representing the level of HCV quasispecies variations. The results are expressed as means ± SD. P < 0.05 was considered significant.
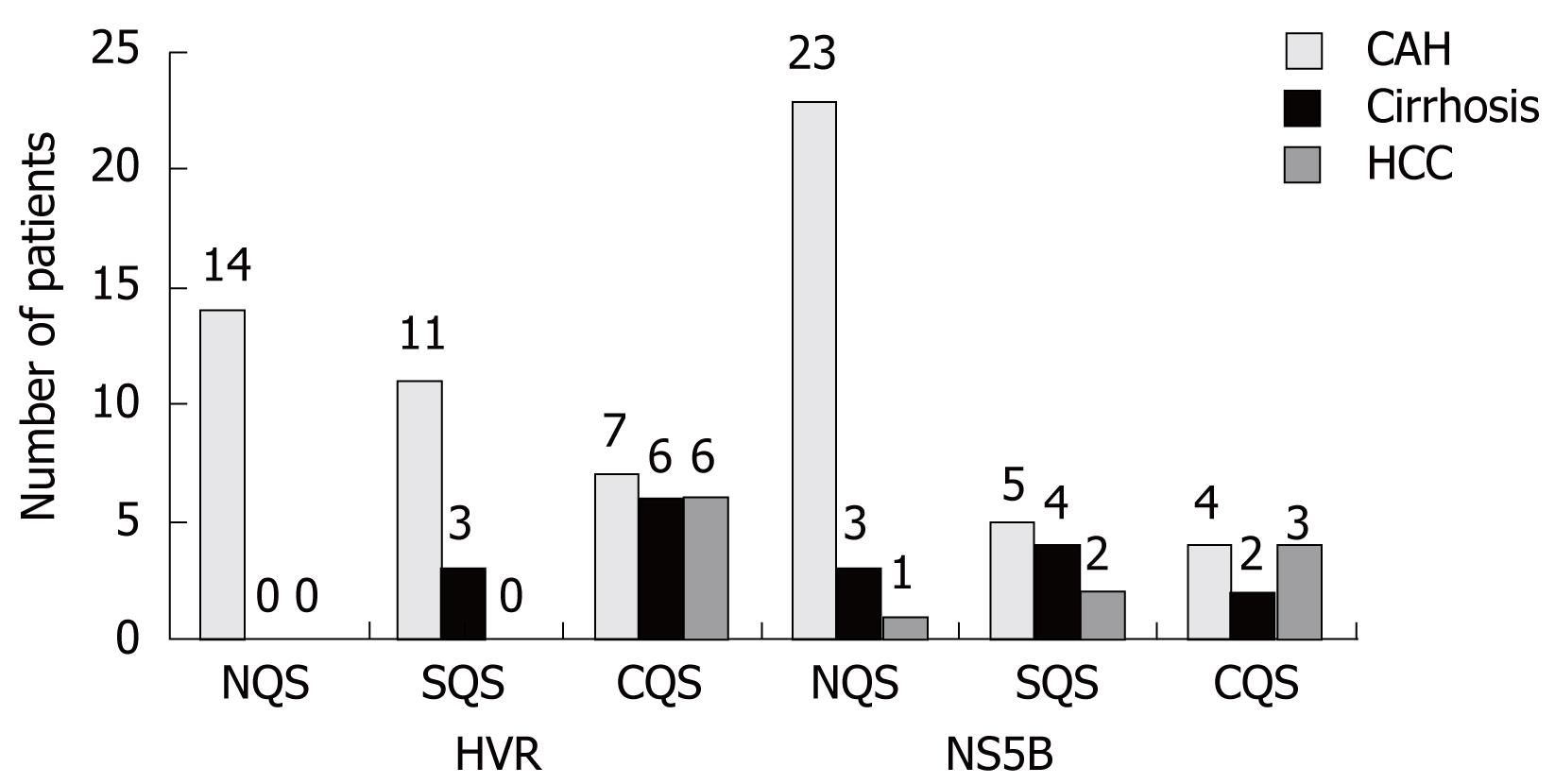
Patients were screened for the presence of quasispecies by SSCP analysis, on the basis of differential migration in PAGE denaturing gels. The appearance of more than 2 bands in gels is associated with the presence of quasispecies. Patients with more than 4 bands in SSCP gels were infected with complex quasispecies (CQS). Complex quasispecies were detected both in the NS5B and HVR regions of HCV, but were relatively more abundant in the HVR (n = 19) than the NS5B (n = 9) region. Among the 32 patients with chronic active hepatitis, 18 (56.3%) showed presence of quasispecies (CQS-7, SQS-11, NQS-14) when the HVR gene was used for the analysis, while 9 (28.2%) showed presence of quasispecies (CQS-4, SQS-5, NQS-23) when the NS5B gene was used for the analysis. All 9 cirrhosis patients showed presence of quasispecies (CQS-6, SQS-3) when the HVR gene was used for the analysis, while only 6 (66.7%) showed presence of quasispecies (CQS-2, SQS-4, NQS-3) when the NS5B gene was used (Figure 2). It was interesting to note that all 6 HCC patients were positive for complex quasispecies when the HVR was used for the analysis, while 5 out of the 6 (83.3%) showed presence of quasispecies (CQS-3, SQS-2, NQS-1) on the basis of the NS5B gene (Figure 2). These results showed the influence of the occurrence of HVR quasispecies on the disease severity in hepatitis C infected patients as all cirrhosis and HCC patients showed evolution of quasispecies in this region. However, NS5B quasispecies also showed their influence on disease severity. No difference was observed in the SSCP band patterns of the patients after 24 wk of therapy, confirming the accuracy of the PCR reaction and SSCP profiles. The genotypic classification of quasispecies was not feasible from this data due to sample size. However, the occurrence of quasispecies in both the genotypes (genotypes 1 and 3) was observed.
