Published online Jun 20, 2023. doi: 10.5662/wjm.v13.i3.46
Peer-review started: March 22, 2023
First decision: April 17, 2023
Revised: May 2, 2023
Accepted: May 31, 2023
Article in press: May 31, 2023
Published online: June 20, 2023
Processing time: 89 Days and 19.3 Hours
Despite the development of newer oncological treatment, the survival of patients with pancreatic cancer (PC) remains poor. Recent studies have identified exosomes as essential mediators of intercellular communications and play a vital role in tumor initiation, metastasis and chemoresistance. Thus, the utility of liquid biopsies using exosomes in PC management can be used for early detection, diagnosis, monitoring as well as drug delivery vehicles for cancer therapy. This review summarizes the function, and clinical applications of exosomes in cancers as minimally invasive liquid biomarker in diagnostic, prognostic and therapeutic roles.
Core Tip: The determination and identification of biomarkers using liquid biopsy can enable the early detection, monitoring, therapeutic interventions, risk of relapse, therapeutic targets and identification of resistance mechanisms in pancreatic cancer (PC). There has been a recent interest in use of exosomes as biomarker in PC management. Exosomes loaded with multiple diagnostic molecules can be isolated from different body fluids and can be used for making the exosome markers-based liquid biopsy more attractive for initial tumor detection, monitoring, and prognostic assessment of PC.
- Citation: Anoop TM, Basu PK, Chandramohan K, Thomas A, Manoj S. Evolving utility of exosomes in pancreatic cancer management. World J Methodol 2023; 13(3): 46-58
- URL: https://www.wjgnet.com/2222-0682/full/v13/i3/46.htm
- DOI: https://dx.doi.org/10.5662/wjm.v13.i3.46
Pancreatic cancer (PC) is associated with poor survival outcome with a 5-year survival of 5%-10%, and a short median survival of 6-8 months after cancer diagnosis[1]. Most individuals diagnosed with advanced disease are symptomatic whereas early stages of the cancer are generally asymptomatic and often undiagnosed. Hence diagnosis is often made after dissemination. Surgery is the only curative treatment[2]. Regrettably, a large number of patients present with unresectable or metastatic disease at the time of diagnosis. Early detection of PC is essential for treatment with curative intent, typically by surgical resection in combination with neoadjuvant or adjuvant chemotherapy and chemo radiation. The majority of patients can have local recurrence or systematic metastasis even after resection. Screening methods for PC often relies on carbohydrate antigen 19-9 (CA 19-9). CA 19-9 demonstrates relatively low sensitivity and specificity in diagnosing PC[3]. Hence a diagnostic test with high sensitivity and specificity and capable of distinguishing PC at early stages from benign diseases is highly recommended. Table 1 shows common circulating biomarkers for PC[4-19]. Comparison of usefulness of various Liquid biopsies used in PC is shown in Table 2[20-30].
| Biomarker | Type | Role in pancreatic cancer |
| CA19-9 | Protein | Widely used biomarkers to aid in the diagnosis[4] |
| Poor screening tool in asymptomatic patients | ||
| Elevated in many benign gastrointestinal conditions as well as other malignancies, including pancreatitis, cirrhosis, cholangitis, and colorectal cancer[5] | ||
| 5%-10% of the caucasian population possesses a Lewis a-/b- genotype and thus does not express CA19-9 | ||
| CEA | Glycoprotein | Elevated across several cancers[6] |
| Non specific | ||
| Inferior sensitivity of CEA compared to ca19-9[7] | ||
| CA125 | Glycoprotein | Associated with ovarian cancer, CRC and cholangiocarcinoma[8] |
| Superiority to CA19-9 in predicting resectability of PC, along with correlating with metastasis-associated disease burden | ||
| Anti-MUC1 antibody | Antibody | Anti-MUC1 antibody assays showed a sensitivity and specificity of 77% and 95%, respectively, in discriminating pancreatic cancer from pancreatitis[9] |
| CTCs | Tumour cells | CTCs had moderate diagnostic value in PC[10] |
| Several studies have demonstrated isolation of CTCs regardless of stage among localized, locally advanced, or metastatic patients | ||
| Conflicting evidence on CTC positivity is correlated with survivability | ||
| In ombination with CA19-9, it was reported to have a superior sensitivity and specificity of 97.8% and 83.3% respectively, compared to when used in isolation[11] | ||
| The presence of CTCs in 54/72 patients with confirmed PDAC (sensitivity = 75.0%, specificity = 96.4%, AUROC = 0.867, 95%CI: 0.798-0.935, and P < 0.001)[12] | ||
| A cut-off of ≥ 3 CTCs in 4 mL blood could differentiate between local/regional and metastatic disease (AUROC: 0.885; 95%CI: 0.800-0.969; and P < 0.001) | ||
| cfDNA | DNA | Plasma ctDNA quantification of hot-spot mutations in KRAS and GNAS are useful in predicting tumor burden in patients diagnosed with PC[13] |
| Digital PCR provided accurate tumor-derived mutant KRAS detection in plasma in resectable PC and improved post-resection recurrence prediction compared to CA19-9[14] | ||
| Detection of plasma cfDNA mutations and copy number alterations may be helpful in pancreatic cancer prognosis and diagnosis | ||
