Published online Mar 26, 2013. doi: 10.5662/wjm.v3.i1.11
Revised: March 8, 2013
Accepted: March 18, 2013
Published online: March 26, 2013
AIM: To develop protocols for isolation of exosomes and characterization of their RNA content.
METHODS: Exosomes were extracted from HeLa cell culture media and human blood serum using the Total exosome isolation (from cell culture media) reagent, and Total exosome isolation (from serum) reagent respectively. Identity and purity of the exosomes was confirmed by Nanosight® analysis, electron microscopy, and Western blots for CD63 marker. Exosomal RNA cargo was recovered with the Total exosome RNA and protein isolation kit. Finally, RNA was profiled using Bioanalyzer and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) methodology.
RESULTS: Here we describe a novel approach for robust and scalable isolation of exosomes from cell culture media and serum, with subsequent isolation and analysis of RNA residing within these vesicles. The isolation procedure is completed in a fraction of the time, compared to the current standard protocols utilizing ultracentrifugation, and allows to recover fully intact exosomes in higher yields. Exosomes were found to contain a very diverse RNA cargo, primarily short sequences 20-200 nt (such as miRNA and fragments of mRNA), however longer RNA species were detected as well, including full-length 18S and 28S rRNA.
CONCLUSION: We have successfully developed a set of reagents and a workflow allowing fast and efficient extraction of exosomes, followed by isolation of RNA and its analysis by qRT-PCR and other techniques.
Core tip: Exosomes are small vesicles (30-150 nm) perceived to be carriers of the unique effector or signaling macromolecules (miRNA, ncRNA, mRNA and protein) between very specific cells within our body. The spectrum of current scientific interests ranges from studying the functions and pathways of exosomes to utilizing them in diagnostics and therapeutics development. Here we describe a complete exosome workflow solution: fast and efficient isolation of exosomes; extraction of their cargo; characterization of exosomal RNA content using quantitative reverse transcription-polymerase chain reaction and other techniques.
- Citation: Zeringer E, Li M, Barta T, Schageman J, Pedersen KW, Neurauter A, Magdaleno S, Setterquist R, Vlassov AV. Methods for the extraction and RNA profiling of exosomes. World J Methodol 2013; 3(1): 11-18
- URL: https://www.wjgnet.com/2222-0682/full/v3/i1/11.htm
- DOI: https://dx.doi.org/10.5662/wjm.v3.i1.11
Cells are known to secrete a large variety of vesicles, macromolecular complexes, and smaller molecules like salts and cofactors, into the extracellular space. The types of vesicles secreted are diverse and depend on the origin of the cells and their current state, for example, transformed, differentiated, stimulated, or stressed. Exosomes are a type of microvesicle, 30-150 nm in size, that have received increased attention over the past decade[1-5]. Exosomes are secreted by all cell types in culture, and also found naturally in body fluids including blood, saliva, urine, and breast milk, in very high numbers (108-1011 per mL)[6,7]. Depending on the cell/tissue of origin, many different roles and functions have been attributed to exosomes: Facilitators of the immune response[8], antigen presentation[6], programmed cell death, angiogenesis, inflammation, coagulation[9], morphogen transporters in the creation of polarity during development and differentiation[4], and mediation of nontargeted effect of ionizing radiation[10]. Recent studies have demonstrated that exosomes are not only specifically targeted to recipient cells to exchange proteins and lipids or to trigger downstream signaling events, but also to deliver specific nucleic acid cargo for cell communication purposes. Valadi et al[11] demonstrated that MC/9 and human mast cell line-1 mast cells secrete exosomes that contain mRNA from approximately 1300 genes and small RNAs, including 121 unique microRNAs. The transfer of exosomes to a donor cell showed that at least some mRNAs were full-length, as they were translated in the recipient cell. Glioblastoma cells also secrete exosomes and microvesicles containing mRNA, miRNA and angiogenic proteins[12] - when taken up by host human brain microvascular endothelial cells, mRNA molecules were translated and tubule formation by the target endothelial cells was stimulated. The spread of oncogenes by exosomes and microvesicles secreted by tumor cells has also been reported[13]. Exosomes seem to play a crucial role in spreading pathogens such as prions and viruses from one cell to another[14-16]. Interest towards exosomes, from their function in the body to more practical applications, such as use in diagnostics and therapeutics development, has grown exponentially in the last few years[17-19].