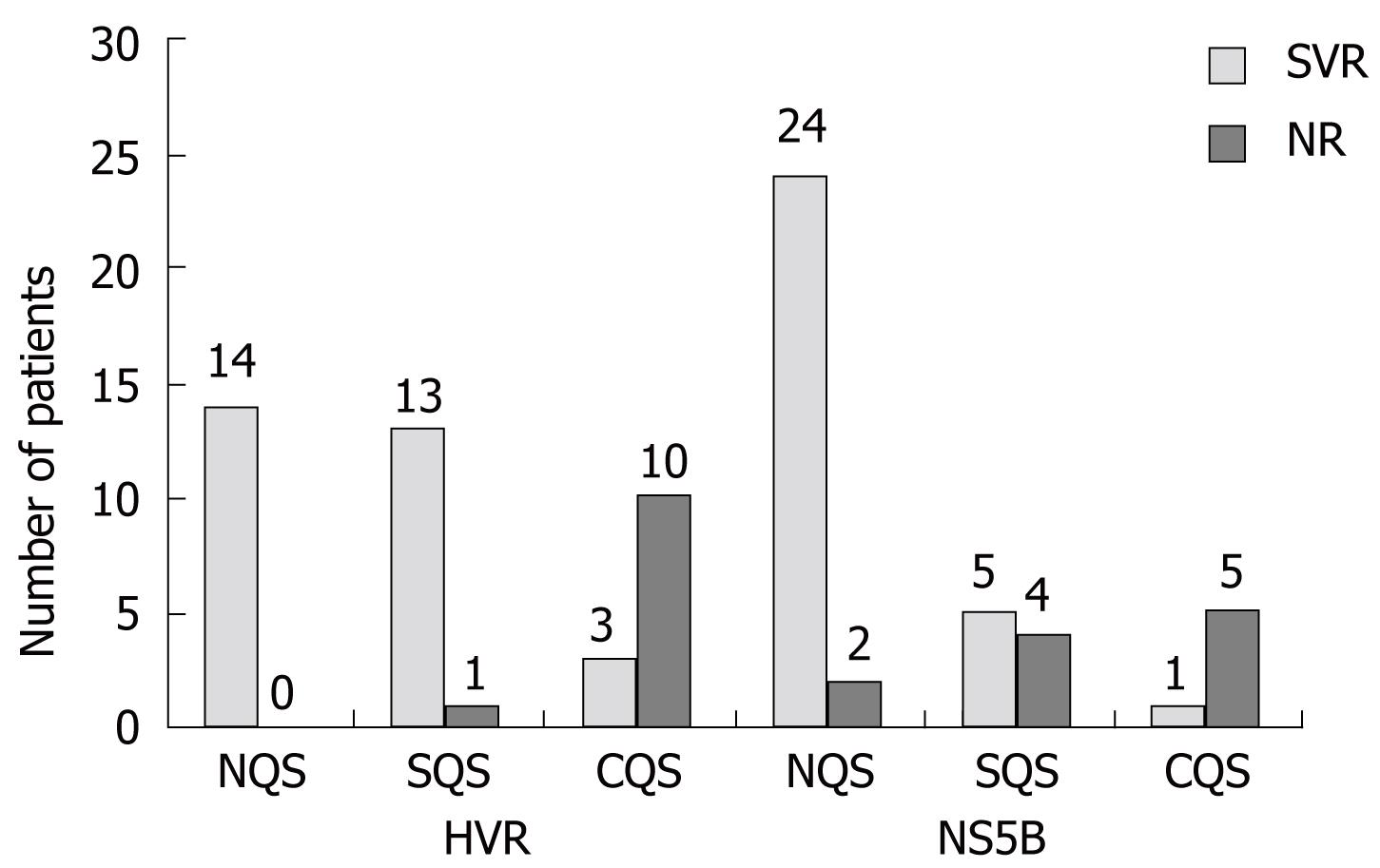
Among the 41 patients on therapy, 30 had sustained virological responses, whereas 11 were non-responders. HVR quasispecies were detected in 27 out of 41 (65.9%) patients (CQS-13, SQS-14, NQS-14), while NS5B quasispecies were detected in 15 out of 41 (36.6%) (CQS-6, SQS-9, NQS-26) (Figure 3). Out of the 11 non-responders, 10 were infected with complex quasispecies (> 4 bands in the SSCP) and 1 was infected with simple quasispecies, based on analysis of the HVR gene; when the NS5B gene was analyzed, among the 11 non-responders, 5 were infected with complex quasispecies, 4 with simple quasispecies and 2 with no quasispecies. Among the 30 SVR patients, 16 (53.3%) were infected with HVR quasispecies (CQS-3, SQS-13, NQS-14), whereas 6 (20.0%) were infected with NS5B quasispecies (CQS-1, SQS-5, NQS-24) (Figure 3). These results suggest that the presence of complex quasispecies with variations in the HVR gene has an impact on virological response, as 90.9% (10/11) of patients infected with complex quasispecies with variations in this region did not respond to combination therapy, whereas only 10.0% (3/30) of patients infected with complex quasispecies were responders.
