Published online Apr 12, 2016. doi: 10.5528/wjtm.v5.i1.1
Peer-review started: October 2, 2015
First decision: November 10, 2015
Revised: November 21, 2015
Accepted: March 7, 2016
Article in press: March 9, 2016
Published online: April 12, 2016
Processing time: 69 Days and 0.3 Hours
Gene editing has recently emerged as a promising technology to engineer genetic modifications precisely in the genome to achieve long-term relief from corneal disorders. Recent advances in the molecular biology leading to the development of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) and CRISPR-associated systems, zinc finger nucleases and transcription activator like effector nucleases have ushered in a new era for high throughput in vitro and in vivo genome engineering. Genome editing can be successfully used to decipher complex molecular mechanisms underlying disease pathophysiology, develop innovative next generation gene therapy, stem cell-based regenerative therapy, and personalized medicine for corneal and other ocular diseases. In this review we describe latest developments in the field of genome editing, current challenges, and future prospects for the development of personalized gene-based medicine for corneal diseases. The gene editing approach is expected to revolutionize current diagnostic and treatment practices for curing blindness.
Core tip: Gene editing technology including Clustered Regularly Interspaced Short Palindromic Repeats/Clustered Regularly Interspaced Short Palindromic Repeats associated protein 9, zinc finger nucleases, or transcription activator like effector nucleases has great potential for generating in vitro and in vivo models of corneal diseases including keratoconus and Fuchs’ endothelial corneal dystrophy. Furthermore, gene editing is a powerful tool for studying molecular mechanisms mediating corneal development, pathogenesis and developing next generation innovative gene therapies including the patient-specific personalized medicine for curing corneal diseases. This review discusses current status and latest developments in the field of gene editing. Gene editing based molecular therapy has the potential to revolutionize current practices in ophthalmology clinic for curing corneal blindness.
- Citation: Raikwar SP, Raikwar AS, Chaurasia SS, Mohan RR. Gene editing for corneal disease management. World J Transl Med 2016; 5(1): 1-13
- URL: https://www.wjgnet.com/2220-6132/full/v5/i1/1.htm
- DOI: https://dx.doi.org/10.5528/wjtm.v5.i1.1
According to World Health Organization ocular diseases affect about 285 million people worldwide. It is estimated that over 39 million people suffer from blindness and 246 million people have low or impaired vision worldwide. In the United States, vision impairment is among the top ten disabilities according to the Centers for Disease Control and Prevention. According to the National Eye Institute, approximately 38 million people have vision impairment in the United States with an annual cost of over $68.8 billion for medical care. If the present increasing trend in eye disease continues, it is estimated that by 2050 the patient volume with blindness will increase by 150% with a corresponding increase of 250% in direct medical costs leading to an economic burden of $717 billion. To break this increasing trend and fulfill unmet clinical needs, it is imperative to develop novel next generation gene-based molecular therapies for ocular disease.
Cornea is the transparent tissue in front of the eye. It provides two thirds of refractive power and protection to the eye[1]. Trauma, injury and/or infection to the eye are known to compromise corneal transparency and cause corneal fibrosis and/or neovascularization. Corneal diseases are the second leading cause of blindness globally with an estimated 23 million patients and nearly 80% of all corneal blindness is preventable. Corneal defects are one of the most prevalent reasons for vision impairment worldwide. About 4% of the United States population has corneal disorders and approximately 1.5 million additional people experience corneal blindness each year. It is more pronounced in developing countries especially among children due to trachoma which alone causes blindness in 4.9 million people worldwide[2,3]. The current treatments for corneal blindness offer only short-term relief, require repeated drug application, meticulous patient compliance, cause side effects, and often fail. The surgical corneal transplantation is typically used to restore vision, requiring donor corneas which are not available in many countries, and their availability in America is sharply declining due to laser surgeries, hepatitis, human immunodeficiency virus (HIV), etc. Therefore there is an urgent need to develop novel corneal disease models and therapeutic strategies to treat corneal diseases. Over the past several years, the major focus of our research has been on the development of novel strategies for gene therapy to treat corneal diseases using adeno-associated virus (AAV) and nanoparticles[1,4-11]. Our lab has demonstrated that various AAV serotypes could be successfully used to deliver therapeutic genes to treat corneal diseases with varying transduction efficiency without major side effects. Our ongoing research suggests that AAV and nanoparticle vectors are essential for achieving intended gene editing in the cornea.
