Published online Dec 20, 2023. doi: 10.5662/wjm.v13.i5.492
Peer-review started: July 21, 2023
First decision: August 31, 2023
Revised: September 7, 2023
Accepted: October 23, 2023
Article in press: October 23, 2023
Published online: December 20, 2023
Processing time: 151 Days and 21.2 Hours
Exosomes are 30-150 nm nanovesicles with sophisticated nucleic acids cargo, actively secreted by all cells within human body, and found in abundance in all body fluids, including urine. These extracellular vesicles have tremendous potential for next generation diagnostics, theoretically enabling noninvasive assessment of organ and tissue function via liquid biopsy analysis.
Recently, feasibility of an exosomal molecular test was demonstrated for post-organ transplant monitoring: Analysis of urine-derived exosomal mRNA cargo allowed early detection of kidney allograft rejection. Here, we further studied urine-derived exosomes and their mRNA content as a highly promising dia
The urine specimens were stored at various conditions and pre-processed in different ways. Next, samples were passed through the columns to capture all extracellular vesicles, the vesicles were lysed to release their content and the exosomal RNA was purified on the mini-columns, reverse transcription was performed, next pre-amplification, followed by a qPCR analysis for a panel of mRNA markers.
To ensure exosomal RNA integrity, the harvested urine specimens should be shipped refrigerated, by overnight delivery. Urine can next be stored at the test site for up to 1 wk at 4 °C, and long term should be frozen at -80 °C. Urine specimens must be centrifuge at low G-force to deplete cells and debris, to ensure consistent top results in downstream molecular assays. All commonly used medications (tacrolimus, cyclosporin A, mycophenolic acid, everolimus, sirolimus, ascomycin, teriflunomide) were tested and confirmed that they do not cause assay interference.
mRNA from urine-derived exosomes was shown to be stable across a broad range of conditions and produced accurate results when analyzed via qPCR assay for detection of kidney allograft rejection. We identified the most optimal conditions for every step of the process, ensuring pre-analytical sample integrity and robust qPCR results.
Core Tip: Recently, it was demonstrated that analysis of urine-derived exosomal mRNA allows early detection of kidney allograft rejection. We further studied exosomes and their mRNA content as a highly promising diagnostic modality. This included stability studies of urine samples and exo-mRNA upon transportation from the point of collection to centralized testing facility, short-term storage of urine at different conditions upon receipt till the point molecular assay is performed, and effects of various interfering substances on the quantitative polymerase chain reaction assay. mRNA from the urine-derived exosomes was proven to be stable across broad range of conditions and produce robust results in post-transplant monitoring assays.
- Citation: McFaul M, Ventura C, Evans S, Dundar H, Rumpler MJ, McCloskey C, Lowe D, Vlassov AV. Urine exosome mRNA-based test for monitoring kidney allograft rejection: Effects of sample transportation and storage, and interference substances. World J Methodol 2023; 13(5): 492-501
- URL: https://www.wjgnet.com/2222-0682/full/v13/i5/492.htm
- DOI: https://dx.doi.org/10.5662/wjm.v13.i5.492
There is an urgent need for advanced post-transplant monitoring molecular tests, allowing to rapidly determine allograft rejection at the earliest stage using non-invasively collected patients’ samples. At the moment one of the most widely utilized tests is serum creatinine, which has a number of limitations[1,2]. The next gen molecular assay, now rather widely used in laboratories world-wide is based on cell free DNA[3,4]. This test analyses percentage of the donor-derived (dd) cfDNA in the blood of organ recipient, and if the fraction increases over time and crosses a predetermined threshold (typically 1% for kidney transplants)- this indicates organ rejection. Although this test was already commercialized a few years ago, its specificity is suboptimal, as other-than-allograft rejection events can also trigger increase in dd cfDNA percentage. This test does not work well for detection of the early stage organ rejection (TCMR1a). Also, this approach is minimally invasive, rather than non-invasive- as it requires the collection of a patient’s blood (which is an inconvenience for immunosuppressed individuals). Consequently, clinicians still need sensitive and specific biomarkers that overcome the above listed limitations of cfDNA-based tests for post-transplant monitoring.
Exosomes are recently discovered nanovesicles secreted by all cells within human body and found in abundance in all body fluids. They contain very sophisticated cargo of nucleic acids and proteins, reflecting the content of parental cells[5-9]. Exosomes and other extracellular vesicles have tremendous potential as biomarkers for disease diagnosis, as they theoretically should allow access to the health state of all organs and tissues via liquid biopsy analysis, using not only blood, but also urine, saliva, and other non-invasively collected samples[10-13]. The value of urinary exosomes in post-transplant diagnostics was demonstrated in a recent study for early detection of kidney transplant rejection[14]. Notably, urine collections are better suited for active surveillance monitoring compared to frequent blood draws and urinary exosomes exhibit promising clinical utility for detecting early kidney allograft rejection and stratifying rejection etiology.
In this study, we further characterized the aforementioned assay to understand the preanalytical limitations for an eventual migration into the clinical diagnostic domain. An emphasis was placed on the viability of urine as a matrix to facilitate at home collections from post-kidney transplant patients and correlate expected outcomes for the advanced exosome assay. This included the stability of exosomal mRNA in urine upon transportation from the point of collection to a centralized testing facility, the storage of urine samples at different conditions upon receipt until the point molecular assay is performed, and the effect of various potentially interfering substances on the downstream quantitative polymerase chain reaction (qPCR) assay. mRNA from the urine-derived exosomes was shown to be stable in a broad range of conditions and produced accurate results when analyzed via qPCR assay for detection of kidney allograft rejection. We identified the most optimal conditions for every step of the process, ensuring preanalytical sample integrity and robust qPCR results.