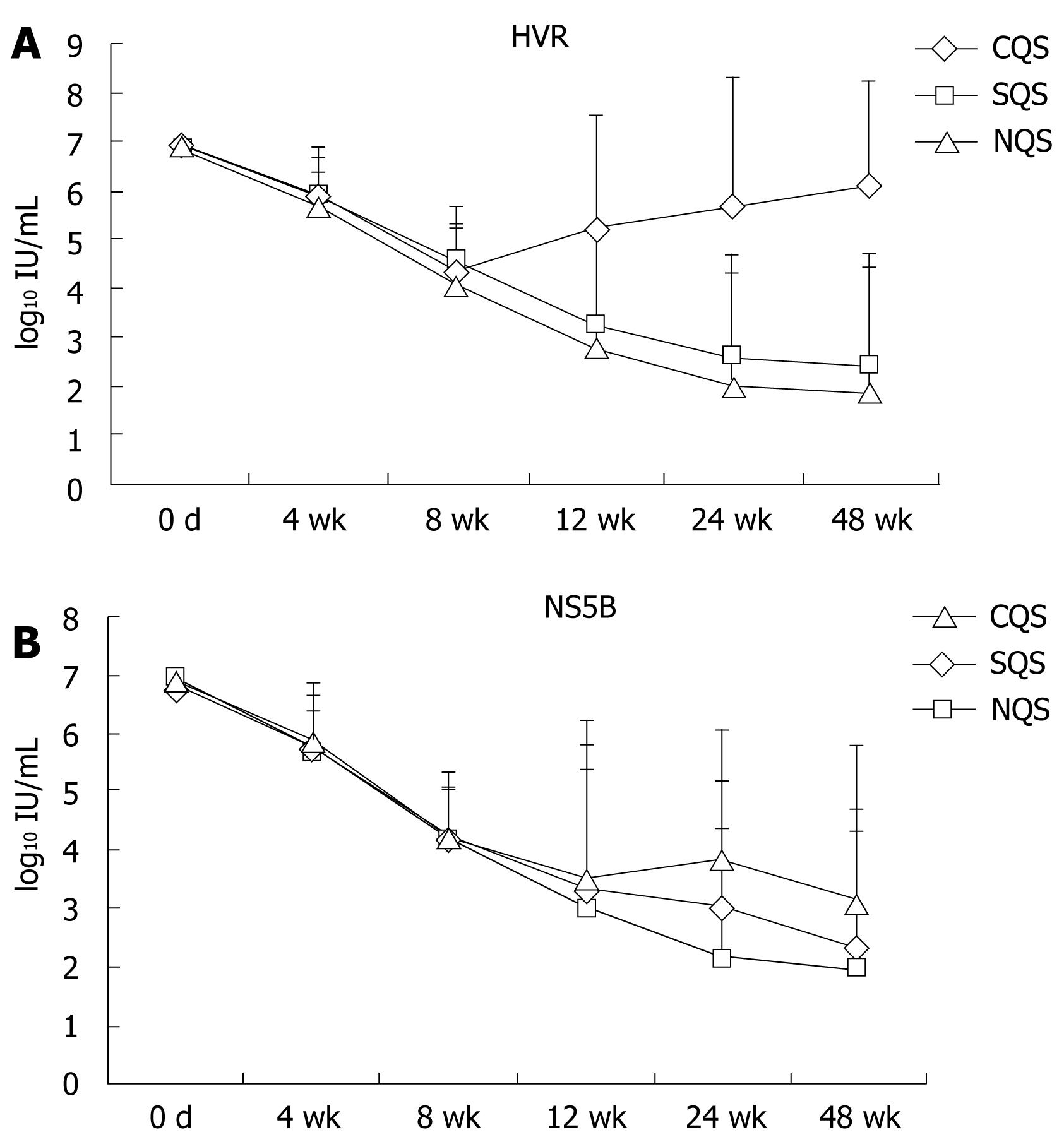
Out of 41 patients on therapy, HVR quasispecies were seen in 27 patients. No statistical difference was observed in viral load of patients with CQS, SQS and NQS (mean ± SD log10 IU/mL) from baseline (CQS-6.9 ± 0.29, SQS-6.8 ± 0.26, and NQS-6.9 ± 0.22) to 8 wk (CQS-4.3 ± 1.05, SQS-4.5 ± 1.2, NQS-4.1 ± 1.1) of combination therapy with minimum standard error. A statistically significant difference in the viral load (P < 0.05; CQS vs SQS, NQS) was observed after 12 wk (CQS-5.2 ± 2.3, SQS-3.2 ± 1.9, NQS-2.8 ± 2.4) of therapy in patients with HVR region quasispecies of HCV. A statistically significant difference in viral load (P < 0.05) with high standard error was observed in comparisons of CQS with SQS and NQS after 24 wk (CQS-5.7 ± 2.6, SQS-2.6 ± 2.1, NQS-2.0 ± 2.33) and 48 wk (CQS-6.1 ± 2.1, SQS-2.4 ± 2.3, NQS-1.9 ± 2.5) of therapy (Figure 4A). No significant difference (P > 0.05) was observed in the mean viral load (log10 IU/mL) from baseline (CQS-6.9 ± 0.29, SQS-6.8 ± 0.3, NQS-6.94 ± 0.22) to 12 wk (CQS-3.5 ± 2.7, SQS-3.3 ± 2.5, NQS-3 ± 2.4) in comparisons of patients with different levels quasispecies (CQS, SQS and NQS) in NS5B region. However, the standard error was almost negligible at baseline, but an increased standard error in viral load was observed after 12 wk of therapy. A statistically significant difference (P < 0.05) with high standard error was observed between the mean viral loads of patients infected with CQS and those infected with NQS after 24 wk (CQS-3.9 ± 2.2, SQS-3.0 ± 2.2, and NQS-2.1 ± 2.3) and 48 wk (CQS-3.1 ± 2.7, SQS-2.3 ± 2.4, NQS-2.0 ± 2.3) of therapy (Figure 4B). A continuous decline in mean viral load was observed in patients without complex quasispecies infection, suggestive of an influence of the presence of quasispecies on viral kinetics and therapeutic outcome during combination therapy.