Gene targeting by homologous recombination has been the gold standard for generating germ-line targeted gene knockout and knock-in mice[12,13]. Ocular cells represent a unique platform to investigate emerging technologies to gain an insight in to the precise molecular mechanisms underlying the disease as well as to develop novel personalized therapeutic strategies. According to clinicaltrials.gov there are currently multiple clinical studies on gene therapy and stem cell based regenerative medicine for ocular diseases. However, gene-targeting strategies in human embryonic stem (hES) and human induced pluripotent stem (hiPS) cells are relatively more cumbersome, inefficient, time consuming, expensive and challenging[14]. As a result, several studies have utilized small interfering RNA and short hairpin RNA to knockdown multiple genes. There are several major caveats of this approach including non-specificity, off target effects, altered cellular physiology, toxicity and only a transient reduction in gene expression leading to an incomplete or partial knockdown effect[15-19]. To overcome these limitations, it is imperative to modify the host genome precisely. The recent advances in gene editing have led to a widespread enthusiasm and significant improvements in this direction. In this review, we describe the current and emerging tools for gene editing, and their potential applications in the treatment of ocular diseases.
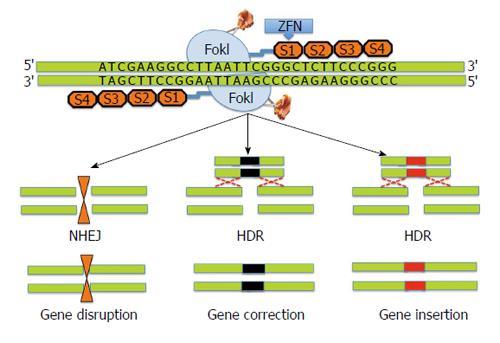
Zinc finger nucleases (ZFNs)’s belong to the first generation of gene editing tools based on the pioneering work of Kim et al[20-23]. ZFNs are designer nucleases that combine the DNA binding domain of eukaryotic transcription factors-zinc finger proteins with the nuclease domain of the FokI restriction enzyme[24,25]. In ZFNs, tandem arrays of Cys2His2 zinc fingers provide DNA binding specificity through recognition of approximately 3 base pairs of the target DNA. The catalytic domain of FokI requires dimerization to cleave the DNA at the targeted site and two adjacent ZFNs to independently bind to a specific codon with correct orientation and spacing. ZFNs work by introducing site-specific DNA double strand breaks (DSB) at a predetermined genomic locus. The DSB introduced by ZFNs undergo repair in the eukaryotic cells by either homology directed repair (HDR) process or non-homologous end joining pathway (NHEJ)[26-28]. DNA repair by homologous recombination leads to preservation of the original DNA sequence in the targeted cells rendering them vulnerable to re-cutting by ZFNs. In contrast, NHEJ can potentially lead to random insertion or deletion of nucleotides at the target break site thereby causing permanent disruption of the original DNA sequence. Figure 1 shows schematic representation of ZFN technology.
A previous study by Urnov et al[29] has demonstrated that ZFNs designed against X-linked severe combined immune deficiency (SCID) mutation in the IL2R gamma gene yielded > 18% gene-modified human cells with about 7% cells exhibiting desired genetic mutation on both X chromosomes. It has been previously demonstrated that HIV-1 uses the co-receptor CCR5, a validated target for HIV therapy[30,31]. Surprisingly, allogeneic stem cell transplant of a naturally occurring homozygous CCR5 deletion mutant (CCR5Δ32/Δ32) led to the elimination of HIV-1 in a patient[32]. Despite the low frequency of naturally occurring CCR5Δ32/Δ32 mutation, researchers have successfully harnessed the potential of ZFNs to disrupt CCR5 gene expression in hematopoietic stem and progenitor cells using a recombinant adenoviral vector encoding CCR5-specific ZFNs[33]. Recently, ZFNs have shown potential therapeutic benefits in clinical trials[34-36]. In a recent open-label phase I clinical study, HIV patient-derived autologous CD4 T cells were subjected to ZFN-mediated gene editing to render them resistant to HIV by knocking out CCR5 gene[36].