| Its sensitivity and specificity in identification of clinically relevant KRAS mutations was 87% and 99% respectively[15] | ||
| Cell-free RNA | RNA | Higher expression of lncRNA MALAT1 has been shown to correlate with poorer PDAC survival[16] |
| Several microRNAs have also been associated with PDAC (i.e., miR-21 and miR-155), and correlate with tumor stage or prognosis[17] | ||
| EVs | Exosomes | KRAS G12D mutations were identified in 7.4% of control patients, 67% of localized PDAC, 80% of locally advanced PDAC, and 85% of metastatic PDAC patients[18] |
| GPC1 EVs could be detected in both pancreatic precursor lesions and pancreatic cancer, and could distinguish between any evidence of malignancy and healthy patients with an AUC of 1 (100% sensitivity, 100% specificity)[19] | ||
| miRNA isolated from EVs revealed a cocktail of miRNAs (miR-1246, 4644, 3976, 4306) upregulated in 83% of pancreatic cancer derived EV | ||
| Glypican-1 exosomes are a potential biomarker for PC |
| Item | CTC[20-22]
| Ct DNA[21,23,24]
| Exosomes[20,25-28]
| CA 19-9[20,21,28-30]
|
| Origin | Viable tumor cells | cfDNA, viable tumor cells, CTCs | DNA, proteins, lipids, RNAs metabolites, and tumor cells | Ductal cells in the pancreas, biliary system, and epithelial cells in the stomach, colon, uterus, and salivary glands |
| Samples used | Plasma | Frozen plasma, urine and other biofluids | Frozen plasma, urine and other biofluids | Plasma |
| Methods | CellSearch, MACS, Dynabeads, microfluidic, SE-iFISH, CD45/CEP8/DAPI staining-FISH, anti-EpCAM Portal-vein blood | Real-time quantitative PCR, digital PCR, droplet digital PCR, next-generation sequencing; commercial liquid biopsy platforms: GuardantTM (breast, colon, and lung cancers and multi-cancer detection) FoundationOne® (multi-cancer detection); signateraTM (colorectal cancer), Galleri (multi-cancer detection), CancerSEEK (multi-cancer detection), TempusTM (multi-cancer detection), Caris (bioinformatics testing of both circulating DNA and RNA) | Ultracentrifugation, ExoChip, precipitation, size-based isolation immunoaffinity-based isolation microfluidics-based isolation | Radio immuno assay |
| Mutation analysis | Yes | Yes | Yes | No |
| Drug delivery vehicle | No | No | Yes | No |
| Sensitivity | 76.0% | 65.0% | 50.0%-85.0% | 78.2% |
| Specificity | 68.0% | 75.0% | 90.0% | 82.8% |
| Usage in clinics | Diagnosis of PDAC, prognosis/prediction of PDAC | Diagnosis of PDAC; monitoring treatment efficacy; monitoring of disease progression | Diagnosis and prognosis of PDAC; prognosis/prediction of PDAC | Combining ct DNA with CA 19-9 levels could improve diagnostic sensitivity to 98%, and specificity to 97%; monitoring treatment efficacy; monitoring of disease progression |
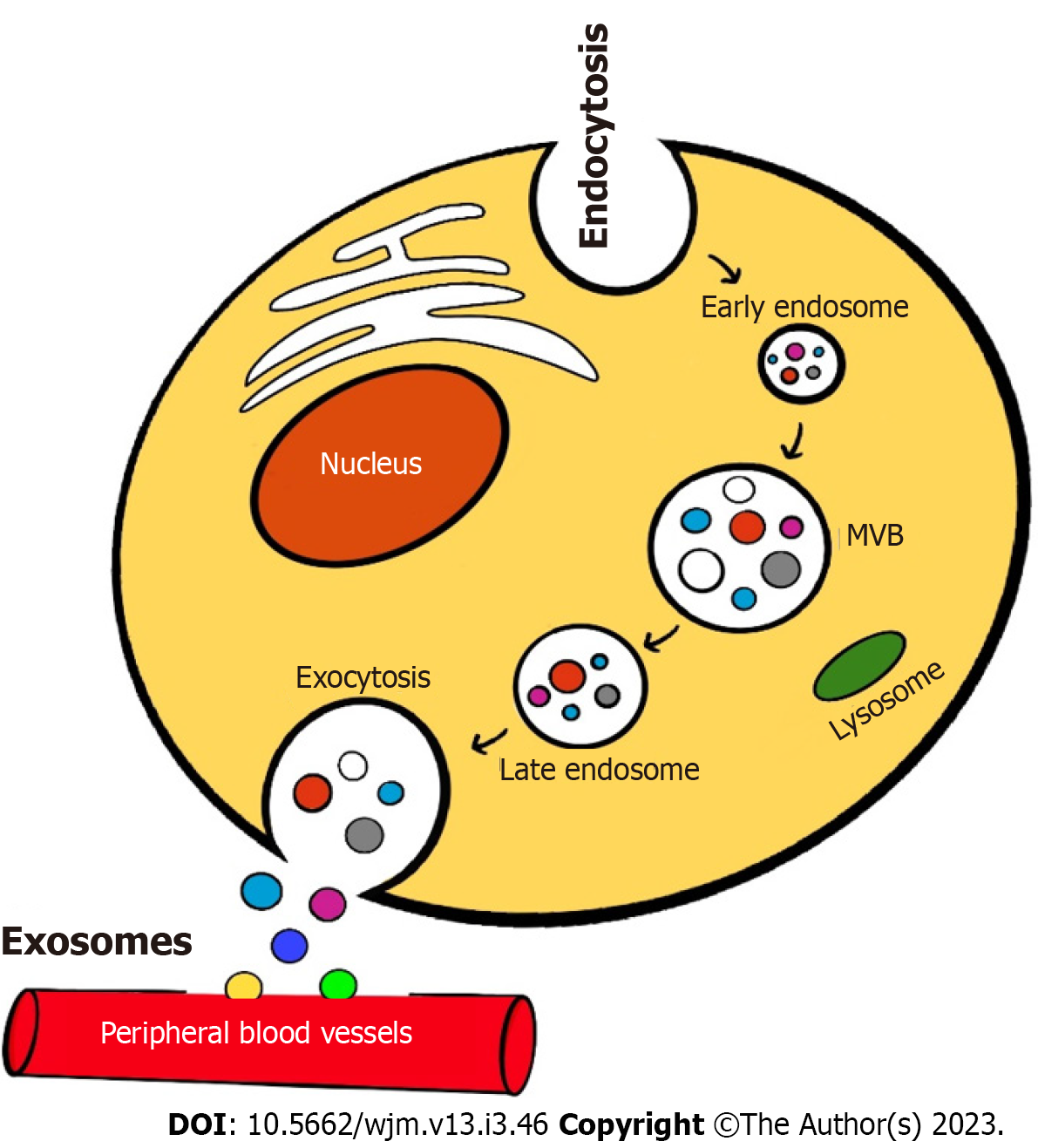
There is an emerging role of molecular profiling of liquid biopsies for cancer diagnosis and prognostication. Extracellular vesicles (EVs) derived from various body fluids and serum. There are four subclasses of EV based on different sizes like Exosomes (30-150 nm), Oncosomes (100-1000 nm), Ectoderms (100-1000 nm) and Apoptotic bodies (200-2000 nm). Exosome or Exosomes derived proteins, etc. are believed to serve as reliable molecular biomarkers. The circulating vesicles in the blood that originate from tumor cells contains immense proteomic and genetic information to monitor cancer progression, metastasis, and drug efficacy[31-33]. Exosomes were originally introduced during the culture of sheep reticulocytes in vitro by Johnstone et al[34]. Exosomes are EVs that are endosomal in origin with a diameter of 40-160 nm (average, 100 nm). The formation of cancer cell derived exosomes is depicted in Figure 1. Initially, exosomes are formed by inward invagination of plasma membrane to form an early endosome. These endosomes form nano-sized vesicles resulting in formation of multi vesicular body (MVB) that contain intraluminal vesicles which contain cytoplasmic components including various nucleic acids and soluble proteins[35]. These intra luminal vesicles are released to the extracellular environment by fusing the MVBs with the plasma membrane. Then with the help of exocytosis, exosomes are released in to circulation.
Exosomes contains many molecules like heat shock proteins, RNAs, DNAs, GTPase, CD63, CD81, CD9, CD82, cholesterol, sphingomyelin, and ceramides. Exosomes facilitate both the transport of essential substances like nucleic acids and proteins into various recipient cells and the communications between cells. The main sources of exosomes are plasma, serum, urine, bile, saliva and breast milk. The secreted exosomes have various cellular functions in cell-to-cell interactions and might be pivotal in the occurrence and development of tumour progression and metastasis[36]. Exosomes have definite role in inflammation, coagulation, and embryo implantation in pregnancy. However, cancer cells are capable of secreting 10 times than normal cells. Hence tumour derived exosomes can provide vast information on cancer. Furthermore, exosomes are potential surrogates of the original cells, hence they are useful for understanding cell biology.