The histological activity index (HAI) score is an important factor for the assessment of disease severity in CAH patients. It was interesting to note that mean HAI scoring (mean ± SD) was comparable among groups based on the degree of quasispecies diversity in the HVR at baseline (CQS-6.64 ± 2.9, SQS-6.72 ± 1.2, NQS-6.9 ± 0.22). Patients grouped on the basis of genetic variability in the NS5B gene showed a statistically significant difference (CQS vs NQS) in the mean HAI scores at baseline (CQS-7.6 ± 3.3, SQS-6.7 ± 2.2, NQS-5.86 ± 2.08). An improvement in the severity of disease was observed in 80.0% (24/30) of the SVR (sustained virological response) patients. Bridging of fibrosis was observed in all NR (non-responders) and 13.3% (4/30) of the SVR patients. No statistical significant difference in the results of liver function tests was observed among patients with different degrees of HVR and NS5B quasispecies, at baseline and on follow-up every 4 wk until 48 wk.
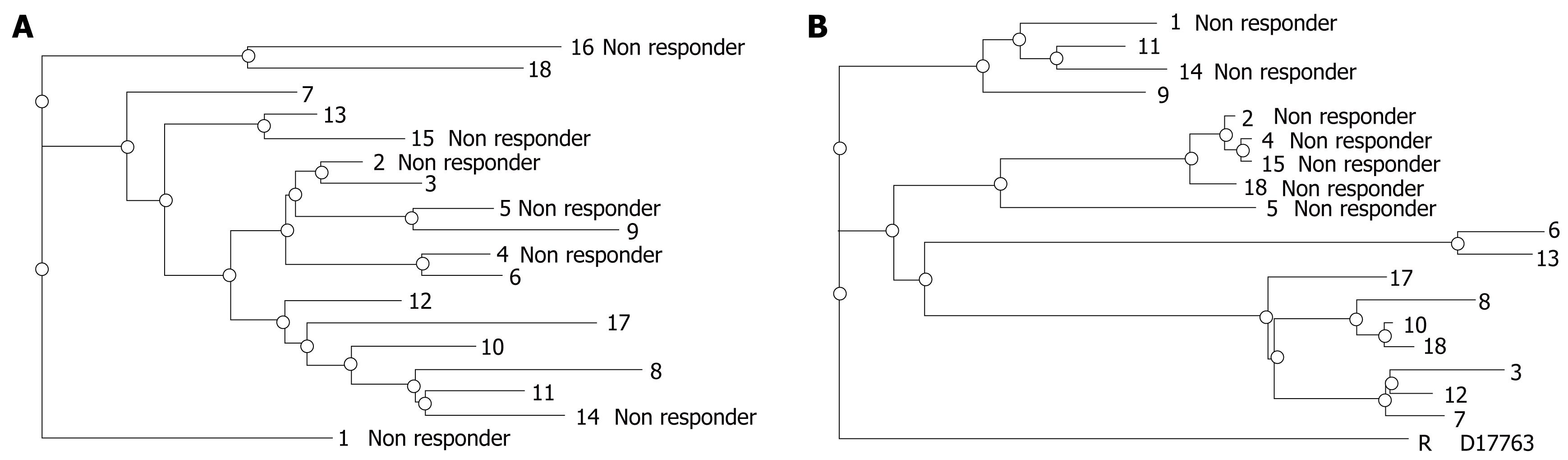
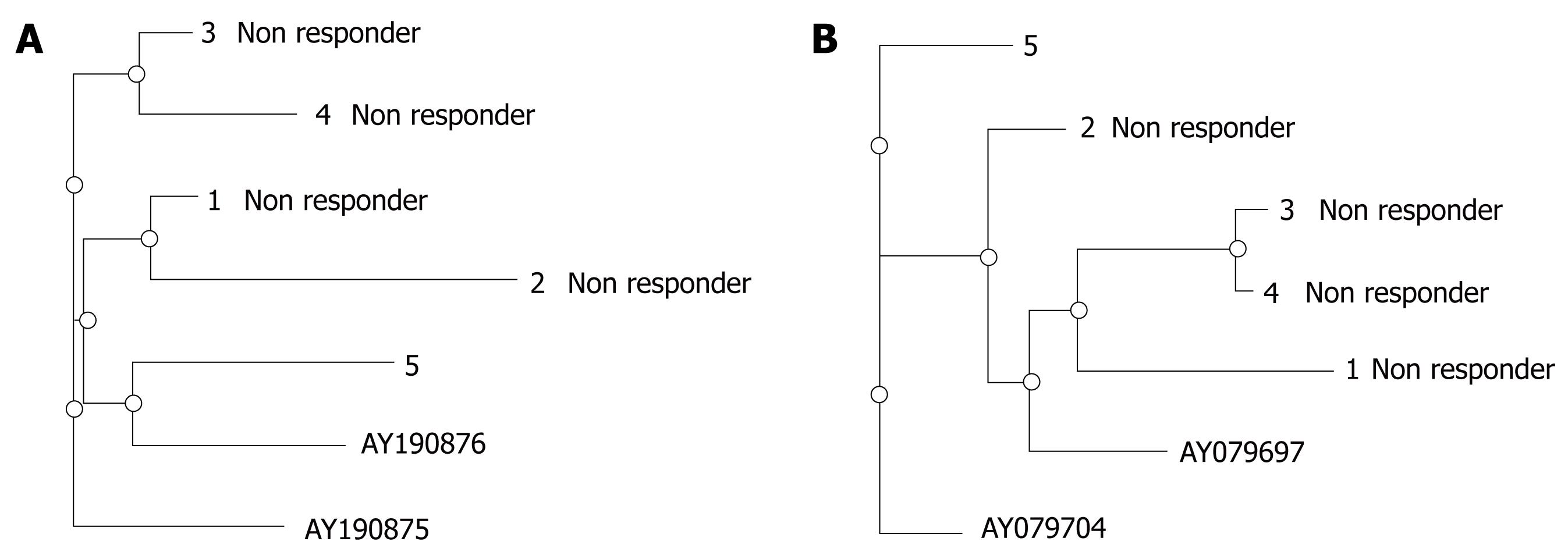
Amino acid variation in the NS5B gene plays a major role in the replication of the virus and may be important for the virological response during combination therapy. It was interesting to note that the number of mutations was higher in genotype 1 as compared to genotype 3 in the HVR region, suggesting an influence of genotype on the rate of mutations in the selected region of HCV and on the evolution of quasispecies. Amino acid variations in the HVR and NS5B regions of different genotypes among patients (n = 23, Genotype 1 = 5, Genotype 3 = 18) during different phases of therapy are suggestive of the occurrence of mutations during the course of therapy. The sequences of virus strains during the course of therapy showed 2-5 amino acid substitutions and synonymous mutations at the nucleotide level in both HVR and NS5B regions, compared with the sequences of virus strains before therapy (data not shown as no specific mutation was observed to be responsible for the persistence of infection during therapy). Phylogenetic analysis of sequenced virus isolated from patient samples showed different distance patterns between responders and non-responders in HVR and NS5B regions, as shown in Figures 5 and 6. The non-responders were on a different branch, suggestive of no close pairing between these non-responder strains of genotype 3 in the HVR region. However, closing pairing was observed between non-responder strains of genotype 3 in the NS5B region: 5 non responder patients were on the same branch and 3 patients were on the other branch. However, all four non-responder were on the same branch in patients of genotype 1. Strains appearing on the same branch (close pairing) during phylogenetic tree analysis possess significant homology in their nucleotide sequences. These results suggest that either the selection of resistant strains or the development of quasispecies may be the cause of unresponsiveness to combination therapy in HCV patients; however, these mechanisms require further analysis.
Diversifications in the HCV genome are the results of viral RNA polymerase infidelity, immune-driven selection and exertion of therapy pressure or a combination of these processes. PCR-SSCP analysis is a well established technique to study genetic variability, including the presence of mutations and quasispecies[10–13]. Millions of viruses are produced via replication and eliminated each day due to exertion of immune pressure and interferon therapy[16]. Ribavirin does not have any effect on the viremia level of HCV-infected patients, but it reduces the ALT level, increases the likelihood of sustained virological response and reduces the probability of relapse[16].
The HVR and NS5B genes were selected in the present study because the persistence of infection may be associated with positive selection of HVR quasispecies by the host immune system and because of the major role of NS5B in the replication of hepatitis C. Factors like age, sex and duration of infection may be possible causes of the occurrence of quasispecies; in this study, we studied virological responses in patients infected with quasispecies before the initiation of therapy and looked for alterations in the SSCP patterns during therapy. Several conflicting reports about a possible correlation of therapy response and disease severity with the presence of quasispecies have been published[152024–27]. The genes encoding HVR protein exhibit a high degree of variability, giving rise to differing phenotypic traits, including alterations in receptor-binding affinity and escape from immune recognition[28]. Substitutions in the NS5B gene of HCV may correlate with the virological response to interferon-based therapy[29–31].
The occurrence of quasispecies was proportionately higher in patients with HCC and cirrhosis than in CAH patients, revealing a significant correlation between the molecular evolution of quasispecies and the severity of disease in hepatitis C patients in this study. The genetic diversity at baseline can be predictive of distinct virological behavior during the course of combination therapy. Among the 13 patients infected with complex quasispecies with variations in the HVR gene, 10 were non-responders at baseline. This correlates significantly with the persistent of infection as the response rate against interferon may be due to two mutually compatible approaches; the infecting virus might have become naturally resistant to interferon or the virus could have evolved into quasispecies. The non-responders had normal signal transduction pathways required for the intracellular activity of interferon[32]. The recognition of the HVR epitope (envelope) by immune cells and the exertion of interferon-based therapy play a major role in the evolution of quasispecies.