Critical to further our understanding of exosomes, is the development of reagents, tools and protocols for their isolation, characterization and analysis of their RNA and protein contents. Several reports have been published to date, using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and next gen sequencing for initial characterization of the RNA content of exosomes derived from human acute monocytic leukemia cell line, human umbilical vein endothelial cells, dendritic cells and human embryonic stem cell-derived mesenchymal stroma cells, as well as serum, saliva, placenta and breast milk[14,20-22]. All these studies utilize ultracentrifugation protocols[23] which are proven methods, producing clean exosome preparations. However, the ultracentrifugation approach has numerous drawbacks: the method is highly labor intensive and time consuming (up to two days per preparation, for a protocol with sucrose gradient); one can not process more than 6 samples at a time (due to rotor limitation); it requires a large amount of starting material; exosome yields are typically low; extensive training for personnel is needed; and the method overall is not very reliable.
Here we describe a novel approach for fast and efficient isolation of exosomes from cell culture media and blood serum, followed by recovery of RNA cargo and its analysis by qRT-PCR. The procedure is completed in a fraction of the time, compared to the current standard protocols utilizing ultracentrifugation, and allows to recover fully intact exosomes in higher yields.
Cell culture media: Fresh cell media was harvested from HeLa cells, grown initially in the presence of 10% fetal bovine serum (FBS), and [after two phosphate buffered saline (PBS) washes] without FBS for the last 12 h. The cell media samples were then centrifuged at 2000 g for 30 min to remove cell debris. The supernatant containing the cell-free cell media was transferred to a fresh container and held on ice until use. Next, each sample was combined with 1/2th volume of Total exosome isolation (from cell culture media) reagent (Invitrogen) and mixed well by vortexing until a homogenous solution was formed. The samples were incubated at 4 °C overnight, then centrifuged at 4 °C at 10000 g for 1 h. The supernatant was aspirated and discarded, and the exosome pellet was resuspended in PBS buffer, then stored at 4 °C short term (1-7 d) and -20 °C long term.
Human blood serum: Frozen serum samples from different donors were thawed in a water bath at room temperature until samples were completely liquid, then centrifuged at 2000 g for 30 min to remove any cellular debris. The supernatant containing the cell-free serum was transferred to a fresh container and briefly held on ice until use. Next, each serum sample was combined with 1/5th volume of Total exosome isolation (from serum) reagent (Invitrogen) and then mixed well by vortexing until a homogenous solution was formed. The samples were incubated at 4 °C for 30 min, then centrifuged at room temperature at 10000 g for 10 min. The supernatant was aspirated and discarded, and the exosome pellet was resuspended in PBS buffer, then stored at 4 °C short term (1-7 d) and -20 °C for long term.
Cell culture media: Using the same HeLa cell media that was prepared for the extraction of exosomes using the Total exosome isolation reagent, above, exosomes were isolated according to the differential ultracentrifugation Basic Protocol 1 as described by Théry et al[23]. Briefly, HeLa cell media was centrifuged at 4 °C at 2000 g for 10 min and then 10000 g for 30 min to produce a cell-free conditioned medium. The pooled media was divided equally into each of 6 polyallomer tubes and centrifuged at 4 °C at 100000 g for 70 min. The subsequent pellet was re-suspended and washed with PBS followed by a second 100000 g centrifugation. A low volume of PBS was used to re-suspend the washed pellets and then they were combined in pairs to result in three concentrated exosomes samples. These samples were then processed by isopycnic centrifugation using continuous sucrose gradients. The gradients were prepared using solutions of 0.25 mol/L and 2 mol/L sucrose in 20 mmol/L 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.3 as described by Abe et al[24].