Oncosomes are tumor derived cells and they contain different oncogenic molecules that can modify the cells to encourage cancer growth. Tumor cell-secreted exosomes are responsible for paracrine signalling during tumor progression, tumor-stromal interactions, proliferative pathway activation, and immunosuppression[37]. Tumor derived exosomes enters the cells by a variety of methods, depending on the cells that secrete them and the target cells. Metastatic breast cancer derived exosomes use transcytosis to cross the brain endothelial cells, while the “CDC42-dependent endocytic pathway” was utilized to enter astrocytes during brain metastasis[38].
There are increasing evidence that exosomes are involved in the pathogenesis of development of pancreatic inflammation as well as related cancer initiation. Repeated episodes of pancreatitis are a strong risk factor which can eventually increase the risk of PC. The pathogenesis and evolution of many pancreases precancerous conditions, including diabetes mellitus and pancreatitis, have been linked to crucial involvement of exosomes[39]. Exosomes can participate in promoting the transformation of various precancerous like intraductal papillary malignant neoplasm to PDAC. Exosomes are a key factor in initiating angiogenesis, cell migration, and epithelial-mesenchymal transition[40]. Cancer-associated fibroblasts, tumor-associated macrophages and pancreatic stellate cells can promote exosomes, that could promote growth, proliferation, drug resistance, epithelial mesenchymal transition, migration, invasion and metastasis of PC[41]. Interestingly, exosomes initiated from PC cells contains tumor suppressor components which can inhibit the cancer cell proliferation and this could open the pandora box of potential therapeutic value of exosomes[42]. Exosomes promote cancer cell proliferation and initiate metastasis by delivering carcinogenic proteins, cytokines, adhesion molecules and miRNA. Thus, initiate proliferation of tumour by activation of different pathways like phosphoinositide 3-kinase/AKT serine/threonine kinase 1 (Akt) and mitogen-activated protein kinase pathways[43]. The features like weight loss and new-onset diabetes are characteristics of the paraneoplastic effect of PC which mostly precede the diagnosis of the PC. The biological reason of PC-associated diabetes is due to exosomal adrenomedullin, endoplasmic reticulum stress which may result in β-cell dysfunction and diabetes[44]. There is emerging evidence that suggest role of exosomemediated immunosuppression in PC. The exosomes have clear role in communications between tumor and immune cells and supposed to have a dynamic role in tumor immunity regulation. Gemcitabine chemotherapy is considered a standard treatment for PC either in combination or monotherapy, based on evidence from many studies which shown a better survival rate and more clinical benefits with median overall survival (OS) of 5 mo to 7 mo[45]. Most of patients with PC ultimately present with rapid disease progression even following chemotherapy with gemcitabine. Tumor derived exosomes can induce the progression of chemotherapy resistance in cancer cells. Chemotherapy agents could also be secreted from the extracellular matrix by exosomes is another reason for chemotherapy resistance. When exposed to gemcitabine, exosomal CAFs which are inherently insensitive to gemcitabine may also leads to chemotherapy resistance. CAF exosome secretion inhibition could decrease proliferation and drug resistance[46].
There are various methods to isolate and characterize exosomes based on their physical and chemical properties. Most popular methods are ultracentrifugation, size exclusion chromatography, magnetic activated cell sorting, membrane filtration and various commercial kits[47]. Western blotting and flow cytometry can be used to analyze and detect exosome markers. Transmission electron microscopy and nanoparticle tracking analysis are other methods to detect exosome.
Liquid biopsy to analyze exosome biomarkers could guide the diagnosis and prognosis of PC. Therefore, the identification of reliable predictive biomarkers for diagnosis and prognosis is an unmet need in PC management. The most common methods for isolation for exosome are summarized in Table 3[48-56] and quantifying methods for exosome are presented in Table 4[48-64]. Methods like Western blotting and enzyme linked immunosorbent assays needs large amounts of sample and extensive technical steps for detection. The comparison of various isolation methods used for exosomes are given in Table 5[65-76].
| Method | Sample volume | Time | Ref. |
| Ultracentrifugation | Low | Approximately 5 h | [48,49] |
| Density-gradient | Low | Approximately 5 h | [50] |
| Nanopillar | 30 μL | Approximately 10 min | [51] |
| Acoustic-based | 0.4-0.7 μL/min | < 30 min | [52] |
| Inertial lift force-based | 70 μL/min | > 4 h | [53] |
| Surface-modified | 4-16 μL/min | < 1 h | [53-55] |
| Nanoshearing | Not mentioned | < 3 h | [56] |
| Method | Size range | Specificity | Time | Ref. |
| Nanoparticle tracking analysis | 10 nm-2 μm | Immunoaffinity | < 1 h | [48] |
| Dynamic light scattering | 10 nm-8 μm | Size | < 1 h | [57] |
| Electron microscopy | 10 nm | Size | < 1 h | [58,59] |
| Nanopore | > 10 nm | Size | < 1 h | [60,61] |
| Magnetic resonance | Wide range | Immunoaffinity | < 10 min | [62] |
| Electrochemical and plasmonic | Depends on binding | Immunoaffinity | < 10 min | [63,64] |
| Conventional isolation of exosomes | ||||
| Methods | Advantages | Disadvantages | Clinical use | Ref. |
| Ultracentrifugation | Widely used; high purity; protein and RNA components are not affected | Highly labour intensive; time-consuming; yields are typically low extensive training of personnel needed; expensive; inappropriate for the extraction of exosomes from a small amount of serum samples | Functional study of exosomes | [65,66] |
| Ultrafiltration | High yield; simple; less time-consuming; do not require the use of special equipment | Low purity, clogging of pores | Study of sample concentration; used in combination with other methods | [67] |
| Precipitation | Widely used; economical | Co-isolation of non-EV particles | For studies with very low purity requirements that do not require omics studies | [68] |
| Size exclusion chromatography, OR, and gel filtration | Fast, reliable, and inexpensive; maintain the biological activity and integrity of exosomes; high purity | Nanoscale contaminants like lipoproteins; extensive laboratory equipment requirements | Suitable for exosome research in those requiring high purity, omics, and large volume samples | [69] |
| Immunoaffinity capture | Convenient; not affected by exosome size; no need for expensive instruments | Expensive; low capacity; low yields | Suitable for the Separation of specific exosome subgroups | [70] |
| Emerging isolation methods | ||||
| Stirred ultrafltration | Do not rely on equipment; less time consuming; reduces the destruction of exosomes during the process | Moderate purity of isolated exosomes; loss of exosomes during the process | Isolating exosomes from culture supernatant of bone marrow mesenchymal stem cells | [71] |
| ExoTIC (exosome total isolation chip) | Simple, easy-to-use, modular, and facilitates high-yield and high-purity EV isolation from biofluids | Special equipment requirements; lack of tests on clinical samples | Efficiently isolate EVs from small sample volumes; EV-based clinical testing from fingerprick quantities (10-100 μL) of blood | [72,73] |