The following box was utilized in studies on transportation of urine specimens: Small box cat #56519 (Therapak); exterior dimensions: 11 × 9 × 11 in; inner dimensions: 8 × 6 × 8 in; wall thickness: 1.5 in. Gel packs (12 oz Gel Pack #PP12 (Sonoco)) were frozen overnight at -20 °C and placed in the boxes, to refrigerate urine specimens. Temperature monitor #40510 (DeltaTrak) was used to monitor temperature inside the box, up to 72 h.
Exosome isolation was performed according to the ExoLution protocol (Exosome Diagnostics, a Bio-Techne brand). In brief, 10 mL urine samples were combined with 2.5 mL of 5×BB buffer in 15 mL tubes. ISS-MHV positive extraction control was added to each sample and loaded onto ExoLution Columns in 50 mL tubes and centrifuged for 5 min at 5000 × g. Flow-through was discarded and columns were washed with 10 mL of wash buffer and re-centrifuged at 5000 × g. ExoLution Columns were then transferred to fresh 50 mL tubes and 550 μL of lysis mix was added directly onto the ExoLution Column membrane surface. Columns were then centrifuged for 5 min at 5000 × g. Flowthrough containing exosomal RNA was collected and transferred to the RNA purification step.
RNA was further purified according to the ExoLution protocol using silica spin columns. Briefly, after a chloroform separation, the aqueous phase was transferred to 1.5 mL reaction tubes containing ethanol and loaded onto silica spin columns. Spin columns were centrifuged for 30 sec at 11000 × g. The flow-through was discarded and the spin columns were re-assembled onto the next set of fresh 2 mL collection tubes. The previous steps were repeated until the entire sample was loaded onto each column.
Next, RNA Prep buffer was added to the silica spin columns, and columns were centrifuged for 30 sec at 11000 × g. The flow-through was discarded, and the spin columns were re-assembled on the next set of fresh 2 mL collection tubes. RNA Wash buffer was next added to the spin columns, and columns were centrifuged for 30 sec at 11000 × g. The flow-through was discarded, and the spin columns were re-assembled on the next set of fresh 2 mL collection tubes. The wash process was then repeated a second time.
The spin columns were centrifuged for 3 min at 16000 × g to dry the membranes. The 2 mL collection tubes were discarded, and the spin columns were placed in fresh 1.5 mL reaction tubes. 15 μL of TE buffer was added directly onto the silica membranes. The columns were incubated for 5 min. Purified exosomal RNA was finally eluted by column centrifugation for 1 min at 16000 × g and immediately carried forward to reverse transcription.
Reverse transcription (RT) reaction mix was prepared using SuperScript® VILO™ cDNA Synthesis Kit (Thermo Fisher Scientific, cat# 11754-250): 10x SuperScript Enzyme Mix 2 μL, 5x VILO Reaction Mix 4 uL, exosomal RNA 14 μL. Samples were placed into thermocycler and were subject to the following cycling conditions: 25 °C: 10 min, 42 °C: 70 min, 85 °C: 5 min, hold: 4 °C.
Pre-Amplification was carried out as follows: TaqMan™ PreAmp Master mix (Thermo Fisher Scientific, 4488593): 12.5 uL, Primer mix 0.5 uL, RT reaction samples 12 uL. Samples were placed into thermal cycler and the following cycling parameters were utilized: Initial denaturation 95 °C for 10 min; 14 cycles: (95 °C for 15 sec, 60 °C for 4 min); hold: 4 °C.
qPCR analysis was performed in triplicate, under the following conditions: TaqMan™ Fast Universal PCR Master Mix (Thermo Fisher Scientific, cat# 4367846): 10 μL, 20x assay mix: 1 μL[11], nuclease-free water: 7 μL, 5x diluted PreAmp sample: 2 μL. qPCR plates were analyzed on QuantStudio 5 (QS5) fast real-time PCR machine with 96-well 0.1 mL Block. Fast cycling conditions were utilized: initial denaturation 95 °C for 20 sec, 40 cycles: (95 °C for 1 sec, 60 °C for 20 sec).
All pure drug compounds were acquired from Fisher Scientific: Teriflunomide (cat# AC467112500), Cyclosporin A (cat# AAJ6319106), Sirolimus (cat# AAJ62473MF), Everolimus (cat# AAJ60139MB), Mycophenolic acid (cat# AAJ6190509), Tacrolimus (cat# AAJ63571MF), Ascomycin (cat# AAJ66751MC), dissolved in DSMO to produce 5000× stocks, and tested for potential interference with assay at concentrations 50× exceeding expected urinary excretion level transplant patients. Expected levels: Tacrolimus: 0.53 μg/mL; Cyclosporin A: 10.5 μg/mL; Mycophenolic acid: 16.67 μg/mL; Everolimus: 0.03 μg/mL; Sirolimus: 0.10 μg/mL; Ascomycin: 0.47 μg/mL; Teriflunomide: 18.33 μg/mL.
Ideally, active surveillance testing for post-transplant monitoring should be facilitated through an at home non-invasive collection of the patients’ specimens. Alternatively, the procedure could be performed with ease in a healthcare provider's office. Regardless of the point of collection, the specimen must be transported to a centralized laboratory, accessioned and temporarily stored prior to analysis.
The objective of this study was to explore stability of urinary exosomes and their diagnostic value with a previously described analytical method[14] for the detection and stratification of kidney allograft rejection. Briefly, in this assay the urine sample is first passed through the column to capture all extracellular vesicles, next the vesicles are lysed to release their content and the exosomal RNA is purified on the mini-column, reverse transcription is performed, next pre-amplification, followed by a qPCR analysis for a panel of mRNA markers. The output Ct values are analyzed by an algorithm that generates the scores that allow determination of rejection for kidney allograft, and type of rejection: TCMR vs ABMR. The entire assay takes less than 6 h to complete, which enables the laboratory to produce rapid “same day” results.