While the promise and feasibility of ZFN technology for gene editing has been demonstrated, multiple challenges remain. For example, ZFNs are relatively difficult to generate and are very expensive. Additionally, ZFNs can be non-specific and may result in off-target cleavage leading to multiple DSBs, which in turn can cause chromosomal rearrangements. These issues were addressed by developing ZFN variants that have ability to reduce off-target non-specific mutagenesis. The ZFN variants include a mix of two distinct ZFNs with different FokI domains that are obligate heterodimers, which introduce DSBs only when two distinct ZFNs are able to bind adjacent DNA regions[37-39].
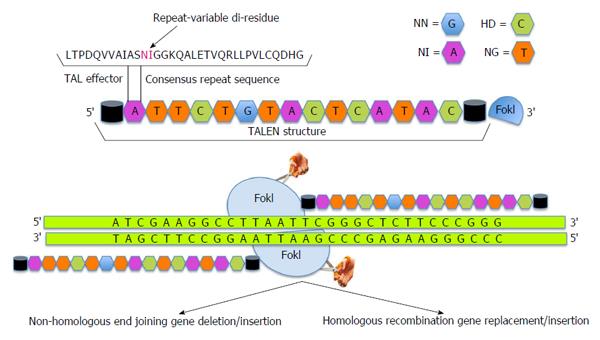
Another approach to administer gene editing has subsequently emerged through the recognition of a new class of designer nucleases termed transcription activator-like effector nucleases (TALENs). The gene editing steps associated with TALEN technology are presented in Figure 2. Transcription activator-like effectors (TALEs) are proteins secreted by Xanthomonas bacteria to subvert the host genome regulatory networks and can be engineered to bind any desired target sequence[40-43]. TALEs have a DNA binding module termed TAL repeat, which is used by each protein in a tandem array with 10-30 repeats to recognize extended DNA sequences with a ratio of 1 TAL repeat to 1 base pair of DNA sequence[43]. Each repeat in turn has about 33-35 amino acids with 2 adjacent amino acids [Repeat Variable Di-residue (RVD)], which confer their specificity for the DNA bases[40,44]. Decoding of the RVD has led to the development of a new class of designer nucleases called TALENs that contain an array of TAL repeats fused to FokI nuclease domain[45-47].
As compared to ZFNs, TALENs are relatively easier to design and generate due to their modular nature[48]. The promise of TALEN approach has been successfully demonstrated through the generation of gene-knockout animal models of C. elegans, rats, mice and zebra fish[49-53]. Deml et al[53] have developed zebrafish mutants carrying MAB21L2 gene to model human ocular coloboma. Homozygous mab21l2Q48Sfs*5 zebrafish mutant embryos exhibit severe lens and retinal defects with complete lethality while mab21l2R51_F52del mutants display a milder lens phenotype and severe coloboma. This study demonstrates the power of genome editing in model organisms for studying molecular mechanisms underlying human ocular diseases. TALENs have recently been exploited to develop genetically engineered hES cell lines, hiPS cells and mouse disease models[45,54-57]. Experimental correction of genetic defects in vitro has been successfully achieved by TALENs in hemophilia[54], mitochondrial diseases[58,59], and Duchenne muscular dystrophy[60]. To demonstrate the potential utility and efficiency of TALENs, Ding et al[61] have successfully generated mutant alleles of 15 genes in cultured somatic cells or human pluripotent stem cells. In an interesting study, Kim and colleagues have generated a library of 18740 TALEN pairs (http://www.talenlibrary.net/) to disrupt or modify every protein-coding gene for the entire human genome using a high throughput Golden-Gate cloning system[62]. In another study, Menon et al[63] utilized iPS cell technology and TALENs to generate a subject-specific mutant gene-corrected iPS cell lines for the treatment of X-linked SCID. It is interesting to note that while the subject derived mutant iPS cells could generate hematopoietic precursors and myeloid cells, only wild-type and gene corrected iPS cells could additionally generate mature cells and T cell precursors expressing the correctly spliced IL2R gamma. The work also suggests that TALEN technology can be employed for the manipulation of immune processes and chronic inflammatory diseases in the eye including corneal inflammatory disorders and diabetic retinopathy. Indeed, scores of further studies are needed to harness the bench-to-bedside potential of this approach and move forward towards the development of an autologous patient-based cell therapy.