| 3D ZnO Nanoarrays | Multifunction; high sensitivity; downstream analysis is possible; enhance the capture of exosomes at a high flow rate | Relatively expensive | Widely used in biosensing and analysis aspects, powerful tools for effective purification and molecular analysis of exosome | [74,75] |
| Nano plasmon-enhanced scattering | Rapid, high-throughput, sensitive, and specifc method for the detection of exosomes from trace samples depending on the amount of scatter area, based on calculation of the proportion of the area that contains scattered light | High reagent cost; complex statistical tools; low capacity | Uses antibodies against the cellular markers CD81, CD63, and CD9, which are enriched on most exosome membranes | [76] |
At present scenario, early diagnosis of PC is very difficult and most are diagnosed at late stage. Mostly CT imaging are used for diagnosis and treatment. CA19-9 which is used in clinical practice has a low specificity and poor ability to distinguish benign pancreatic diseases from PC[77]. Thus, the search for novel early diagnostic markers is a concern for PC diagnosis and treatment. Exosomes are excessively produced in excess by malignant tumours. They also carry information about the tumour genetics and microenvironment, which determines its behaviour and its prognosis[78]. Circulating biomarkers are non-invasive and inexpensive for monitoring disease[79]. The circulating molecular tumor markers are circulating tumor cells, cell-free DNA, cell-free RNA, circulating tumor proteins, and exosomes. When compared to circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA), exosomes are considered as the best diagnostic biomarkers in PC with a sensitivity of 93% and a specificity of 92%[80]. The ctDNA group had similar specificity 0.92 (95%CI 0.88-0.95) but lowest sensitivity. Thus, ctDNA was useful for the diagnosis of PC rather than screening. Whereas CTCs exhibits medium sensitivity and lowest specificity compared to others. The sensitivity, specificity and AUC of ctDNA were 0.6400, 0.9200, and 0.9478 respectively. Glypican 1 (GPC1) is expressed in the serum of patients with PC but not in benign pancreatic disease. Melo et al[19] described that Glypican-1 identifies cancer exosomes and could detects early PC. It was described that GPC1+ circulating exosomes could be used as prognostic biomarker in pre- and post-surgical patients. The GPC1+ circulating exosomes could distinguish PC precursor lesions from healthy individuals and benign pancreatic disease. The circulating GPC1+ exosomes levels were higher in PC precursor lesions than the levels in the healthy donor group and benign pancreatic diseases. There is supporting evidence that there is a potential use in early detection of pancreas cancer. Melo et al[19] study shows that circulating GPC1+ exosomes exhibit a sensitivity of 100% and specificity of 100%; with a positive predictive value of 100% and a negative predictive value of 100% compared to CA 19-9 which was inferior in distinguishing between pancreas cancer and healthy controls.
The functional role of MicroRNAs has a greater opportunity in developing both prognostic and diagnostic markers. MiRNA -based biomarkers can help in the early diagnosis of disease. A recent study evaluated the expression patterns of miR-21, miR-155, miR-17-5p, and miR-196a in circulating exosomes as biomarkers for PC. The expression profile of miR-17-5p and miR-21 were increased in PC patients, The increased expression of miR-17-5p was seen in unresectable pancreatic patients[81]. miR-155 and miR-21 are over-expressed in PDAC, and can distinguish PC from benign lesions[82]. Upregulated miR-221-3p and miR-212 is associated in PDAC and they are responsible for cancer proliferation in PDAC cells. miR-128 expression is decreased in PC while non-cancerous tissue has a normal level of miR-128. Gemcitabine resistance is associated with downregulation of miRNA200b, miRNA-200c, let-7b, let-7c, let-7d, and let-7e. miR-155 and miR-1246 also have been related to gemcitabine resistance. There are miRNA that function as tumor suppressors in pancreatic ductal cancer like miR-99b, miR-100, miR-99a, miR-34a, miR-148a, miR-200a, miR-200b, and miR-200c. MicroRNAs expression profiles showed that miR-143, miR-29c, miR-148b, miR-150, and miR-96, were present in PDAC and chronic pancreatitis whereas miR-196b, miR-203, miR-196a, miR-210, miR-222, miR-210, miR216, miR-375, and miR-217, were expressed only in pancreatic carcinoma[83]. miR-190, miR-196a, miR-222, miR-15b, miR200b, miR-95, and miR-221 are elevated in pancreatic adenocarcinoma[84]. Nakamura et al[85], developed an exosome-based signature for non-invasive and early detection of PDAC. Previous research studies showed that serum Ephrin type-A receptor 2 in exosomes (Exo-EphA2) was expressed highly in PC cells. A study by Wei et al[86] the evaluated role of serum Exo-EphA2 as a potential diagnostic biomarker in PC. Serum Exo-EphA2 were higher in PC than in non-cancer pancreatic disease. Exo-EphA2 in combination with CA 19-9 was more useful to discriminate early stage of PC from non-cancer pancreatic disease. Alkaline phosphatase placental-like 2 presents in PC EVs has a potential application in liquid biopsy-based diagnostic tests. Shin et al[87] developed ALPPL2 direct and sandwich aptamer-linked immobilized sorbent assay for EVs, which could sensitively and specifically detect membrane protein,17 could be a potential biomarker for early diagnosis of PDAC. Recently, there are reports of exosomal migration inhibitory factor (MIF) may be an attractive sensitive biomarker for PC. MIF is an immunostimulatory cytokine associated with tumorigenesis. Costa-Silva et al[88] reported that the pancreatic exosomes are capable of inducing premetastatic niche formation in liver. They demonstrated that the exosome education-induced liver metastasis was abolished by silencing of exosomal MIF. The combined use of exosomal glypican-1 and MIF is a promising tool to identify very early stages of PC. The potential of MIF as a target for the treatment of PDAC should be explored in future.
The level of circulating Exo-EphA2 was higher in PC patients when compared to that of healthy controls, suggesting it could be a diagnostic and prognostic marker for PC. In a study by Wei et al[86] found that high expression of Exo-EphA2 in PC was associated with shorter OS. Exosomal KRAS mutations seems better than CA 19-9 Levels for the prognostic surveillance in PDAC[17]. A study by Tsuchida et al[89] revealed that KRAS mutation detected at baseline with a mutation frequency above 5% indicated poor clinical outcome following monitoring in the treatment course of patients with metastatic PDAC. Costa-Silva et al[88] found that MIF was markedly higher in exosomes from stage I PDAC patients who later developed liver metastasis. Thus, it is suggested that higher exosomal MIF may be a prognostic marker for the development of PDAC liver metastasis. Potential role of exosomal biomarkers for prognosis evaluation in PDAC was evaluated in a systematic review and meta-analysis, involving eleven studies comprising 634 patients and seen that detection of positive exosomal biomarkers increased risk of mortality and progression across disease stages. Positive exosomal biomarkers preoperatively had higher risk of mortality in resectable stages than positive exosomal biomarkers in unresectable stages[90].