Taking into account the novelty of urine as a sample of choice for this molecular assay, and also exosomes, particularly their RNA cargo, as markers of organ rejection, it is of high importance to characterize every step in more detail, to ensure subsequent smooth clinical implementation of this test. Here we explored: (1) Stability of urine specimens and exosomal mRNA upon transportation from the point of collection to testing facilities, (2) storage of urine samples at different conditions upon receipt till the point molecular assay is performed, and (3) effect of various potentially interfering substances on the downstream qPCR assay.
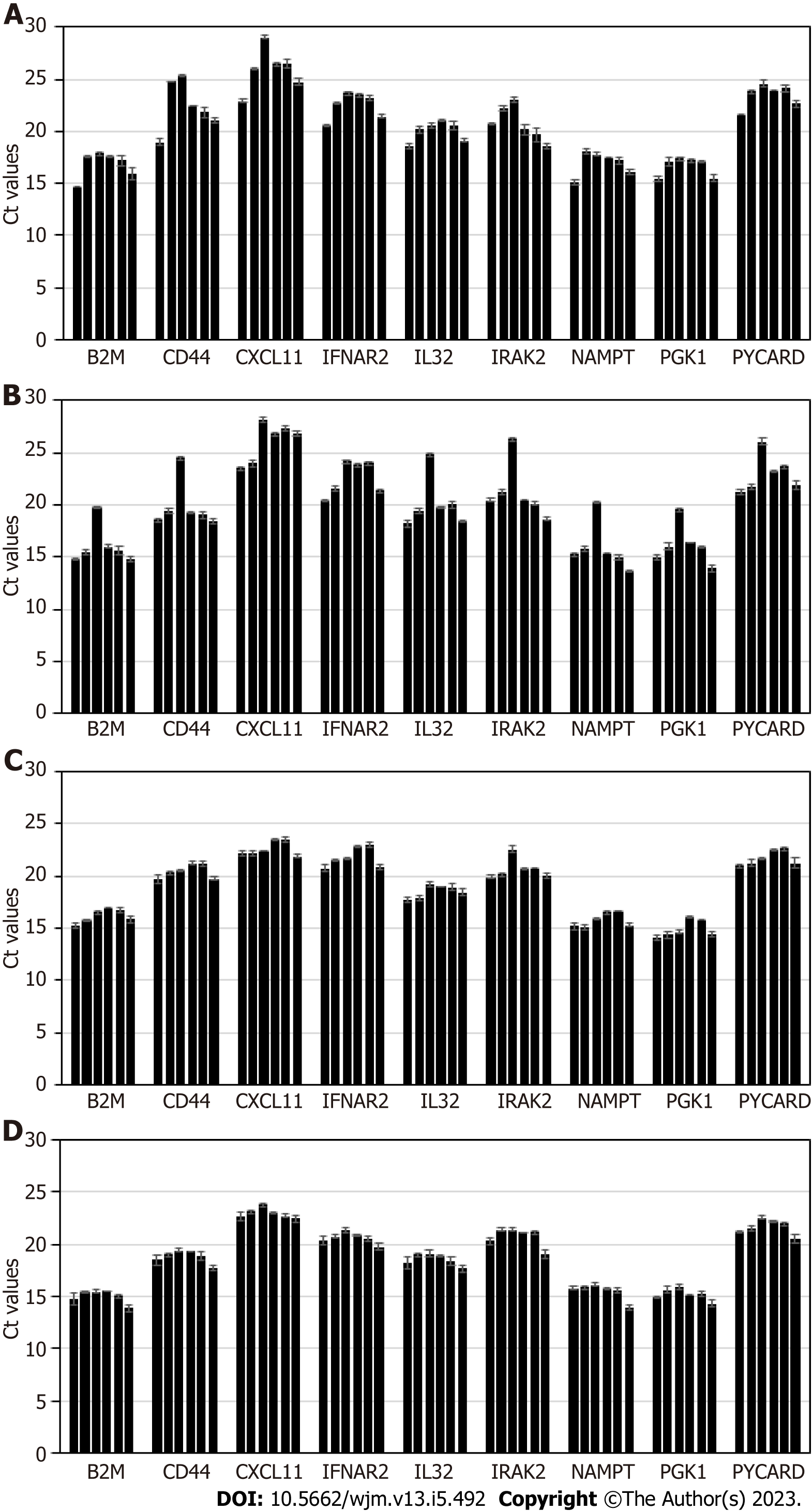
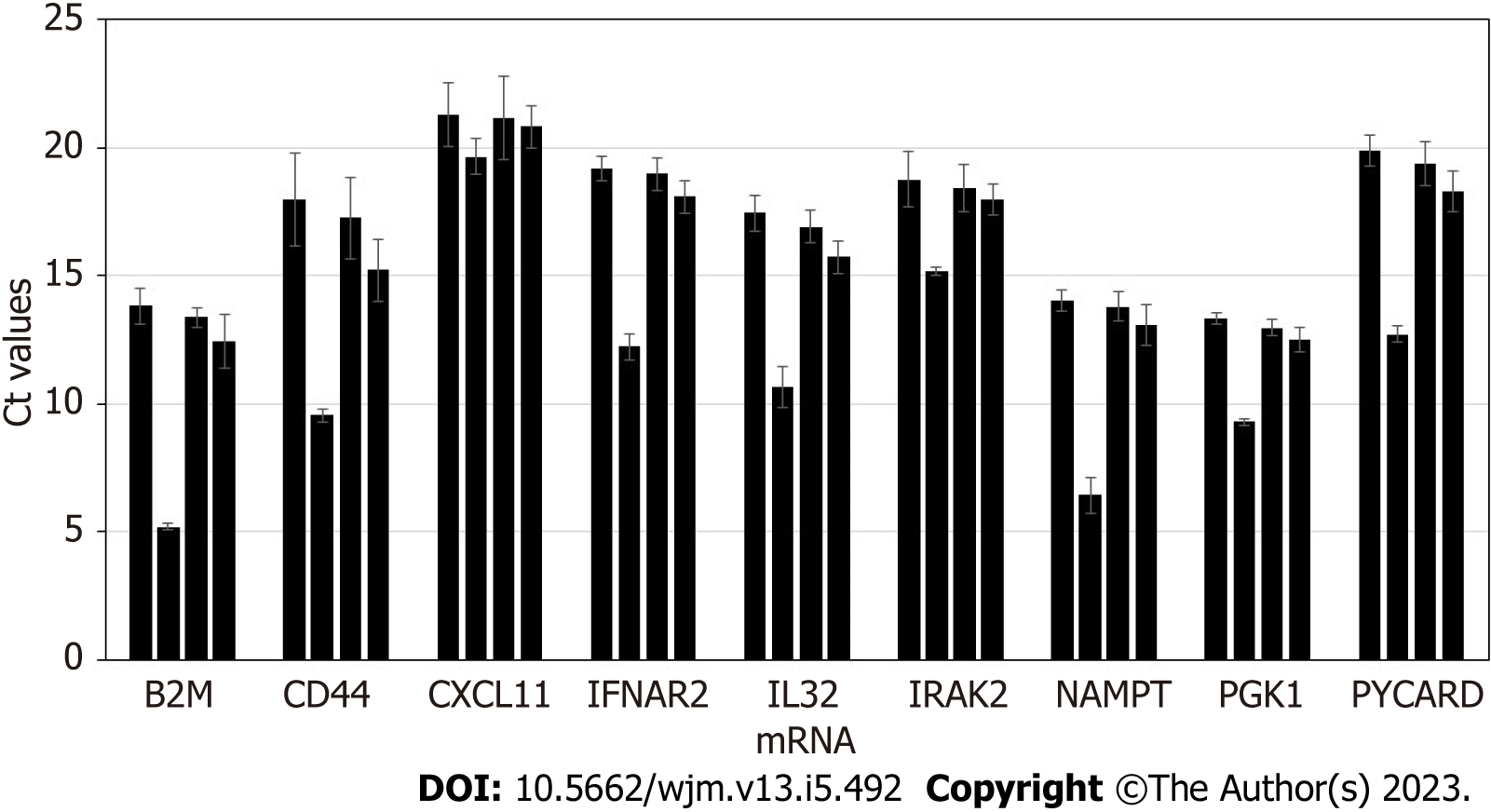
Stability of exosomal mRNA upon preanalytical conditions mimicking urine transportation at different temperatures, and effects of various urine upfront processing techniques were studied in detail. Results are shown in Figure 1. Urine specimens were derived from 4 donors, and processed following the complete workflow, with qPCR assay performed for a panel of 9 mRNA targets. Preanalytical conditions mimicking specimen shipment and urine processing included: 2 d storage at +4 °C; 2 d at +20 °C; 2 d at +40 °C; whole urine frozen and thawed (mimicking transportation in frozen state); urine pre-processed to remove cells and debris by centrifugation for 20 min at 2000 × g, and supernatant subject to single or double freeze/thaw cycles.