The reversal of malignant phenotype via TALEN technology has been recently reported. Hu et al[64] have demonstrated that genome editing of human papilloma virus (HPV) oncogenes E6/E7 by TALENs efficiently reduced viral DNA load, restored the function of tumor suppressor p53/RB1, and reversed the malignant phenotype of host cells both in vitro as well as in vivo. In this study, HPV E6/E7 specific TALENs were effective in inducing apoptosis, inhibiting growth and reducing tumorigenicity in HPV positive cell lines. Further, direct cervical application of HPV E7 targeted TALENs efficiently mutated the E7 oncogene and reversed the malignant phenotype in K14-HPV16 transgenic mice. The study suggested two possible mechanisms for the reversal of the malignant phenotype. Firstly, TALENs specifically recognized and cleaved HPV DNA sequence in host cells leading to DSBs that directly induced apoptosis and suppressed their proliferation. Secondly, the cells that survived genotoxic stress, activated DSB repair via NHEJ pathway causing E6/E7 mutation. This led to the activation of E6/E7-inhibited tumor suppressor p53/RB1 and downregulation of CDK2 and E2F1. The ongoing experiments in our laboratory are attempting to generate in vitro and in vivo models and newer therapeutic approaches for corneal disorders and dystrophies using TALEN technology. This powerful gene editing approach has been particularly useful in studying keratoconus and Fuchs’ endothelial corneal dystrophy.
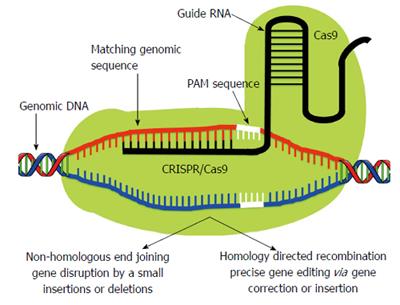
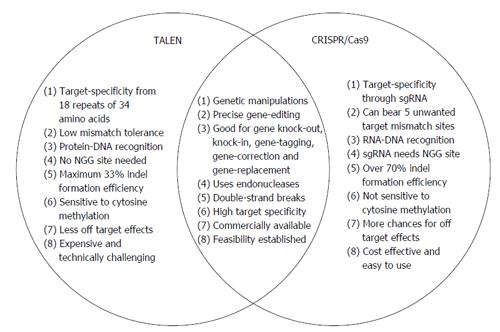
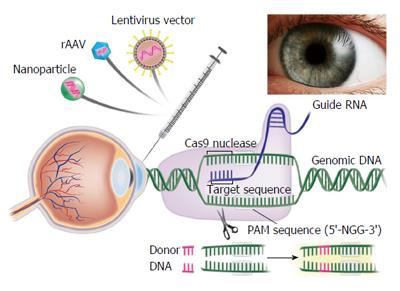
Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/Clustered Regularly Interspaced Short Palindromic Repeat Associated Systems (Cas9), derived from the bacterial adaptive immune system, has tremendous potential for achieving precise in vitro and in vivo gene editing[65-69]. Figure 3 depicts the core principle of this approach for obtaining intended gene editing in the genome. For the sake of convenience, Figure 4 provides a side-by-side comparison between TALENs and CRISPR/Cas9 systems. CRISPR/Cas9 based gene editing relies on co-expression of the bacterial Cas9 endonuclease and a short guide RNA (sgRNA) sequence to generate DNA DSBs in eukaryotic cells. The excision occurs at genomic sites that have a short homologous sequence to the 5’ end of the sgRNA followed by an NGG sequence called protospacer-adjacent motif (PAM)[66,70]. Since DNA DSB are primarily repaired through the error-prone NHEJ pathway in eukaryotes via small indels generated at the target sites. Therefore, CRISPR/Cas9 system provides a simple and cost-effective approach to simultaneously disrupt the open reading frames of multiple coding genes to produce loss/gain of function alleles at a high versatility[71-78]. CRISPR/Cas9 system has been successfully used for genome editing in C. elegans, Drosophila, mosquito, zebrafish, mouse, rat and human[79-90]. Cas9 nucleases cleave the double stranded DNA through the activity of their RuvC and HNH nuclease domains to generate DSBs. Cas9 can be engineered to cut only one strand of the DNA by catalytically inactivating either the RuvC or HNH nuclease domains[66,91,92]. These newly designed Cas9 nickases offer a unique approach to gene editing with high fidelity and specificity.