Once prepared, the sucrose gradients were overlaid with one of the three concentrated exosome samples and then centrifuged at 4 °C at 110000 g for 16 h. Immediately after centrifugation, each of the gradients was fractionated using a peristaltic pump and fraction collector and each fraction was measured on a refractometer to identify those that contain exosomes based on the density range of 1.13-1.19 g/mL as defined by Record et al[25]. The resulting set of fractions were pooled within the respective sample and diluted 3-fold with 20 mmol/L HEPES, pH 7.3 then centrifuged in polyallomer konical™ tubes at 4 °C at 110000 g for 1 h to produce a final pellet consisting of purified exosome that was re-suspended in 200 μL PBS.
Human blood serum: Using the same human serum that was thawed for the extraction of exosomes using the Total exosome isolation reagent, above, exosomes were isolated according to the differential ultracentrifuge Basic Protocol 2 as described by Théry et al[23]. Briefly, serum was diluted with an equal volume of PBS and gently mixed until homogenous. Then the mixture was centrifuged at 4 °C at 2000 g for 30 min and 12000 g for 45 min to produce a cell-free serum. The pooled serum was divided equally into each of the 6 polyallomer tubes and centrifuged at 4 °C at 110000 g for 2 h. The subsequent pellet was re-suspended and washed twice with PBS followed each time by centrifugation at 4 °C at 110000 g for 70 min. A low volume of PBS was used to re-suspend the washed pellets, resulting in three concentrated exosomes samples.
Exosome samples isolated from cell media or blood serum (1-5 μL) were mixed with 2 × non-reducing Tris-glycine SDS sample buffer (Novex), then heated at 75 °C for 5 min and loaded onto a 1.5 mm × 15 mm well 4%-20% Tris-Glycine gel (Novex). Benchmark prestained protein ladder (Invitrogen) was added to one well as a control to monitor the molecular weight of the protein samples. The gel was run under denaturing conditions at 150 V for 1.5 h then transferred to a membrane using the iBlot instrument (Life Technologies). After transfer, the membranes were processed on the BenchPro 4100 (Life Technologies) with CD63 antibody diluted 100 μg × to 20 mL (Abcam). The WesternBreeze Chemiluminescence kit was utilized to label the membrane. Membranes were then exposed to X-ray film for 1-10 min and the film was analyzed.
The Total exosome RNA and protein isolation kit (Invitrogen) was utilized for recovery of RNA from both the concentrated exosome samples (reagent and ultracentrifugation) and control samples for each sample type - HeLa cell pellets (1 × 106 cells/pellet) for the HeLa cell culture and cell-free serum for the serum samples. Two hundred microlitre of each sample (brought up to volume with PBS if necessary) was combined with 205 μL of 2× denaturing solution, vortexed to lyse, and then incubated on ice for 5 min. After incubation, 410 μL of Acid-Phenol: Chloroform was added to the mixture and vortexed for 30-60 s to mix. Samples were then centrifuged for 5 min at 10000 g at room temperature to separate the mixture into aqueous and organic phases. Once centrifugation was complete, the aqueous (upper) phase was carefully removed without disturbing the lower phase or the interphase, and transferred to a fresh tube. One point twenty-five volumes of 100% EtOH was added to the aqueous phase for each sample then vortexed to mix. About 700 μL of volume was placed onto spin column in a collection tube then spun at 10000 g for 15 s to move the sample through the filter cartridge. Samples were then washed once with 700 μL Wash Solution 1 × and 2 × with 500 μL wash solution 2/3 (centrifuged at 10000 g for 15 s for each wash). After washing, filter was dried by spinning for an additional 1 min at 10000 g. The filter cartridge was transferred into a fresh collection tube and 50 μL of preheated (95 °C) nuclease-free water was applied to the center of the filter. Samples were centrifuged for 30 s at 10000 g to recover the RNA, then a second 50 μL volume of preheated (95 °C) nuclease-free water was applied to the center of the filter and centrifuged for 30 s at 10000 g. After the second spin, the eluate containing the RNA was collected and stored at -20 °C. A DNase treatment was performed on RNA extracted from the HeLa cell pellet using the DNase-free kit (Ambion) to remove any contaminating DNA. DNase treatment was not performed on other samples as they had a much smaller sample input. After treatment, the sample was diluted to 2 ng/μL and 1 μL of each RNA sample was analyzed on the Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Pico kit (Series II) to determine the mass of RNA going into downstream analysis.