Storage of urine samples at elevated temperature, +20 °C and especially +40 °C caused gradual degradation of mRNA, as indicated by Ct (threshold cycles) shift that varied among targets and donors (as expected). While for some donors and certain mRNA targets Ct shift was minimal (< 0.5 Ct), for other donors and certain mRNA targets effects were substantial (> 5 Ct). Importantly, variable increases in Ct values introduce unpredictable changes to the outputs of algorithms analyzing levels of multiple mRNA targets and as a consequence will decrease accuracy of calculated outcomes. Normalization to PGK1 or other RNA largely addresses this issue. However, overall it is recommended that urine specimens be transported refrigerated, to ensure temperatures are consistently maintained below +20 °C.
Freezing urine specimens on dry ice and -80 °C, followed by defrosting, caused mild increase in Ct values compared to unfrozen specimens, presumably due to partial mRNA degradation upon sample thawing; 2 freeze/thaw cycles did not have significant further effects on mRNA integrity.
Specimens which were frozen without initial centrifugation at low G-force to deplete cells and debris generated somewhat variable Ct values for some samples and mRNA targets. This indicates the need for preanalytical centrifugation of urine specimens to ensure removal of interfering cellular RNA fraction (including blood-derived: See discussion below), consistent qPCR results, and assay algorithmic output scores.
In summary, for exosome-based molecular assays, the preanalytical process plays an important role in downstream assay integrity. Urine specimens must be transported under refrigeration, and centrifugation of the samples ensures depletion of cells and debris, and consistent, reproducible results. Effects of urine freezing and prolonged storage on the accuracy and reproducibility of the assay will be further discussed below.