The better understanding of the prognostic role of miRNAs in PC can be done by profiling miRNAs at different stages of cancer. In a study by Takahasi et al[91], authors suggest that plasma exosomal miR-451a may be useful to predict recurrence and prognosis in PDAC patients. The miR-451a had a significant association with tumor, stage and showed the highest upregulation in the stage II patients who showed recurrence after surgery. In a retrospective clinical study by Namkung et al[92] comprising 200 pancreatic ductal adenocarcinoma tissue samples, miRNA-574-5p, miRNA-1244, miRNA-145, miRNA-328, miRNA-26b, and miRNA4321 showed association with OS and disease-free survival. Poor survival outcomes have been seen in PDAC with lower expression of miR-183 and miR-34a as well as high expression of miR-1290, miR-155, miR-203, miR-222, and miR-10b[93]. Similarly, Microarray-based expression profiling of miRNAs derived from exosomes study revealed that miR-451a was the highest upregulated miRNA in stage II patients who developed recurrence after surgery. It was seen that survival rates of the high exosomal miR-451a patients were significantly worse than those of the low miR-451a patients[94].
Currently, one of the most common biomarkers used for so long to monitor the therapeutic responses in PC is CA 19-9. Exosomes have a significant role in monitoring response to therapy and disease progression. Melo et al[19] clearly demonstrated that all 190 patients with PDAC serum had higher GPC1+ exosomes than healthy individuals and was an independent prognostic marker for disease specific survival. In view of the fact that CA19-9 is not a reliable marker that correlates with clinical evolution of PC, a combination of CA19-9 together with exosome derived GPC1 could be explored for treatment monitoring and disease progression. Besides early diagnosis and prognosis, clinical utility of exosome proteins is evolving for personalized and posttreatment disease monitoring[95]. Circulating exosomal PD-L1 is an attractive option in disease monitoring. Recently, Chen et al[96] study explains the rationale for the application of exosome PD-L1 as a predictor for anti-PD-1 therapy.
Currently, innovators are exploring the utility of exosomes for biomedical applications. Many advanced drug delivery systems that used to deliver various anticancer and antiviral agents explore the use of polymeric nanoparticles and liposomes to encapsulate drug and thus utilize for drug delivery.
Exosomes can be used as therapeutic drugs carriers because of favourable bioavailability, biocompatibility, ability to penetrate biological membranes and immunogenicity[97]. Exosomes can be used as transporters, therapeutic targets and therapeutic drugs.
Due to the favorable bioavailability and biocompatibility with the characteristics of exosomes, there appears a greater future of exosomes used either as parental exosomes or artificially modified exosomes for drug delivery vehicle. To avoid systemic toxicity, drugs can be encapsulated in exosomes and transferred to target cells[98]. Exosomes possess better biocompatibility as drug carriers. It is generally considered that injected exosomes shed from endogenous cells are tolerated with minimal immune reaction. The cargos can be delivered into the tumor microenvironment with the utility of exosomes[99]. Kamerkar et al[100] studied modified exosomes for cancer prevention and treatment and revealed that exosomes had a longer retention time in the circulation. Engineered exosomes specialized for malignant KRAS G12D were more successful in targeting oncogenic KRAS. Recent evidence suggests that safety and efficacy of exosomes in treating PC is not far. Exosome-based therapies for cancers have been developed due to the easy permeability of the exosome membrane, low toxic side effects and low immunogenicity. Paclitaxel -loaded exosomes have shown a great potential for delivery of che
Even though there are several limitations in implementing exosome analysis clinically, it is a promising diagnostic and therapeutic tool for PC. The role of exosomes in cancer treatment continues to evolve.
I would like to convey my gratitude to Malavika Anoop, Standard X of ST Thomas Central School, Mukkolakkal, Thiruvananthapuram, Kerala for her invaluable assistance in completing my digital art work for this article.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Feliu J, Spain; Gao W, China; Zhao CF, China S-Editor: Chen YL L-Editor: A P-Editor: Ju JL
| 1. | Rawla P, Sunkara T, Gaduputi V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J Oncol. 2019;10:10-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 893] [Cited by in RCA: 1520] [Article Influence: 253.3] [Reference Citation Analysis (1)] |
| 2. | Barros AG, Pulido CF, Machado M, Brito MJ, Couto N, Sousa O, Melo SA, Mansinho H. Treatment optimization of locally advanced and metastatic pancreatic cancer (Review). Int J Oncol. 2021;59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 3. | Poruk KE, Gay DZ, Brown K, Mulvihill JD, Boucher KM, Scaife CL, Firpo MA, Mulvihill SJ. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med. 2013;13:340-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 185] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 4. | Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol. 2012;3:105-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 352] [Reference Citation Analysis (3)] |
| 5. | Kriz D, Ansari D, Andersson R. Potential biomarkers for early detection of pancreatic ductal adenocarcinoma. Clin Transl Oncol. 2020;22:2170-2174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Zhang X, Shi S, Zhang B, Ni Q, Yu X, Xu J. Circulating biomarkers for early diagnosis of pancreatic cancer: facts and hopes. Am J Cancer Res. 2018;8:332-353. [PubMed] |
| 7. | Brand RE, Nolen BM, Zeh HJ, Allen PJ, Eloubeidi MA, Goldberg M, Elton E, Arnoletti JP, Christein JD, Vickers SM, Langmead CJ, Landsittel DP, Whitcomb DC, Grizzle WE, Lokshin AE. Serum biomarker panels for the detection of pancreatic cancer. Clin Cancer Res. 2011;17:805-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 190] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 8. | Liu L, Xu HX, Wang WQ, Wu CT, Xiang JF, Liu C, Long J, Xu J, Fu de L, Ni QX, Houchen CW, Postier RG, Li M, Yu XJ. Serum CA125 is a novel predictive marker for pancreatic cancer metastasis and correlates with the metastasis-associated burden. Oncotarget. 2016;7:5943-5956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 9. | Gold DV, Modrak DE, Ying Z, Cardillo TM, Sharkey RM, Goldenberg DM. New MUC1 serum immunoassay differentiates pancreatic cancer from pancreatitis. J Clin Oncol. 2006;24:252-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Martini V, Timme-Bronsert S, Fichtner-Feigl S, Hoeppner J, Kulemann B. Circulating Tumor Cells in Pancreatic Cancer: Current Perspectives. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 11. | Tjensvoll K, Nordgård O, Smaaland R. Circulating tumor cells in pancreatic cancer patients: methods of detection and clinical implications. Int J Cancer. 2014;134:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 101] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | Ankeny JS, Court CM, Hou S, Li Q, Song M, Wu D, Chen JF, Lee T, Lin M, Sho S, Rochefort MM, Girgis MD, Yao J, Wainberg ZA, Muthusamy VR, Watson RR, Donahue TR, Hines OJ, Reber HA, Graeber TG, Tseng HR, Tomlinson JS. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J Cancer. 2016;114:1367-1375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 125] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 13. | Okada T, Mizukami Y, Ono Y, Sato H, Hayashi A, Kawabata H, Koizumi K, Masuda S, Teshima S, Takahashi K, Katanuma A, Omori Y, Iwano H, Yamada M, Yokochi T, Asahara S, Kawakubo K, Kuwatani M, Sakamoto N, Enomoto K, Goto T, Sasajima J, Fujiya M, Ueda J, Matsumoto S, Taniue K, Sugitani A, Karasaki H, Okumura T. Digital PCR-based plasma cell-free DNA mutation analysis for early-stage pancreatic tumor diagnosis and surveillance. J Gastroenterol. 2020;55:1183-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Takai E, Totoki Y, Nakamura H, Morizane C, Nara S, Hama N, Suzuki M, Furukawa E, Kato M, Hayashi H, Kohno T, Ueno H, Shimada K, Okusaka T, Nakagama H, Shibata T, Yachida S. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci Rep. 2015;5:18425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 149] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 15. | Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 16. | Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 831] [Article Influence: 75.5] [Reference Citation Analysis (0)] |