Reverse Transcription (RT) Master Mixes were prepared for each sample using either the High Capacity cDNA Reverse Transcription kit and protocol with random primers for mRNA or the TaqMan® MicroRNA Reverse Transcription Kit reagents and protocol (Applied Biosystems) with gene specific RT primers for five miRNA targets (let7e, miR26a, miR16, miR24 and miR451). Ten microlitre of the RT Master Mix was added to corresponding wells in a 96-well plate, and 5 μL of each sample was added to the master mix. Plates were covered with adhesive (non-optical) cover and spun down to remove air bubbles, and then placed into a 9700 thermocycler and incubated as follows: for mRNA - 25 °C for 10 min; 37 °C for 120 min; and 85 °C for 5 min; for miRNA - 4 °C for 5 min; 16 °C for 30 min; 42 °C for 30 min; and 85 °C for 5 min. Reactions were kept at 4 °C until use.
qPCR master mixes were prepared for each of five microRNAs (let7e, miR26a, miR16, miR24 and miR451) and two mRNAs [glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and 18S] by combining 5 μL of AB Universal PCR Master Mix II, 2.5 μL; of nuclease-free water, and 0.5 μL of the 20 × Taqman Assay. After mixing, 8 μL of each master mix was placed into wells in a 384-well plate (enough for triplicate reactions for each isolation replicate). Two microlitre of each RT reaction was added in triplicate to the master mix of each target and the plates were sealed with optical adhesive cover. Plates were spun down to remove air bubbles then placed into a 7900HT instrument and run using the following thermocycler protocol: 95 °C for 10 min; (95 °C for 15 s; 60 °C for 60 s) 40 cycles. Once the run was complete, automatic Ct analysis was performed with SDSv2.3 software and average and standard deviation was calculated for each set of isolations and qPCR reactions for each target.
Isolation of exosomes from cell culture media and body fluids is presently a tedious, non-specific, and difficult process. The widely used approach is based on ultracentrifugation in combination with sucrose density gradients or sucrose cushions to float the relatively low-density exosomes away from other vesicles and particles[23]. The protocols range in time from 8 to 30 h and require an ultracentrifuge and extensive training to ensure successful isolation of exosomes.