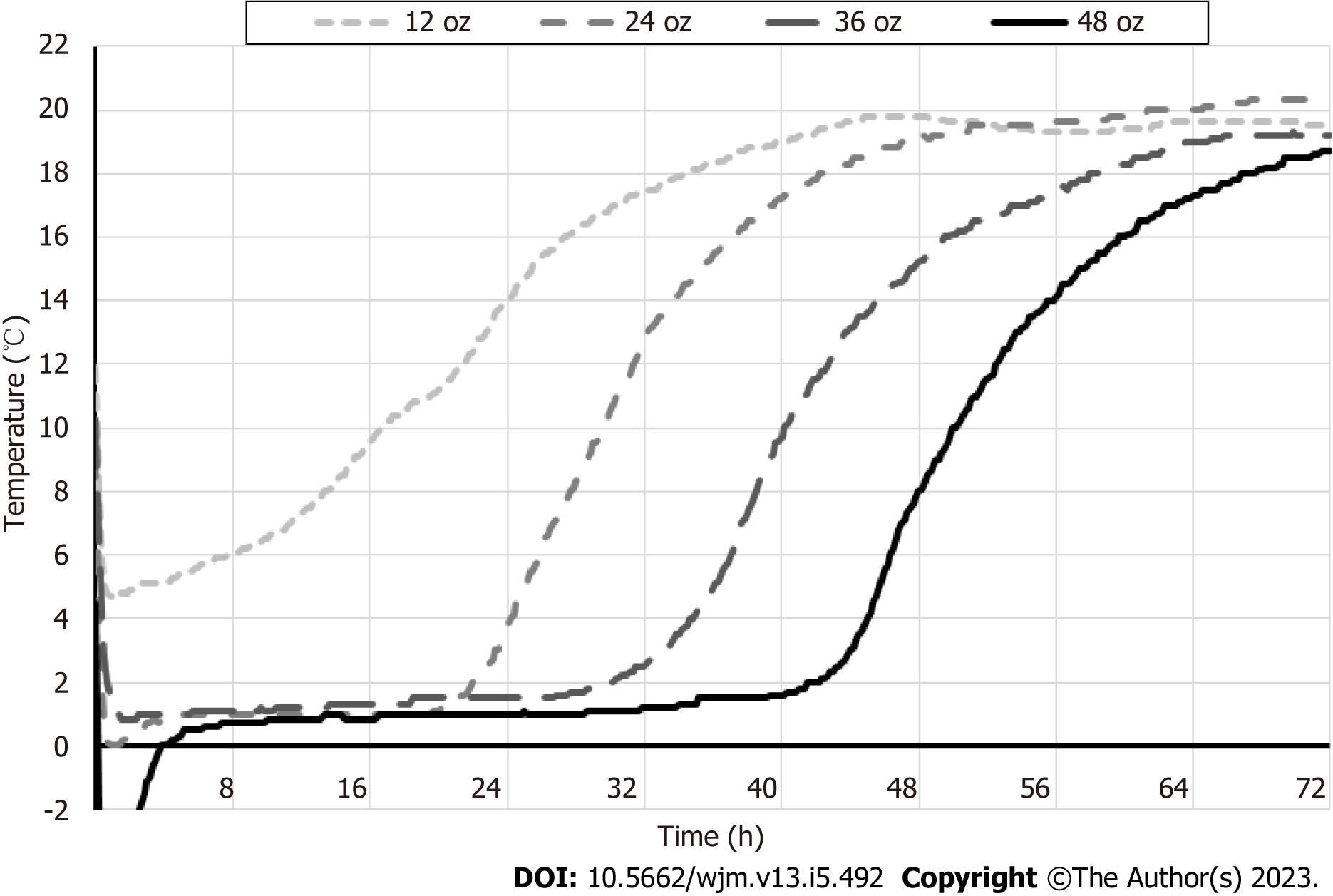
Next, temperature of the urine specimens was monitored during conditions mimicking transportation, to identify the optimal gel pack volume that would ensure sample refrigeration (Figure 2). One, two, three or four 12 oz gel packs were frozen overnight at -20 °C and placed in the 1.5-inch thick styrofoam box, that was stored at ambient temperature. Results are shown in Figure 2.
At 24 h, which is typical for overnight sample shipment, temperature inside the box with single gel pack (12 oz) was approximately 14 °C; two, three or four gel packs successfully maintained temperature below 4 °C. By 48 h, temperature inside the boxes with 1-3 gel packs (12 oz) was > 15 °C; only four gel packs secured temperature of approximately 8 °C. By 72h, temperature inside all boxes with 1-4 gel packs (12 oz) was > 18 °C.