| 17. | Allenson K, Castillo J, San Lucas FA, Scelo G, Kim DU, Bernard V, Davis G, Kumar T, Katz M, Overman MJ, Foretova L, Fabianova E, Holcatova I, Janout V, Meric-Bernstam F, Gascoyne P, Wistuba I, Varadhachary G, Brennan P, Hanash S, Li D, Maitra A, Alvarez H. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28:741-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 379] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 18. | Lorenzon L, Blandino G. Glypican-1 exosomes: do they initiate a new era for early pancreatic cancer diagnosis? Transl Gastroenterol Hepatol. 2016;1:8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2108] [Cited by in RCA: 2203] [Article Influence: 220.3] [Reference Citation Analysis (0)] |
| 20. | Raufi AG, May MS, Hadfield MJ, Seyhan AA, El-Deiry WS. Advances in Liquid Biopsy Technology and Implications for Pancreatic Cancer. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 21. | Yadav DK, Bai X, Yadav RK, Singh A, Li G, Ma T, Chen W, Liang T. Liquid biopsy in pancreatic cancer: the beginning of a new era. Oncotarget. 2018;9:26900-26933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 22. | Han L, Chen W, Zhao Q. Prognostic value of circulating tumor cells in patients with pancreatic cancer: a meta-analysis. Tumour Biol. 2014;35:2473-2480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 23. | Ma M, Zhu H, Zhang C, Sun X, Gao X, Chen G. "Liquid biopsy"-ctDNA detection with great potential and challenges. Ann Transl Med. 2015;3:235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 68] [Reference Citation Analysis (0)] |
| 24. | Yi X, Ma J, Guan Y, Chen R, Yang L, Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int J Cancer. 2017;140:2642-2647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 25. | Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, Asaoka T, Noda T, Wada H, Kawamoto K, Gotoh K, Takeda Y, Tanemura M, Mori M, Doki Y. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci Rep. 2017;7:42339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 212] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 26. | Ohuchida K, Mizumoto K, Kayashima T, Fujita H, Moriyama T, Ohtsuka T, Ueda J, Nagai E, Hashizume M, Tanaka M. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol. 2011;18:2381-2387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 27. | van den Boorn JG, Dassler J, Coch C, Schlee M, Hartmann G. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev. 2013;65:331-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 204] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 28. | Heredia-Soto V, Rodríguez-Salas N, Feliu J. Liquid Biopsy in Pancreatic Cancer: Are We Ready to Apply It in the Clinical Practice? Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 29. | Arneth B. Update on the types and usage of liquid biopsies in the clinical setting: a systematic review. BMC Cancer. 2018;18:527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 30. | Makler A, Asghar W. Exosomal biomarkers for cancer diagnosis and patient monitoring. Expert Rev Mol Diagn. 2020;20:387-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 31. | Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017;7:789-804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 870] [Cited by in RCA: 1387] [Article Influence: 173.4] [Reference Citation Analysis (1)] |
| 32. | Paolini L, Zendrini A, Di Noto G, Busatto S, Lottini E, Radeghieri A, Dossi A, Caneschi A, Ricotta D, Bergese P. Residual matrix from different separation techniques impacts exosome biological activity. Sci Rep. 2016;6:23550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 136] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 33. | Dreyer F, Baur A. Biogenesis and Functions of Exosomes and Extracellular Vesicles. Methods Mol Biol. 2016;1448:201-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 104] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 34. | Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412-9420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1577] [Cited by in RCA: 1990] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 35. | Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1075] [Cited by in RCA: 1383] [Article Influence: 125.7] [Reference Citation Analysis (0)] |
| 36. | Nuzhat Z, Kinhal V, Sharma S, Rice GE, Joshi V, Salomon C. Tumour-derived exosomes as a signature of pancreatic cancer - liquid biopsies as indicators of tumour progression. Oncotarget. 2017;8:17279-17291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 37. | Patel G, Agnihotri TG, Gitte M, Shinde T, Gomte SS, Goswami R, Jain A. Exosomes: a potential diagnostic and treatment modality in the quest for counteracting cancer. Cell Oncol (Dordr). 2023;1-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 38. | Chen H, Chengalvala V, Hu H, Sun D. Tumor-derived exosomes: Nanovesicles made by cancer cells to promote cancer metastasis. Acta Pharm Sin B. 2021;11:2136-2149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 39. | Ruze R, Song J, Yin X, Chen Y, Xu R, Wang C, Zhao Y. Mechanisms of obesity- and diabetes mellitus-related pancreatic carcinogenesis: a comprehensive and systematic review. Signal Transduct Target Ther. 2023;8:139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 40. | Sun W, Ren Y, Lu Z, Zhao X. The potential roles of exosomes in pancreatic cancer initiation and metastasis. Mol Cancer. 2020;19:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 41. | Zhang YF, Zhou YZ, Zhang B, Huang SF, Li PP, He XM, Cao GD, Kang MX, Dong X, Wu YL. Pancreatic cancer-derived exosomes promoted pancreatic stellate cells recruitment by pancreatic cancer. J Cancer. 2019;10:4397-4407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 42. | Beloribi S, Ristorcelli E, Breuzard G, Silvy F, Bertrand-Michel J, Beraud E, Verine A, Lombardo D. Exosomal lipids impact notch signaling and induce death of human pancreatic tumoral SOJ-6 cells. PLoS One. 2012;7:e47480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 43. | Liu H, Qiao S, Fan X, Gu Y, Zhang Y, Huang S. Role of exosomes in pancreatic cancer. Oncol Lett. 2021;21:298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Sagar G, Sah RP, Javeed N, Dutta SK, Smyrk TC, Lau JS, Giorgadze N, Tchkonia T, Kirkland JL, Chari ST, Mukhopadhyay D. Pathogenesis of pancreatic cancer exosome-induced lipolysis in adipose tissue. Gut. 2016;65:1165-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 186] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 45. | Ouyang G, Wu Y, Liu Z, Lu W, Li S, Hao S, Pan G. Efficacy and safety of gemcitabine-capecitabine combination therapy for pancreatic cancer: A systematic review and meta-analysis of randomized controlled trials. Medicine (Baltimore). 2021;100:e27870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 46. | Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36:1770-1778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 593] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 47. | Lan B, Zeng S, Grützmann R, Pilarsky C. The Role of Exosomes in Pancreatic Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 48. | Ko J, Carpenter E, Issadore D. Detection and isolation of circulating exosomes and microvesicles for cancer monitoring and diagnostics using micro-/nano-based devices. Analyst. 2016;141:450-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 167] [Article Influence: 18.6] [Reference Citation Analysis (1)] |