As a way to simplify and shorten exosome isolation, we developed two Total exosome isolation reagents that enable straightforward and reliable concentration of intact exosomes from cell culture media and blood serum samples. By tying up water molecules, the reagents force less-soluble components, such as vesicles, out of solution. When the reagent is added to the biological sample, and solution is incubated at 4 °C, the precipitated exosomes can be recovered by standard centrifugation at 10000 g. The pellet is then resuspended in PBS or similar buffer and exosomes are ready for downstream end-point analysis or biological studies on their pathways, functions and trafficking.
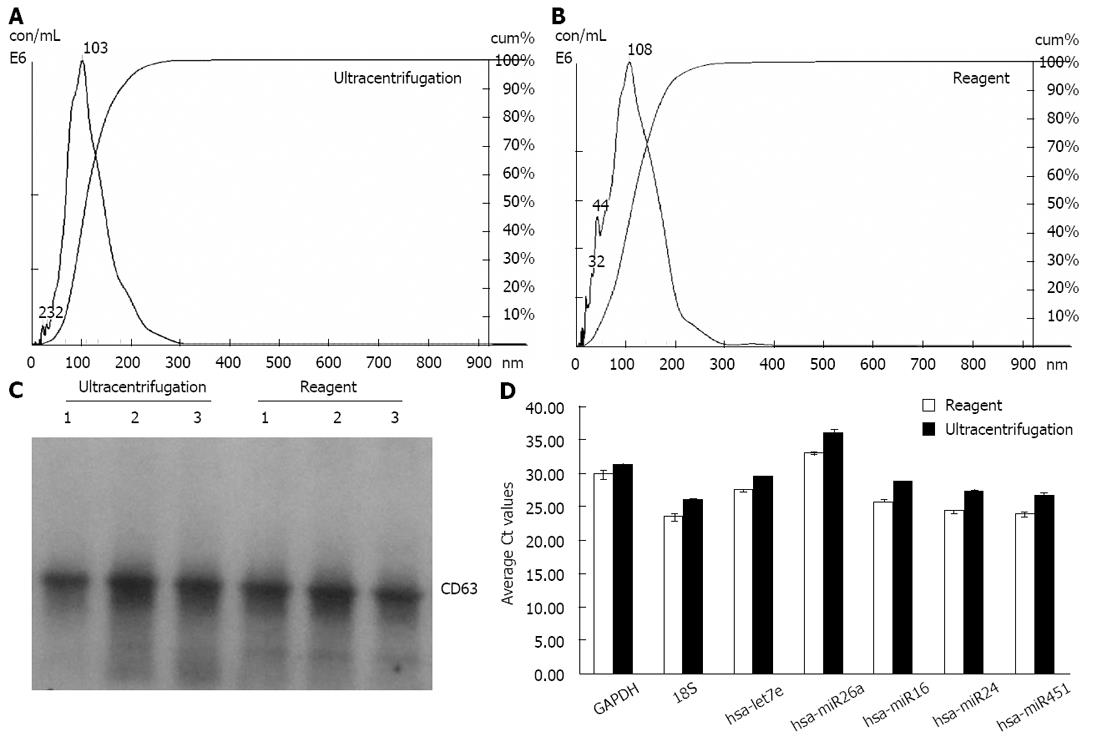
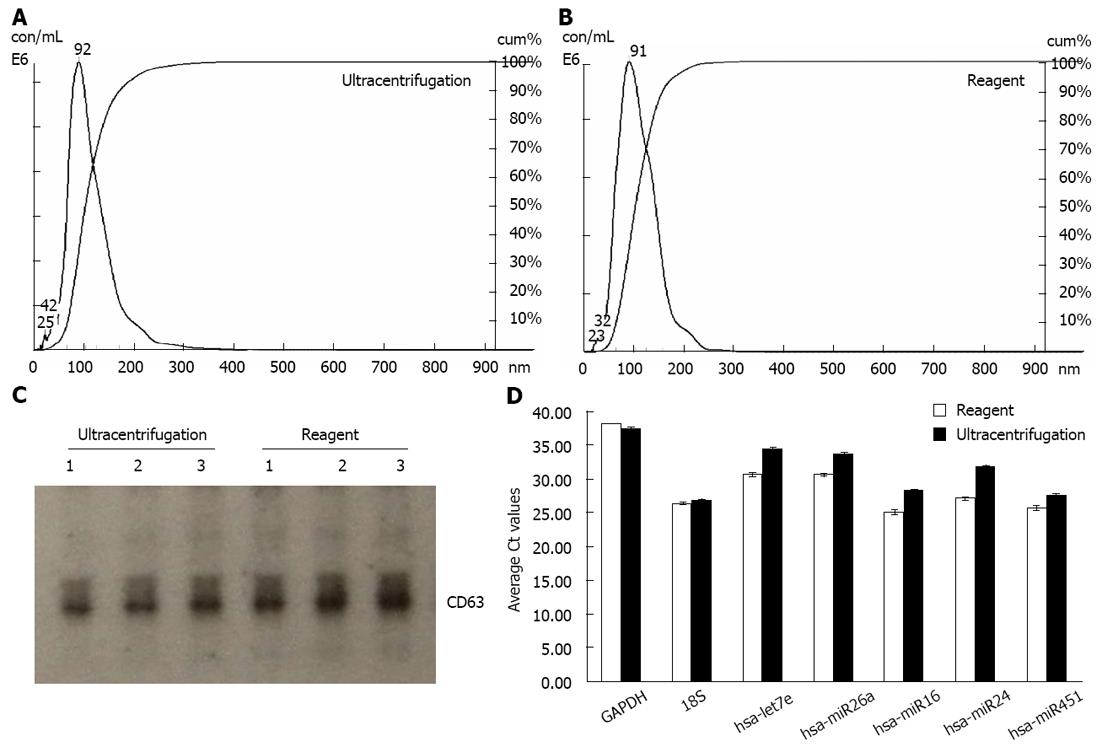
We extracted exosomes from HeLa cell culture media and blood serum samples (derived from healthy human donors) using the Total exosome isolation reagents as well as the ultracentrifugation procedure[23], for comparison purposes. Sizing and quantification of exosomes was performed with the NanoSight® LM10 instrument, following the manufacturer’s protocol. This instrument uses a laser light source to illuminate nano-scale particles (10-1000 nm) which are seen as individual point-scatters moving under Brownian motion. The paths of the point scatters, or particles, are calculated over time to determine their velocity which can be used to calculate their size independent of density. The image analysis NTA software compiles this information and allows one to automatically track the size and number of the nanoparticles. Results are shown in Figure 1A and B for HeLa cell culture media, and Figure 2A and B for serum. The reagent method recovered a significant number of nanovesicles, in comparable or higher yields vs the ultracentrifugation procedures; All nanovesicles were smaller than 300 nm, most of them being in the typical exosome size range of 30-150 nm. Similar results were obtained for cell media derived from THP-1 and Jurkat cell lines, 1-10 mL sample volume input (data not shown).