Overall, for molecular tests, harvested urine specimens should be shipped by next day overnight delivery, in Styrofoam boxes ≥ 1.5 inch thick with gel packs totaling 24-48 oz, to ensure optimal refrigeration and specimen tem
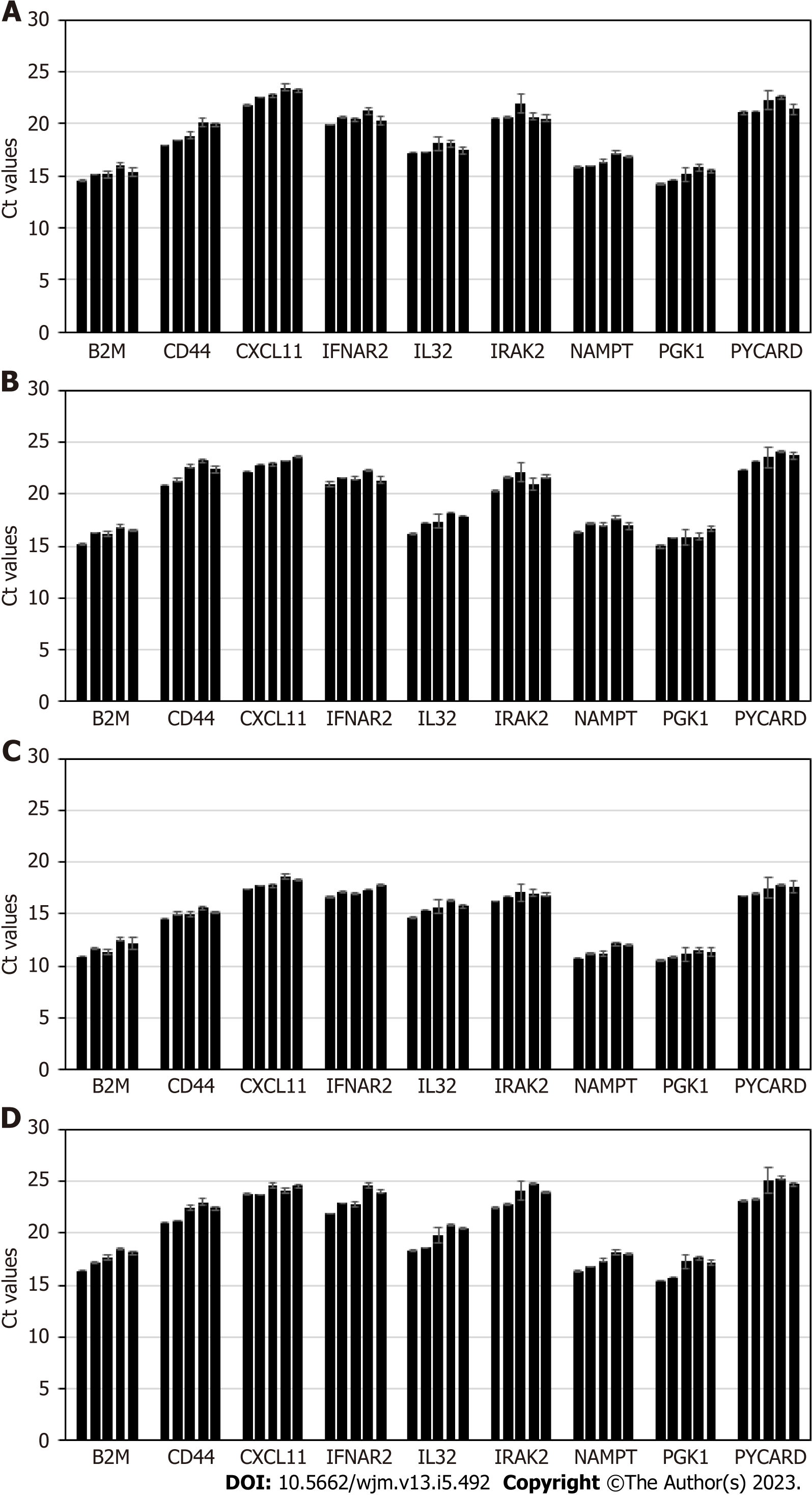
Stability of exosomal mRNA was next studied upon prolonged urine storage at different temperatures. Results are shown in Figure 3. Urine samples were derived from 4 donors, and processed following the complete workflow, with qPCR assay performed for a panel of 9 mRNA targets. Conditions included: 2 d at +4 °C; 7 d at +4 °C; 14 d at +4 °C; 4 d at -80 °C; 30 d at -80 °C.
Storage of urine samples at +4 °C for up to one week did not affect mRNA integrity and assay outcome; at 2 wk of storage gradual degradation of mRNA is occurring, as indicated by Ct shift that varied for different targets and donor-to-donor (as expected). Thus, unfrozen urine specimens should ideally be tested within 7 d of specimen receipt and refrigerated storage.
Once samples were frozen at -80 °C, and then defrosted and tested in assay, Ct values typically shift up compared to fresh samples, and for some donors and certain mRNA targets the change is significant (> 2 Ct). Prolonged sample storage at -80 °C does not cause any further Ct increase. Thus, for the purpose of consistency and reproducibility, all urine samples should be either processed unfrozen (within a week, as stated above) or all subject to centrifugation at low G-force, frozen, then defrosted and tested at any time point. As mentioned above, normalization to PGK1 or other RNA, will ensure consistent assay performance.
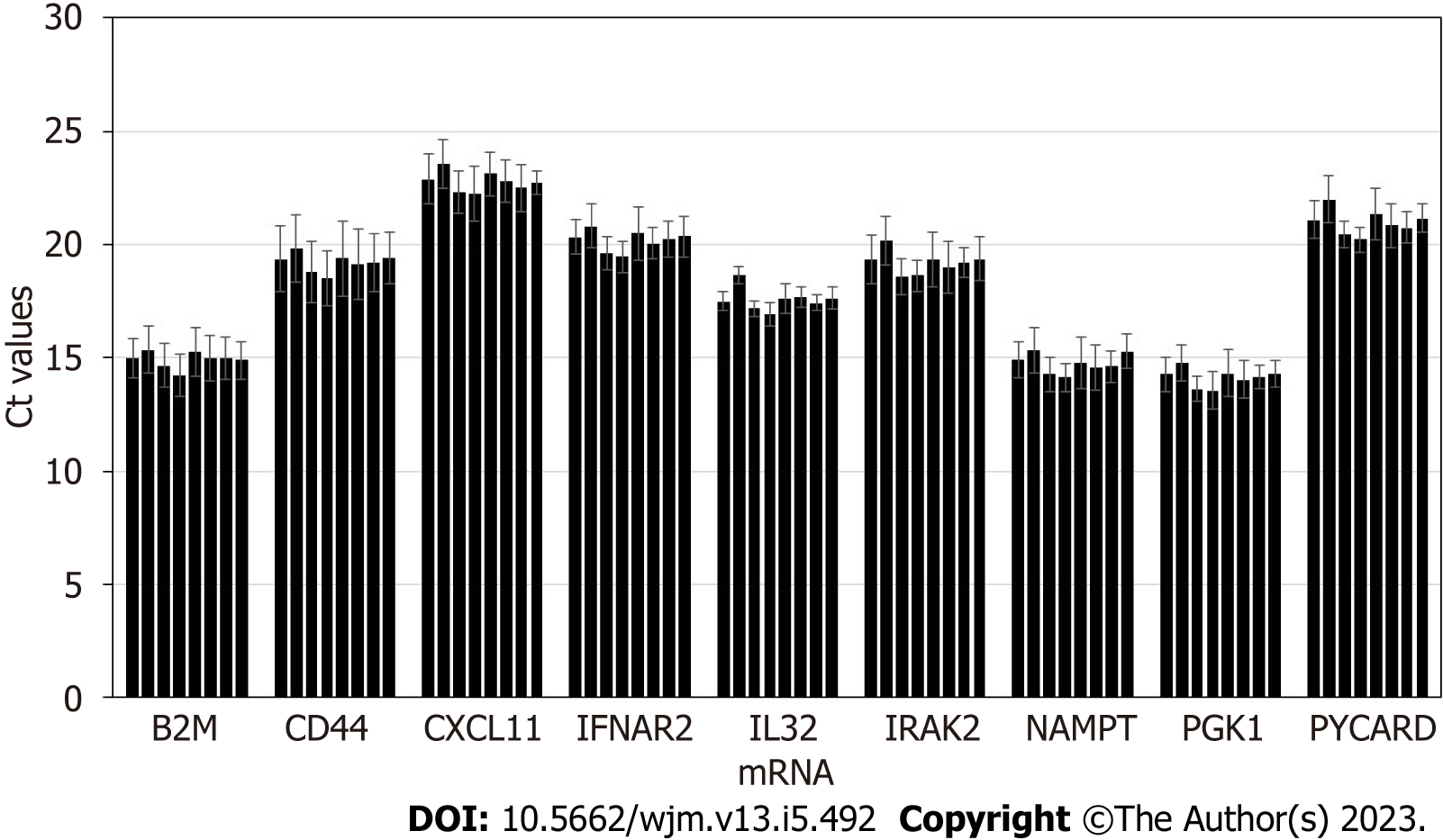
First, medications commonly prescribed to transplant patients, were studied for potential interference with exosome molecular assay. Urine samples derived from 4 donors were utilized, and various parent drugs and metabolites were spiked in before exosome purification, at concentrations exceeding 50-fold the expected urinary excretion levels for transplant patients. Expected urine levels for immunosuppressants and other common medications were: Tacrolimus: 0.53 μg/mL; Cyclosporin A: 10.5 μg/mL; Mycophenolic acid: 16.67 μg/mL; Everolimus: 0.03 μg/mL; Sirolimus: 0.10 μg/mL; Ascomycin: 0.47 μg/mL; Teriflunomide: 18.33 μg/mL.
Samples were processed following the complete workflow, and qPCR assay was performed for a panel of 9 mRNA targets. Results are displayed in Figure 4. None of the drugs caused major interference with assay at the dose significantly exceeding the typical circulating levels in bodily fluids, as indicated by Ct values within +/- 1 compared to controls. This demonstrates that molecular assay is robust, and all common medications are depleted from the mRNA analytes during the exosome and RNA purification workflow, and the trace amounts remaining do not have negative impact on reverse transcription or real-time PCR.