| 49. | Bobrie A, Colombo M, Krumeich S, Raposo G, Théry C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 428] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 50. | Zhang Z, Wang C, Li T, Liu Z, Li L. Comparison of ultracentrifugation and density gradient separation methods for isolating Tca8113 human tongue cancer cell line-derived exosomes. Oncol Lett. 2014;8:1701-1706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 51. | Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, Zhang JX, Liu X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip. 2013;13:2879-2882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 266] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 52. | Lee K, Shao H, Weissleder R, Lee H. Acoustic purification of extracellular microvesicles. ACS Nano. 2015;9:2321-2327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 380] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 53. | Dudani JS, Gossett DR, Tse HT, Lamm RJ, Kulkarni RP, Carlo DD. Rapid inertial solution exchange for enrichment and flow cytometric detection of microvesicles. Biomicrofluidics. 2015;9:014112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 54. | Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4900] [Cited by in RCA: 6160] [Article Influence: 513.3] [Reference Citation Analysis (0)] |
| 55. | Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 456] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 56. | Vaidyanathan R, Naghibosadat M, Rauf S, Korbie D, Carrascosa LG, Shiddiky MJ, Trau M. Detecting exosomes specifically: a multiplexed device based on alternating current electrohydrodynamic induced nanoshearing. Anal Chem. 2014;86:11125-11132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 204] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 57. | Berne BJ, Pecora R. Dynamic light scattering with applications to chemistry, biology, and physics, Courier Corporation. United Kingdom: Dover Publications, 2000. [DOI] [Full Text] |
| 58. | Lyons AB, Parish CR. Determination of lymphocyte division by flow cytometry. J Immunol Methods. 1994;171:131-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1358] [Cited by in RCA: 1369] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 59. | van der Pol E, Hoekstra AG, Sturk A, Otto C, van Leeuwen TG, Nieuwland R. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost. 2010;8:2596-2607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 403] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 60. | McLeod E, Dincer TU, Veli M, Ertas YN, Nguyen C, Luo W, Greenbaum A, Feizi A, Ozcan A. High-throughput and label-free single nanoparticle sizing based on time-resolved on-chip microscopy. ACS Nano. 2015;9:3265-3273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 61. | Platt M, Willmott GR, Lee GU. Resistive pulse sensing of analyte-induced multicomponent rod aggregation using tunable pores. Small. 2012;8:2436-2444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 62. | Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835-1840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 595] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 63. | Zhu L, Wang K, Cui J, Liu H, Bu X, Ma H, Wang W, Gong H, Lausted C, Hood L, Yang G, Hu Z. Label-free quantitative detection of tumor-derived exosomes through surface plasmon resonance imaging. Anal Chem. 2014;86:8857-8864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 201] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 64. | Elshafey R, Tavares AC, Siaj M, Zourob M. Electrochemical impedance immunosensor based on gold nanoparticles-protein G for the detection of cancer marker epidermal growth factor receptor in human plasma and brain tissue. Biosens Bioelectron. 2013;50:143-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 65. | Correll VL, Otto JJ, Risi CM, Main BP, Boutros PC, Kislinger T, Galkin VE, Nyalwidhe JO, Semmes OJ, Yang L. Optimization of small extracellular vesicle isolation from expressed prostatic secretions in urine for in-depth proteomic analysis. J Extracell Vesicles. 2022;11:e12184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 66. | Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, Nolte-'t Hoen EN, Piper MG, Sivaraman S, Skog J, Théry C, Wauben MH, Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1352] [Cited by in RCA: 1779] [Article Influence: 148.3] [Reference Citation Analysis (1)] |
| 67. | Kim K, Son T, Hong JS, Kwak TJ, Jeong MH, Weissleder R, Im H. Physisorption of Affinity Ligands Facilitates Extracellular Vesicle Detection with Low Non-Specific Binding to Plasmonic Gold Substrates. ACS Appl Mater Interfaces. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 68. | Kamei N, Nishimura H, Matsumoto A, Asano R, Muranaka K, Fujita M, Takeda M, Hashimoto H, Takeda-Morishita M. Comparative study of commercial protocols for high recovery of high-purity mesenchymal stem cell-derived extracellular vesicle isolation and their efficient labeling with fluorescent dyes. Nanomedicine. 2021;35:102396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 69. | Guo J, Wu C, Lin X, Zhou J, Zhang J, Zheng W, Wang T, Cui Y. Establishment of a simplified dichotomic size-exclusion chromatography for isolating extracellular vesicles toward clinical applications. J Extracell Vesicles. 2021;10:e12145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 70. | Sidhom K, Obi PO, Saleem A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 316] [Cited by in RCA: 456] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 71. | Hu GW, Li Q, Niu X, Hu B, Liu J, Shen XL, Wang Y, Deng ZF. Stirring ultrafiltration: a new method to isolate exosome. Dier Junyi Daxue Xuebao. 2014;35:598-602. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 72. | Gao J, Li A, Hu J, Feng L, Liu L, Shen Z. Recent developments in isolating methods for exosomes. Front Bioeng Biotechnol. 2022;10:1100892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 89] [Reference Citation Analysis (0)] |
| 73. | Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, Hsu EC, Gowrishankar G, Kanada M, Jokerst JV, Sierra RG, Chang E, Lau K, Sridhar K, Bermudez A, Pitteri SJ, Stoyanova T, Sinclair R, Nair VS, Gambhir SS, Demirci U. The Exosome Total Isolation Chip. ACS Nano. 2017;11:10712-10723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 295] [Article Influence: 36.9] [Reference Citation Analysis (1)] |
| 74. | Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, Hu BY, Qin W, Zou TT, Fu Y, Shen XT, Zhu WW, Geng Y, Lu L, Jia HL, Qin LX, Dong QZ. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 324] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 75. | Chen Z, Cheng SB, Cao P, Qiu QF, Chen Y, Xie M, Xu Y, Huang WH. Detection of exosomes by ZnO nanowires coated three-dimensional scaffold chip device. Biosens Bioelectron. 2018;122:211-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 92] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 76. | Yu LL, Zhu J, Liu JX, Jiang F, Ni WK, Qu LS, Ni RZ, Lu CH, Xiao MB. A Comparison of Traditional and Novel Methods for the Separation of Exosomes from Human Samples. Biomed Res Int. 2018;2018:3634563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 157] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 77. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55839] [Article Influence: 7977.0] [Reference Citation Analysis (132)] |