Samples were next analyzed by Western blots with antibody specific to CD63 - a well characterized exosomal marker[11,20,23]. Results are shown in Figure 1C for HeLa cell culture media and Figure 2C for serum - confirming that clean exosome populations were recovered with both protocols. CD9, TSG101, Annexin II exosomal markers were also confirmed by Western blots (data not shown).
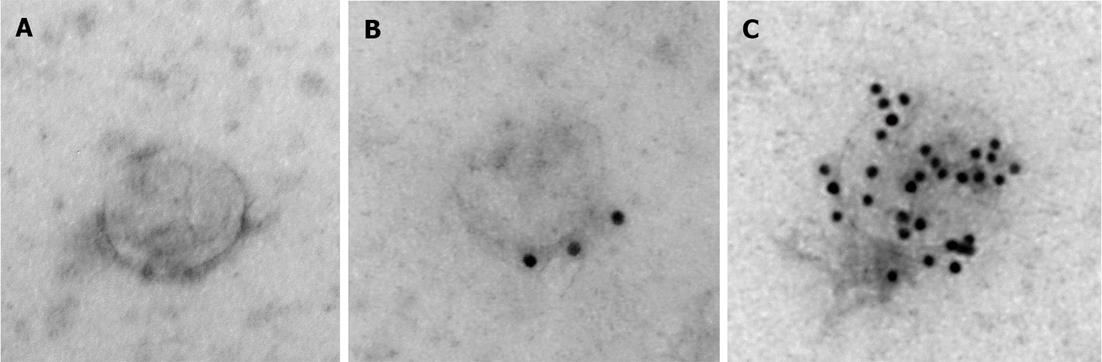
To further characterize samples obtained with the reagent, electron microscopy analysis was performed. Figure 3A shows a representative image of the unlabeled exosome, Figure 3B shows exosome immunolabeled with anti-CD63 antibodies followed by 10 nm protein A gold nanoparticles, and Figure 3C shows exosome immunolabeled with anti-CD81 antibodies followed by 10 nm protein A gold nanoparticles. The exosomes recovered with the reagent have typical appearance and size (about 100 nm[23]), and immunolabeling with anti-CD81 and anti-CD63 antibodies, which are well known exosomal markers[20,23], was very efficient confirming that the nanovesicles recovered with the reagent are exosomes.
Next, we proceeded with the isolation and analysis of the exosomal RNA cargo. The Total exosome RNA and protein isolation kit, developed specifically for this purpose, uses acid-phenol: Chloroform extraction to provide a robust, initial RNA purification step, followed by a final purification over a glass-fiber filter. Ethanol is added to samples that are passed through a filter cartridge containing the glass-fiber filter, which immobilizes the RNA. The filter is washed, and the RNA is eluted with a low ionic-strength solution.
We followed this protocol to isolate RNA from exosomes derived from HeLa cell culture media and blood serum samples using both the Total exosome isolation reagents and ultracentrifugation procedure[23]. Subsequent analysis with Qubit fluorometer has shown that for exosomes isolated from 100 mL of HeLa cell culture media using the ultracentrifugation protocol, it is possible to recover approximately 380 pg RNA. The reagent method resulted in isolation of somewhat more exosomes from the same volume of HeLa cell culture media, containing approximately 520 pg RNA.