Next, effect of trace amount of blood in urine was studied. Hematuria is a fairly common condition, and blood was expected to impact the performance of exosome molecular assay. Urine samples derived from 4 donors were utilized, and blood (0%-1%) was spiked into urine before exosome purification. Results are shown in Figure 5.
Blood in urine did not inhibit mRNA target detection but rather added its own “signal” (originating from blood-derived cellular mRNA) to that of the urinary exosomes. For all mRNA analytes, Ct values significantly decrease indicating several fold increase in mRNA levels due to blood-derived mRNA co-purifying with exosomal RNA. However, once the urine specimens are centrifuged (which is an obligatory part of the exosome assay, as was described above), the bulk of blood cells are successfully depleted and Ct values generated are very similar to normal urine specimens. Spiking in serum instead of blood confirmed the results - assay interference is coming from blood-derived components that can be successfully removed by centrifugation.
In case blood cells are lysed, the contents obviously cannot be easily spotted or removed from urine; however such RNA is rapidly degrading in urine that contains high levels of RNases, and thus interference with assays should be minimal – if any.
Overall, centrifugation of the urine samples ensures depletion of blood-derived cells and large debris, and consistent, reproducible results. As a precaution, urine samples with visible hematuria should be excluded for this molecular test.
Solid organ transplantation has made tremendous progress in the last decade- aided by imaging techniques, donor-recipient human leukocyte antigen matching, and immunosuppressive therapy. In the United States alone, more than 40000 organ transplants are performed annually, with kidney, liver, heart, and lung being among the most common.
Today, major challenges for transplantation are improving long-term graft viability and preserving patient quality of life. Allograft rejection represents the greatest risk of transplant failure among patients. Early identification of subclinical injury, and the subsequent differentiation of injury type, could enable earlier clinician intervention and provide opportunities for personalized treatment. Further, minimally invasive methods of post-transplant monitoring could improve patients’ quality of life while serving as a rejection screening tool and adjunct to histopathology.
One of the more promising discoveries in recent years has been the use of exosomal mRNA signatures to determine allograft health[14]. Urine has long been known as a valuable source of molecules serving as diagnostic markers for renal disease, and urinary exosomes have been shown to be effective screening tools for kidney allograft rejection. With herein demonstrated mRNA stability and overall robustness, urinary exosomal assays could represent a non-invasive and less burdensome approach to monitor graft health and also potentially open the door for at-home sample collections.