| 78. | Chen K, Wang Q, Kornmann M, Tian X, Yang Y. The Role of Exosomes in Pancreatic Cancer From Bench to Clinical Application: An Updated Review. Front Oncol. 2021;11:644358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 79. | Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene. 2016;35:1216-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 377] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 80. | Zhu Y, Zhang H, Chen N, Hao J, Jin H, Ma X. Diagnostic value of various liquid biopsy methods for pancreatic cancer: A systematic review and meta-analysis. Medicine (Baltimore). 2020;99:e18581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 81. | Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 220] [Article Influence: 18.3] [Reference Citation Analysis (1)] |
| 82. | Kabiraj L, Kundu A. Potential role of microRNAs in pancreatic cancer manifestation: a review. J Egypt Natl Canc Inst. 2022;34:26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 83. | Frampton AE, Krell J, Prado MM, Gall TM, Abbassi-Ghadi N, Del Vecchio Blanco G, Funel N, Giovannetti E, Castellano L, Basyouny M, Habib NA, Kaltsidis H, Vlavianos P, Stebbing J, Jiao LR. Prospective validation of microRNA signatures for detecting pancreatic malignant transformation in endoscopic-ultrasound guided fine-needle aspiration biopsies. Oncotarget. 2016;7:28556-28569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 84. | Reese M, Flammang I, Yang Z, Dhayat SA. Potential of Exosomal microRNA-200b as Liquid Biopsy Marker in Pancreatic Ductal Adenocarcinoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 85. | Nakamura K, Zhu Z, Roy S, Jun E, Han H, Munoz RM, Nishiwada S, Sharma G, Cridebring D, Zenhausern F, Kim S, Roe DJ, Darabi S, Han IW, Evans DB, Yamada S, Demeure MJ, Becerra C, Celinski SA, Borazanci E, Tsai S, Kodera Y, Park JO, Bolton JS, Wang X, Kim SC, Von Hoff D, Goel A. An Exosome-based Transcriptomic Signature for Noninvasive, Early Detection of Patients With Pancreatic Ductal Adenocarcinoma: A Multicenter Cohort Study. Gastroenterology. 2022;163:1252-1266.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 92] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 86. | Wei Q, Li Z, Feng H, Ren L. Serum Exosomal EphA2 is a Prognostic Biomarker in Patients with Pancreatic Cancer. Cancer Manag Res. 2021;13:3675-3683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 87. | Shin HS, Jung SB, Park S, Dua P, Lee DK. ALPPL2 Is a Potential Diagnostic Biomarker for Pancreatic Cancer-Derived Extracellular Vesicles. Mol Ther Methods Clin Dev. 2019;15:204-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 88. | Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, García-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1554] [Cited by in RCA: 2061] [Article Influence: 206.1] [Reference Citation Analysis (0)] |
| 89. | Tsuchida K. Evaluation of clinical outcomes of pancreatic cancer patients using circulating nucleic acids. Transl Gastroenterol Hepatol. 2019;4:2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 90. | Bunduc S, Gede N, Váncsa S, Lillik V, Kiss S, Juhász MF, Erőss B, Szakács Z, Gheorghe C, Mikó A, Hegyi P. Exosomes as prognostic biomarkers in pancreatic ductal adenocarcinoma-a systematic review and meta-analysis. Transl Res. 2022;244:126-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 91. | Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, Ikeda Y, Shibuya M, Miura F, Sano K. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci. 2018;25:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 92. | Namkung J, Kwon W, Choi Y, Yi SG, Han S, Kang MJ, Kim SW, Park T, Jang JY. Molecular subtypes of pancreatic cancer based on miRNA expression profiles have independent prognostic value. J Gastroenterol Hepatol. 2016;31:1160-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 93. | Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, Goggins M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600-3610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 241] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 94. | Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ, Jimenez P, Rodriguez J, Garcia-Foncillas J. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281-2290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 284] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 95. | Jafari A, Babajani A, Abdollahpour-Alitappeh M, Ahmadi N, Rezaei-Tavirani M. Exosomes and cancer: from molecular mechanisms to clinical applications. Med Oncol. 2021;38:45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 96. | Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1806] [Cited by in RCA: 2056] [Article Influence: 293.7] [Reference Citation Analysis (0)] |
| 97. | Song H, Liu B, Dong B, Xu J, Zhou H, Na S, Liu Y, Pan Y, Chen F, Li L, Wang J. Exosome-Based Delivery of Natural Products in Cancer Therapy. Front Cell Dev Biol. 2021;9:650426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (1)] |
| 98. | Rajput A, Varshney A, Bajaj R, Pokharkar V. Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives. Molecules. 2022;27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 84] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 99. | Butreddy A, Kommineni N, Dudhipala N. Exosomes as Naturally Occurring Vehicles for Delivery of Biopharmaceuticals: Insights from Drug Delivery to Clinical Perspectives. Nanomaterials (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 126] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 100. | Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498-503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1167] [Cited by in RCA: 1905] [Article Influence: 238.1] [Reference Citation Analysis (1)] |
| 101. | Pascucci L, Coccè V, Bonomi A, Ami D, Ceccarelli P, Ciusani E, Viganò L, Locatelli A, Sisto F, Doglia SM, Parati E, Bernardo ME, Muraca M, Alessandri G, Bondiolotti G, Pessina A. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J Control Release. 2014;192:262-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 680] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 102. | Chen H, Wang L, Zeng X, Schwarz H, Nanda HS, Peng X, Zhou Y. Exosomes, a New Star for Targeted Delivery. Front Cell Dev Biol. 2021;9:751079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 171] [Article Influence: 42.8] [Reference Citation Analysis (1)] |
| 103. | Mittal A, Chitkara D, Behrman SW, Mahato RI. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials. 2014;35:7077-7087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 104. | Masamune A, Kikuta K, Watanabe T, Satoh K, Hirota M, Shimosegawa T. Hypoxia stimulates pancreatic stellate cells to induce fibrosis and angiogenesis in pancreatic cancer. Am J Physiol Gastrointest Liver Physiol. 2008;295:G709-G717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 201] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 105. | Tao J, Yang G, Zhou W, Qiu J, Chen G, Luo W, Zhao F, You L, Zheng L, Zhang T, Zhao Y. Targeting hypoxic tumor microenvironment in pancreatic cancer. J Hematol Oncol. 2021;14:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 254] [Article Influence: 63.5] [Reference Citation Analysis (0)] |