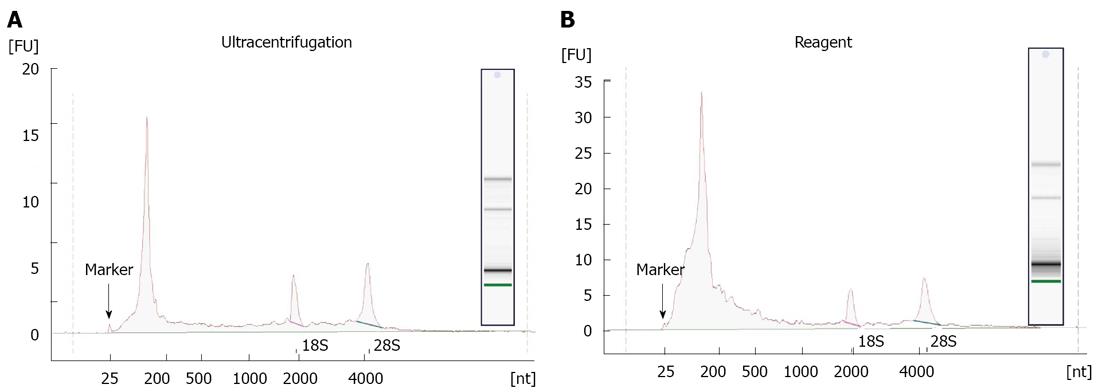
RNA recovered from exosomes with the reagent and ultracentrifugation was next characterized by capillary electrophoresis using the Agilent 2100 Bioanalyzer, and a RNA Pico chip. Results are shown in Figure 4. For both types of samples, profiles were similar: the majority of RNA content was small (< 200 nt), but there were some longer RNA species present including full-length 18S and 28S rRNA. This is in agreement with earlier studies[11,12,20], indicating that exosomes primarily contain short RNA (such as miRNA) and degraded mRNA, but also some full-length molecules including mRNA > 1 kb long. Overall, the amount of total RNA recovered, and specifically the small RNA fraction, is higher for the reagent method compared to ultracentrifugation protocol.
Finally, the levels of five microRNAs (let7e, miR26a, miR16, miR24 and miR451) and two mRNAs (GAPDH and 18S) earlier reported to be present in exosomes[11,20], were analyzed by qRT-PCR. Results are displayed in Figure 1D for HeLa cell culture media and Figure 2D for human blood serum. Based on Ct values, 25-33 for the majority of analytes, RNA isolation was efficient and the amount of material recovered is sufficient for standard PCR analysis - RNA recovered from exosomes derived from 3 μL serum or 30 μL cell media is sufficient for one qPCR reaction. The reagent method recovered somewhat higher levels of exosomes-compared to ultracentrifugation procedure, as indicated by 0.5-2 Ct shift up for different RNAs.
To conclude, we describe here a novel approach for fast and efficient isolation of exosomes from cell culture media and blood serum, that can be followed by recovery of RNA cargo and its analysis by qRT-PCR or sequencing. The procedure is completed in a fraction of the time, compared to the current standard protocols utilizing ultracentrifugation, and allows recovery of fully intact exosomes in higher yields. This is the first step towards developing standardized techniques and protocols for fast, high throughput and robust isolation of exosomes from various sample types and downstream analysis of their constituents. We believe these reagents and workflows will be highly useful to scientists working on the edge of cellular and molecular biology and focusing on analysis of extracellular (circulating) RNA residing within the exosomes, microvesicles and protein complexes.
Exosomes are small (30-150 nm) vesicles containing unique RNA and protein cargo, secreted by all cell types in culture. They are also found in abundance in body fluids including blood, saliva, urine. At the moment, the mechanism of exosome formation, the makeup of the cargo, biological pathways and resulting functions are incompletely understood. One of their most intriguing roles is intercellular communication-exosomes function as the messengers, delivering various effector or signaling macromolecules between specific cells. There is an exponentially growing need to dissect structure and the function of exosomes and utilize them for development of minimally invasive diagnostics and therapeutics.