To conclude, we characterized molecular assay utilizing urine-derived exosomes. This included stability of urine samples upon transportation from the point of collection to a centralized testing facility, storage of urine at different conditions upon receipt till the point molecular assay is performed, upfront processing, and effect of various interference substances on the downstream qPCR assay. mRNA from urine-derived exosomes was shown to be stable across a broad range of conditions and produced accurate results when analyzed via qPCR assay for kidney allograft rejection. We identified the most optimal conditions for every step of the process, ensuring preanalytical sample integrity and robust qPCR results.
Exosomes are nano-sized extracellular vesicles with nucleic acid and protein cargo, actively secreted by all cells within human body, and found in abundance in all body fluids, including urine. These extracellular vesicles have tremendous potential for next generation diagnostics, theoretically enabling noninvasive assessment of organ and tissue function via liquid biopsy analysis.
Recently, feasibility of an exosomal molecular test was demonstrated for post-organ transplant monitoring: analysis of urine-derived exosomal mRNA cargo allowed early detection of kidney allograft rejection. Taking into account the novelty of this approach, urine and in particular extracellular vesicles with their diverse RNA cargo have to be better characterized to ensure robustness of this molecular assay.
We further studied urine-derived exosomes and their mRNA content as a highly promising diagnostic modality. This included stability studies of urine samples and exosomal mRNA upon transportation from the point of collection to a centralized testing facility, short-term storage of urine at different conditions upon receipt till the point molecular assay is performed, and effects of various potentially interfering substances on the downstream quantitative polymerase chain reaction (qPCR) assay.
The urine specimens were stored at various temperatures and conditions and pre-processed in different ways. Next, samples were passed through the columns to capture all extracellular vesicles, the vesicles were lysed to release their content and the exosomal RNA was purified on the mini-columns, reverse transcription was performed, next pre-amplification, followed by a qPCR analysis for a panel of mRNA markers.
To ensure exosomal RNA integrity, the harvested urine specimens should be shipped refrigerated, by overnight delivery. Urine can next be stored at the test site for up to 1 wk at 4 °C, and long term should be frozen at -80 °C. Urine specimens must be centrifuged at low G-force to deplete cells and debris, to ensure consistent top results in downstream molecular assays. All commonly used medications (tacrolimus, cyclosporin A, mycophenolic acid, everolimus, sirolimus, ascomycin, teriflunomide) were tested and confirmed that they do not cause assay interference.
mRNA from the urine-derived exosomes was proven to be stable across a broad range of conditions and produce robust results in molecular post-transplant monitoring assays. We identified optimal conditions for every step of the workflow, ensuring pre-analytical sample integrity and robust downstream qPCR results.
Exosomes and in particular their mRNA cargo have the potential to revolutionize post-transplant monitoring, and detect early rejection events for kidney as well as other allografts- based on molecular analysis of urine, saliva and other body fluids.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Transplantation
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhao CF, China S-Editor: Liu JH L-Editor: A P-Editor: Zhang YL
| 1. | Gwinner W. Renal transplant rejection markers. World J Urol. 2007;25:445-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 2. | Yilmaz S, Yilmaz A, Häyry P. Chronic renal allograft rejection can be predicted by area under the serum creatinine vs time curve (AUCCr). Kidney Int. 1995;48:251-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Knight SR, Thorne A, Lo Faro ML. Donor-specific Cell-free DNA as a Biomarker in Solid Organ Transplantation. A Systematic Review. Transplantation. 2019;103:273-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 4. | Burnham P, Khush K, De Vlaminck I. Myriad Applications of Circulating Cell-Free DNA in Precision Organ Transplant Monitoring. Ann Am Thorac Soc. 2017;14:S237-S241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 5. | Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4254] [Cited by in RCA: 4120] [Article Influence: 179.1] [Reference Citation Analysis (0)] |
| 6. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8246] [Cited by in RCA: 9798] [Article Influence: 544.3] [Reference Citation Analysis (0)] |
| 7. | Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1477] [Article Influence: 113.6] [Reference Citation Analysis (0)] |
| 8. | Schageman J, Zeringer E, Li M, Barta T, Lea K, Gu J, Magdaleno S, Setterquist R, Vlassov AV. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int. 2013;2013:253957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 142] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 9. | Zeringer E, Li M, Barta T, Schageman J, Pedersen KW, Neurauter A, Magdaleno S, Setterquist R, Vlassov AV. Methods for the extraction and RNA profiling of exosomes. World J Methodol. 2013;3:11-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 68] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (2)] |
| 10. | Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, Breakefield XO, Skog JK. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466-477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 557] [Article Influence: 139.3] [Reference Citation Analysis (0)] |
| 11. | Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 465] [Cited by in RCA: 630] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 12. | Li M, Rai AJ, DeCastro GJ, Zeringer E, Barta T, Magdaleno S, Setterquist R, Vlassov AV. An optimized procedure for exosome isolation and analysis using serum samples: Application to cancer biomarker discovery. Methods. 2015;87:26-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 13. | Li M, Zeringer E, Barta T, Schageman J, Cheng A, Vlassov AV. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos Trans R Soc Lond B Biol Sci. 2014;369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 283] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 14. | El Fekih R, Hurley J, Tadigotla V, Alghamdi A, Srivastava A, Coticchia C, Choi J, Allos H, Yatim K, Alhaddad J, Eskandari S, Chu P, Mihali AB, Lape IT, Lima Filho MP, Aoyama BT, Chandraker A, Safa K, Markmann JF, Riella LV, Formica RN, Skog J, Azzi JR. Discovery and Validation of a Urinary Exosome mRNA Signature for the Diagnosis of Human Kidney Transplant Rejection. J Am Soc Nephrol. 2021;32:994-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |