Copyright
©The Author(s) 2015.
World J Gastroenterol. Nov 14, 2015; 21(42): 12157-12170
Published online Nov 14, 2015. doi: 10.3748/wjg.v21.i42.12157
Published online Nov 14, 2015. doi: 10.3748/wjg.v21.i42.12157
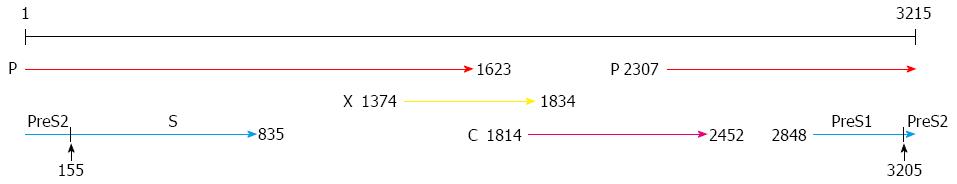
Figure 1 Structure of the hepatitis B virus genome.
The hepatitis B virus genome consists of an incomplete double stranded DNA with four open reading frames: surface proteins encoding gene (PreS1/S2/S), polymerase encoding gene (P), X protein encoding gene (X), and core protein encoding gene (C)[17].
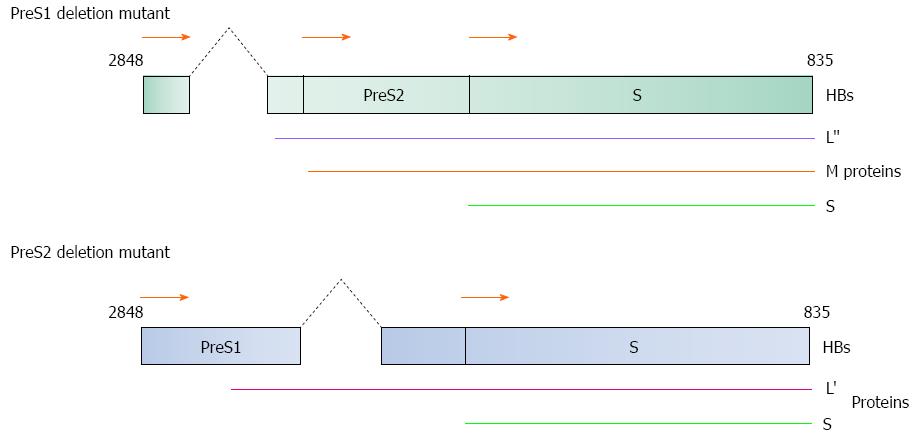
Figure 2 Diagrammatical representation of the PreS1/S2 deletion mutants.
(Top) 3’-end PreS1 deletion mutant and (Bottom) 5’-end PreS2 deletion mutant are two common mutation forms found in HBV-induced HCC patients. Both mutants produce truncated large surface proteins (L’ and L”). M: Middle protein; S: Small protein. HBs gene nucleotide position 2848 to 835 as shown in Figure 1.
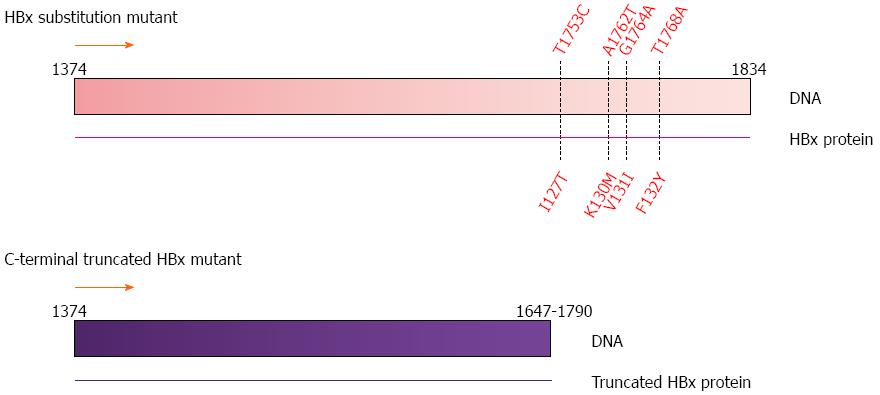
Figure 3 Common mutations in the HBx gene.
Substitution mutations at nucleotide positions T1753C, A1762T, G1764A and T1768A; and C-terminal truncations ranging from nucleotide position 1647 to 1790 observed in human HCC tumor samples[43]. HBx gene nucleotide position 1374 to 1834 as shown in Figure 1.
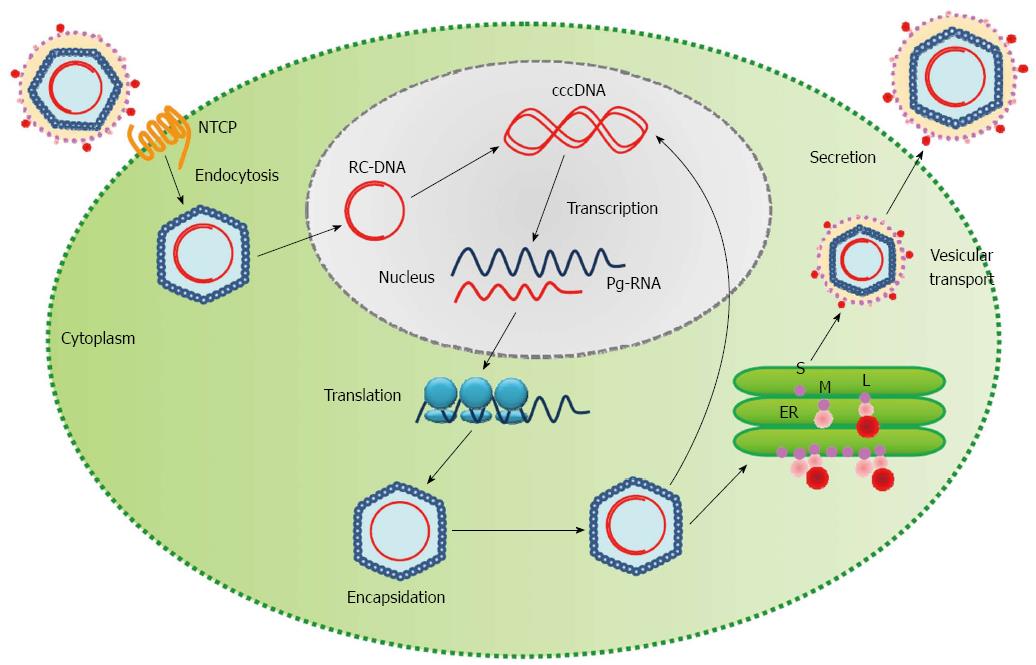
Figure 4 Life cycle of the hepatitis B virus.
HBV enters the hepatocyte and is uncoated in the cytoplasm. The relaxed-circular viral DNA (RC-DNA) is released from the nucleocapsid into the host nucleus and is converted into covalently closed circular DNA (cccDNA). The cccDNA acts as a template for transcription of pregenomic virion RNAs (Pg-RNA). Pg-RNA is then transferred into the cytoplasm, bound by viral polymerase and encapsidated. Inside the nucleocapsid, the Pg-RNA is reverse transcribed into RC-DNA, where either the RC-DNA can be released back into the host nucleus for amplification of viral DNA or the entire nucleocapsid can be transferred into the endoplasmic reticulum (ER) for viral surface protein coating and ultimately released from the cell[13]. S: Small surface protein; M: Middle surface protein; L: Large surface protein; HBV: Hepatitis B viral.
Figure 5 Word cloud of frequent CIS genes from three separate studies using SB transposon forward genetic screening with different genetic backgrounds[63,65,67].
The roles of these frequent CIS gene human orthologs were analyzed using TCGA database. The font size indicates the frequency of genetic alteration reported in human HCC samples. Color of the font indicates the copy number change or expression profile of the gene: red, amplification; blue, deletion; tomato, tends to upregulation; purple, tends to downregulation; yellow, frequently found in all three studies.
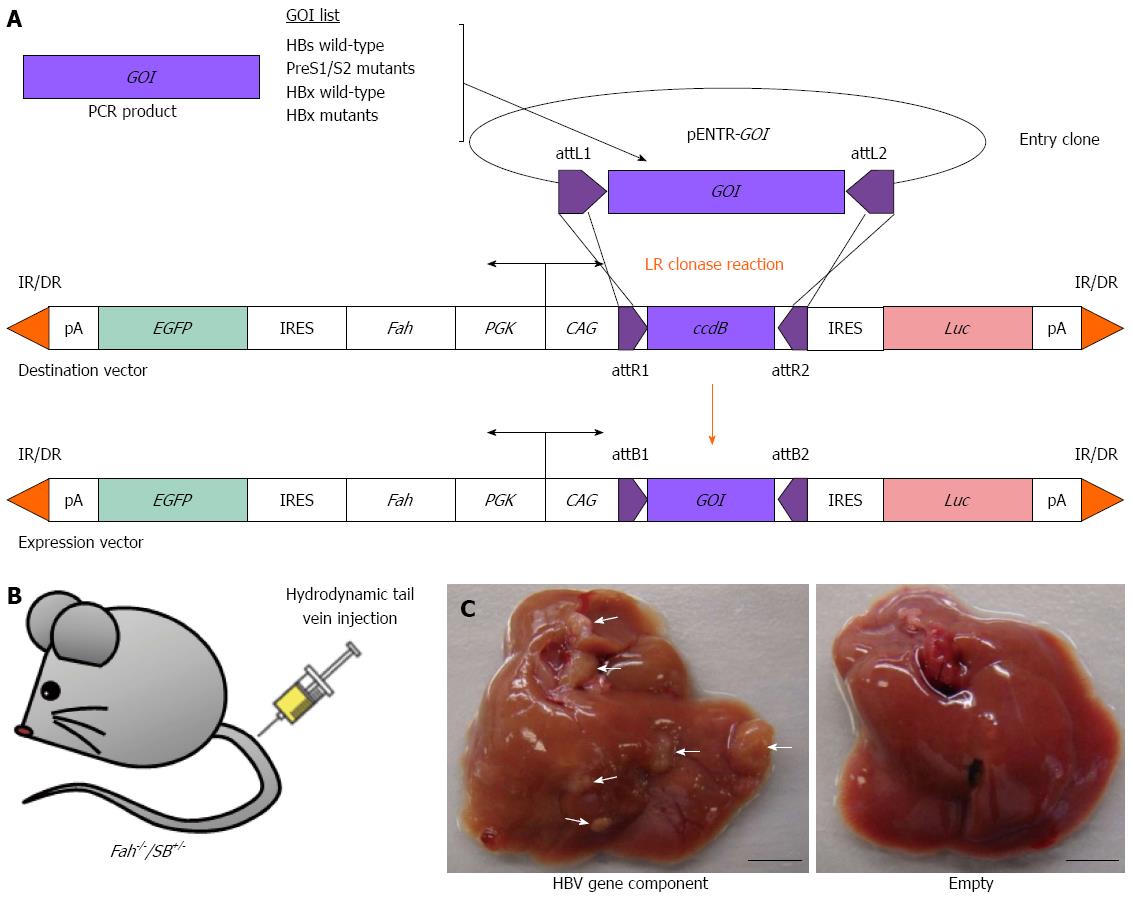
Figure 6 Reverse genetic screening of individual HBV gene components using the Fah-/-/SB+/- transgenic mouse model.
A: Construction of gene delivery expression vectors that carry various individual HBV gene-of-interest (GOI) components by PCR amplification and insertion of GOI into an entry clone (pENTR) and followed by LR clonase reaction (Life Technologies); B: Hydrodynamic tail vein injection of gene delivery expression vector into Fah-/-/SB+/- transgenic mouse; C: Liver tumors indicated by white arrowheads observed in Fah-/-/SB+/- mouse injected with a HBV gene component (left) after 160-d post-hydrodynamic injection. Fah-/-/SB+/- transgenic mouse injected with an empty gene delivery expression vector displayed normal liver morphology (right). EGFP: Enhanced green fluorescent protein gene; Luc: Luciferase gene; IR/DR: Inverted repeat/direct repeat sequences for Sleeping Beauty transposase binding and mobilization; pA: Polyadenalytion signal; IRES: Internal ribosome entry site; PGK: Phosphoglycerate kinase 1 promoter; CAG: Cytomegalovirus enhancer fused to the chicken β-actin promoter; attL/R/B sites: Recombination sites used by the Gateway cloning system (Life Technologies); ccdB: Topoisomerase poison from Escherichia coli. Scale bar: 0.5 cm.
- Citation: Chiu AP, Tschida BR, Lo LH, Moriarity BS, Rowlands DK, Largaespada DA, Keng VW. Transposon mouse models to elucidate the genetic mechanisms of hepatitis B viral induced hepatocellular carcinoma. World J Gastroenterol 2015; 21(42): 12157-12170
- URL: https://www.wjgnet.com/1007-9327/full/v21/i42/12157.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i42.12157