Several reports have been published to date, using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and next gen sequencing for initial characterization of the RNA content of exosomes derived from severals cell lines, as well as serum, saliva, placenta and breast milk. All these studies utilize ultracentrifugation isolation protocols which allow to produce clean exosome preparations, however, suffer from numerous drawbacks. Critical to further our understanding of exosomes, is the development of reagents, tools and protocols for their simple and robust isolation, characterization and analysis of their RNA and protein contents.
The authors developed the workflow allowing fast and efficient extraction of exosomes, followed by isolation of RNA and its analysis by qRT-PCR. The procedure is completed in a fraction of the time, compared to the current standard protocols utilizing ultracentrifugation, and allows to recover fully intact exosomes in higher yields.
The workflow presented here allows fast isolation of exosomes and downstream analysis of their constituents, thus enabling basic exosome research as well as development of minimally invasive diagnostic alternative to biopsies.
In this study, the authors describe a novel approach for fast and efficient extraction of exosomes from cell culture media and body fluids, and compare the effectiveness of isolation methods with ultracentrifigation protocol. This manuscript is well organized and the experiments are well conducted.
P- Reviewer Chang LS S- Editor Huang XZ L- Editor A E- Editor Xiong L
| 1. | Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nature Reviews. 2012;13:328-335. |
| 2. | Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871-881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 621] [Cited by in RCA: 616] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 3. | Smythies J, Edelstein L. Transsynaptic modality codes in the brain: possible involvement of synchronized spike timing, microRNAs, exosomes and epigenetic processes. Front Integr Neurosci. 2012;6:126. [PubMed] |
| 4. | Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18:199-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 313] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 5. | Xu D, Tahara H. The role of exosomes and microRNAs in senescence and aging. Adv Drug Deliv Rev. 2012;20:1. |
| 6. | Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569-579. [PubMed] |
| 7. | Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1477] [Article Influence: 113.6] [Reference Citation Analysis (0)] |
| 8. | Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2703] [Cited by in RCA: 3108] [Article Influence: 194.3] [Reference Citation Analysis (0)] |
| 9. | Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, Ratajczak MZ. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 575] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 10. | Al-Mayah AH, Irons SL, Pink RC, Carter DR, Kadhim MA. Possible role of exosomes containing RNA in mediating nontargeted effect of ionizing radiation. Radiat Res. 2012;177:539-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 135] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 11. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8246] [Cited by in RCA: 9797] [Article Influence: 544.3] [Reference Citation Analysis (0)] |
| 12. | Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4100] [Cited by in RCA: 3943] [Article Influence: 231.9] [Reference Citation Analysis (0)] |
| 13. | Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1347] [Cited by in RCA: 1551] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 14. | Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937-10949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 354] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 15. | Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328-6333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1168] [Cited by in RCA: 1307] [Article Influence: 87.1] [Reference Citation Analysis (0)] |
| 16. | Leblanc P, Alais S, Porto-Carreiro I, Lehmann S, Grassi J, Raposo G, Darlix JL. Retrovirus infection strongly enhances scrapie infectivity release in cell culture. EMBO J. 2006;25:2674-2685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 104] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Spielmann N, Wong DT. Saliva: diagnostics and therapeutic perspectives. Oral Dis. 2011;17:345-354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 18. | Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603-1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 529] [Cited by in RCA: 605] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 19. | Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1730] [Cited by in RCA: 1858] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 20. | Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241-D1244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 697] [Cited by in RCA: 839] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 21. | Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, ‘t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272-9285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 481] [Cited by in RCA: 584] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 22. | Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248-7259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 750] [Cited by in RCA: 813] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 23. | Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2443] [Cited by in RCA: 3727] [Article Influence: 196.2] [Reference Citation Analysis (0)] |
| 24. | Abe S, Davies E. How to make sucrose density gradients. College of Agriculture, Ehime University, Japan. Available from: http://web-mcb.agr.ehime-u.ac.jp/english/methods/gradient.htm. |
| 25. | Record M, Subra C, Silvente-Poirot S, Poirot M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol. 2011;81:1171-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 414] [Article Influence: 29.6] [Reference Citation Analysis (0)] |