Several studies have shown an association of epidemiological factors and treatment regime with the presence of quasispecies[13141933]. Complex quasispecies with variations in the HVR and NS5B genes of HCV were significantly associated with virological response during wk 48 of therapy. Virus quasispecies evolution within patients increases the pathogenicity of HCV. The resulting viral quasispecies are heterogeneous and sensitive to selection pressures. Complex variants escape from immune pressure and the antiviral spectrum of the combination therapy[34]. No mutant strains have the capacity for replication, but only selective mutants can replicate, which may be responsible for persistence of infection. The circulating viremia in blood and liver cells is significantly associated with disease prognosis[35]. There was no statistically significance difference in the HAI scores among patients with different degrees of HVR quasispecies at baseline. However, there was a significant difference in the HAI scores at baseline between patients infected with different genetic variants (CQS vs NQS) of the NS5B gene. Selective adaptive mutations in the resistant strains occurring during replication influence the RDRP activity and lead to increases in viremia level and HAI scoring[7–9]. These results suggest the significance of evolution of quasispecies on virological responses and disease severity.
The amino acid variability in HVR and NS5B was evaluated from cDNA-derived sequences. The occurrence of HVR quasispecies influences the therapeutic outcome; because no strains isolated from non-responder patients showed close pairing, it is possible that some other factors may be responsible for the persistence of infection. However, the possibility of resistant strains can not be denied. The NS5B sequences of non-responder strains were on the same branch, showing the possibility of resistant strain infection in these patients. However, adaptive mutations in NS5B lead to persistence of infection and a severe course of liver disease in patients. These findings reveal that the occurrence of HVR quasispecies, including the presence of resistant strains, is a major factor influencing the therapeutic outcome and disease severity in chronic hepatitis C patients. Genetic variability was not uniformly distributed throughout the HCV genome.
In conclusion, viral quasispecies heterogeneity can be an important marker to predict the response of combination therapy in patients with chronic hepatitis C. Quasispecies complexity on the basis of SSCP pattern could be helpful in differentiation of SVR and NR patients. Complex quasispecies heterogeneity causes high HCV viremia, which was significantly correlated with the severity of liver disease and prognosis during the course of combination therapy. However, there was no significant alteration in SSCP profile during therapy. The occurrence of resistant strains may be another factor contributing to the persistence of HCV infection. Quasispecies in both HVR and NS5B genes may have a major impact on treatment and be a good predictor of virological response. Complex quasispecies are resistant to combination therapy and influence disease prognosis during therapy. Further evaluation of viral quasispecies complexity in the HVR and NS5B genes, as well as other HCV genes (whole genome), is required for a better understanding of the influence of quasispecies on the persistence of infection and disease prognosis during the course of therapy in HCV-infected patients.
The hepatitis C virus (HCV) continues to persist unless patients are put on interferon therapy. Treatment with interferon-based therapy and error prone replication of the viral genome via RNA-dependent RNA polymerase (RDRP) accelerates the evolution of HCV variants during the course of infection. The highly heterogeneous population in HCV-infected patients, consisting of quasispecies, developed because of interferon therapy, immune pressure and replication strategy. Quasispecies are one of the major virological factors other than genotypes that can influence the response to therapy in chronic hepatitis C patients; their role in this needs to be evaluated.
HCV quasispecies can be detected by cloning of reverse transcription-polymerase chain reaction products; however, sequencing of 15-20 independent clones is expensive and laborious, and not practical for a large number of clinical samples. For large numbers of clinical samples, faster methodologies like single-strand conformational polymorphism (SSCP), heteroduplex analysis (HDA) and gel shift assay (GSA) have been developed based on the differential migration of amplified DNA in acrylamide gel electrophoresis. In the present study, we used the established and reliable method of SSCP for the detection of quasispecies with variations in the HVR and NS5B genes. The patients were grouped on the basis of the number of bands in the SSCP gel. Patients with > 4 SSCP bands were considered to be infected with complex quasispecies (CQS) of HCV. Patients with 3 or 4 bands were considered to be infected with simple quasispecies (SQS); and those with 2 bands were considered to be infected with no quasispecies (NQS). Our interest was to evaluate whether or not the occurrence of quasispecies at baseline influences the virological response and disease severity in chronic hepatitis C patients.
The significance of disease severity on the occurrence of HVR quasispecies in serum of hepatitis C infected patients was observed as all of the cirrhosis and HCC patients showed evolution of quasispecies in this region. However, a major role of NS5B quasispecies was also observed with regard to disease severity. No difference was observed in the SSCP patterns of patients after 24 wk of therapy, confirming the validity of the PCR reaction and SSCP profiles. These results suggest an effect of complex quasispecies with variations in the HVR gene on virological response, as 90.9% (10/11) of patients infected with complex quasispecies with variations in this region did not respond to combination therapy, whereas only 10.0% (3/30) patients infected with complex quasispecies showed SVR. A continuous decline of mean viral load was observed in the patients without complex quasispecies viral infection, suggestive of an influence of quasispecies on viral kinetics and therapeutic outcome during combination therapy.
The study suggests that quasispecies complexity in both HVR and NS5B genes had a major impact on treatment and is a good predictor of virological responses. However, the existence of quasispecies based on other HCV genes should be further evaluated for a better understanding of the influence of quasispecies on virological responses and disease severity in patients with chronic hepatitis C.
Chronic active hepatitis (CAH): anorexia, weight loss, jaundice, ascites, edema present singly or in combination, persistently or intermittently and biochemical features (SGOT, SGPT more than twice normal and globulin 1.5 times normal) of hepatocellular disease for more than 6 months duration. Liver biopsy is necessary for the assessment of disease severity. A diagnosis of cirrhosis of the liver was considered on the basis of the following criteria: ascites, splenomegaly, shrunken liver, endoscopy showing esophageal varices and biochemical features of hepatocellular failure characterized by low serum albumin level (< 2.5 gm). Hepatocellular Carcinoma (HCC): patients were evaluated on the basis of history, clinical examination, liver function tests and serological tests for hepatitis B & C, serum alpha-feto protein levels and liver imaging [ultrasound sonography (US), computed tomography (CT), magnetic resonance imaging (MRI)], and final diagnosis of HCC was made on FNAC (fine needle aspiration cytology). Quasispecies: The origination of a highly heterogeneous population in hepatitis C-infected patients as a result of interferon therapy, immune pressure and replication strategies.
In this study the authors assessed whether the occurrence of HCV quasispecies influence responses to antiviral therapy and the severity of chronic hepatitis C. Quasispecies were assessed on the basis of SSCP patterns. The results are interesting and suggest that complex quasispecies are a major factor in non-response to combination therapy. The SSCP pattern remains unaltered during the course of therapy. The concept of this study is interesting; evaluation of other HCV genes should be performed in future work.
| 1. | Kim WR. The burden of hepatitis C in the United States. Hepatology. 2002;36:S30-S34. [Cited in This Article: ] |
| 2. | Watanabe K, Yoshioka K, Yano M, Ishigami M, Ukai K, Ito H, Miyata F, Mizutani T, Goto H. Mutations in the nonstructural region 5B of hepatitis C virus genotype 1b: their relation to viral load, response to interferon, and the nonstructural region 5A. J Med Virol. 2005;75:504-512. [Cited in This Article: ] |
| 3. | Panigrahi AK, Panda SK, Dixit RK, Rao KV, Acharya SK, Dasarathy S, Nanu A. Magnitude of hepatitis C virus infection in India: prevalence in healthy blood donors, acute and chronic liver diseases. J Med Virol. 1997;51:167-174. [Cited in This Article: ] |
| 4. | Durante Mangoni E, Forton DM, Ruggiero G, Karayiannis P. Hepatitis C virus E2 and NS5A region variability during sequential treatment with two interferon-alpha preparations. J Med Virol. 2003;70:62-73. [Cited in This Article: ] |
| 5. | Martell M, Esteban JI, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225-3229. [Cited in This Article: ] |
| 6. | Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, Coiana A, Peddis G, Usai F, Serra G, Chessa L. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci USA. 2002;99:3081-3086. [Cited in This Article: ] |
| 7. | Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967-972. [Cited in This Article: ] |
| 8. | Bartenschlager R, Frese M, Pietschmann T. Novel insights into hepatitis C virus replication and persistence. Adv Virus Res. 2004;63:71-180. [Cited in This Article: ] |
| 9. | Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946-952. [Cited in This Article: ] |
| 10. | Gomez J, Martell M, Quer J, Cabot B, Esteban JI. Hepatitis C viral quasispecies. J Viral Hepat. 1999;6:3-16. [Cited in This Article: ] |
| 11. | Laskus T, Wilkinson J, Gallegos-Orozco JF, Radkowski M, Adair DM, Nowicki M, Operskalski E, Buskell Z, Seeff LB, Vargas H. Analysis of hepatitis C virus quasispecies transmission and evolution in patients infected through blood transfusion. Gastroenterology. 2004;127:764-776. [Cited in This Article: ] |
| 12. | Takeda T, Tatsumi N, Nakayama Y, Yasuda T, Nishiguchi S, Seki S. Mode of HCV infection examined by polymorphism of hypervariable region-1 in cases of acute hepatitis C after accidental exposure to blood-borne pathogens. J Med Virol. 2005;75:35-41. [Cited in This Article: ] |
| 13. | Chen S, Wang YM. Multigene tracking of quasispecies in viral persistence and clearance of hepatitis C virus. World J Gastroenterol. 2005;11:2874-2884. [Cited in This Article: ] |
| 14. | Vuillermoz I, Khattab E, Sablon E, Ottevaere I, Durantel D, Vieux C, Trepo C, Zoulim F. Genetic variability of hepatitis C virus in chronically infected patients with viral breakthrough during interferon-ribavirin therapy. J Med Virol. 2004;74:41-53. [Cited in This Article: ] |
| 15. | Tao Q, Wei L, Chang J, Wang H, Sun Y. Relationship between interferon therapy and variability in nonstructural gene 5b of hepatitis C virus. J Gastroenterol. 1998;33:684-693. [Cited in This Article: ] |
| 16. | Davis GL, Esteban-Mur R, Rustgi V, Hoefs J, Gordon SC, Trepo C, Shiffman ML, Zeuzem S, Craxi A, Ling MH. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493-1499. [Cited in This Article: ] |
| 17. | Gerotto M, Sullivan DG, Polyak SJ, Chemello L, Cavalletto L, Pontisso P, Alberti A, Gretch DR. Effect of retreatment with interferon alone or interferon plus ribavirin on hepatitis C virus quasispecies diversification in nonresponder patients with chronic hepatitis C. J Virol. 1999;73:7241-7247. [Cited in This Article: ] |
| 18. | Fan X, Di Bisceglie AM. Genetic complexity and serum reactivity of HVR1 quasispecies of hepatitis C virus in patients with cirrhosis. Am J Gastroenterol. 2002;97:1489-1495. [Cited in This Article: ] |
| 19. | Chambers TJ, Fan X, Droll DA, Hembrador E, Slater T, Nickells MW, Dustin LB, Dibisceglie AM. Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection. J Virol. 2005;79:3071-3083. [Cited in This Article: ] |
| 20. | Gupta R, Subramani M, Khaja MN, Madhavi C, Roy S, Habibullah CM, Das S. Analysis of mutations within the 5' untranslated region, interferon sensitivity region, and PePHD region as a function of response to interferon therapy in hepatitis C virus-infected patients in India. J Clin Microbiol. 2006;44:709-715. [Cited in This Article: ] |
| 21. | Knodell RG, Ishak KG, Black WC, Chen TS, Craig R, Kaplowitz N, Kiernan TW, Wollman J. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. 1981;1:431-435. [Cited in This Article: ] |
| 22. | Child CG, Turcotte JG. Surgery and portal hypertension. Major Probl Clin Surg. 1964;1:1-85. [Cited in This Article: ] |
| 23. | Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60:646-649. [Cited in This Article: ] |
| 24. | Thelu MA, Baud M, Leroy V, Seigneurin JM, Zarski JP. Dynamics of viral quasispecies during interferon therapy in non responder chronic hepatitis C patients. J Clin Virol. 2001;22:125-131. [Cited in This Article: ] |
| 25. | Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339-344. [Cited in This Article: ] |
| 26. | Zeuzem S. Heterogeneous virologic response rates to interferon-based therapy in patients with chronic hepatitis C: who responds less well? Ann Intern Med. 2004;140:370-381. [Cited in This Article: ] |
| 27. | Abbate I, Lo Iacono O, Di Stefano R, Cappiello G, Girardi E, Longo R, Ferraro D, Antonucci G, Di Marco V, Solmone M. HVR-1 quasispecies modifications occur early and are correlated to initial but not sustained response in HCV-infected patients treated with pegylated- or standard-interferon and ribavirin. J Hepatol. 2004;40:831-836. [Cited in This Article: ] |
| 28. | Brown RJ, Juttla VS, Tarr AW, Finnis R, Irving WL, Hemsley S, Flower DR, Borrow P, Ball JK. Evolutionary dynamics of hepatitis C virus envelope genes during chronic infection. J Gen Virol. 2005;86:1931-1942. [Cited in This Article: ] |
| 29. | Nelson DR, Gray AH, Kolberg JA, Joh J, Urdea MS, Mizokami M, Davis GL, Lau JY. Variations of hepatitis C virus NS5B sequence (nucleotides 8261-8566) do not correlate with response to interferon-alpha therapy. J Viral Hepat. 1995;2:285-292. [Cited in This Article: ] |
| 30. | Duffy M, Salemi M, Sheehy N, Vandamme AM, Hegarty J, Curry M, Nolan N, Kelleher D, McKiernan S, Hall WW. Comparative rates of nucleotide sequence variation in the hypervariable region of E1/E2 and the NS5b region of hepatitis C virus in patients with a spectrum of liver disease resulting from a common source of infection. Virology. 2002;301:354-364. [Cited in This Article: ] |
| 31. | Hamano K, Sakamoto N, Enomoto N, Izumi N, Asahina Y, Kurosaki M, Ueda E, Tanabe Y, Maekawa S, Itakura J. Mutations in the NS5B region of the hepatitis C virus genome correlate with clinical outcomes of interferon-alpha plus ribavirin combination therapy. J Gastroenterol Hepatol. 2005;20:1401-1409. [Cited in This Article: ] |
| 32. | Pawlotsky JM, Pellerin M, Bouvier M, Roudot-Thoraval F, Germanidis G, Bastie A, Darthuy F, Remire J, Soussy CJ, Dhumeaux D. Genetic complexity of the hypervariable region 1 (HVR1) of hepatitis C virus (HCV): influence on the characteristics of the infection and responses to interferon alfa therapy in patients with chronic hepatitis C. J Med Virol. 1998;54:256-264. [Cited in This Article: ] |
| 33. | Rothman AL, Morishima C, Bonkovsky HL, Polyak SJ, Ray R, Di Bisceglie AM, Lindsay KL, Malet PF, Chang M, Gretch DR. Associations among clinical, immunological, and viral quasispecies measurements in advanced chronic hepatitis C. Hepatology. 2005;41:617-625. [Cited in This Article: ] |
| 34. | Eckels DD, Zhou H, Bian TH, Wang H. Identification of antigenic escape variants in an immunodominant epitope of hepatitis C virus. Int Immunol. 1999;11:577-583. [Cited in This Article: ] |