Recently, Chen et al[93] have successfully combined tamoxifen-inducible CRISPR/Cas-mediated genome editing with Flp/FRT and Cre/LoxP system to generate inducible gene knockout hPSC lines. They found that targeting dual sgRNA was essential for biallelic knockin of FRT sequences to flank the exon. They further developed a strategy to simultaneously insert an activity controlled recombinase-expressing cassette and removed the drug-resistance gene thereby enhancing the generation of SOX2, PAX6, OTX2 and AGO2 inducible knockout human ES and iPS cell lines. The target genes in these cell lines can be uniformly deleted at any given time by simple application of 4-OHT.
Wu et al[94] have recently reported successful correction of Crygc gene mutation that causes cataracts in mice. In this study, a dominant mutation in Crygc gene could be rescued in mouse zygotes by co-injection of Cas9 mRNA and a sgRNA targeting the mutant allele. Correction in the Crygc gene occurred by HDR based on an exogenously supplied oligonucleotide or the endogenous wild type allele, with only rare evidence of off-target modifications. The resulting mice were fertile and were able to transmit the corrected allele to their progeny. Similarly, Courtney et al[95] have examined the potential of an allele-specific CRISPR/Cas9 system for hereditary corneal dystrophies by specifically focusing on a dominant-negative mutation in KRT12, Leu132Pro which results in Meesmann’s epithelial corneal dystrophy. Further, Zhong et al[96] have utilized the CRISPR/Cas9 system to generate Kcnj13 mutant mice, which mimic human KCNJ13-related Leber congenital amaurosis, an early form of blindness.
The studies discussed above provide proof of principle for the application of CRISPR/Cas9 system in developing models of corneal dystrophies and personalized therapeutics for treating ocular diseases.
Cornea is an ideal target tissue for the development of personalized therapy. Gene editing approaches can successfully be used to develop novel corneal disease models. For example, it is possible to develop disease in a dish model for corneal dystrophies using patient derived corneal tissues. However, there are multiple challenges that need to be overcome before gene editing for corneal disease management becomes a reality. One of the major challenges is the lack of an authentic in vitro corneal endothelial cell culture model. This is because feline and human corneal endothelial cells are extremely difficult to culture. To overcome this major limitation, we have recently established reversibly immortalized feline and human corneal endothelial cell lines using Doxycycline inducible lentiviral vector system expressing human papillomavirus E6/E7 chimeric gene product. These immortalized feline and human corneal endothelial cell lines are valuable to study pathophysiology as well as molecular mechanisms regulating dystrophies and wound healing in the cornea. Currently there is no in vivo model for Fuchs’ endothelial corneal dystrophy. We are attempting to develop novel Fuchs’ endothelial corneal dystrophy models employing CRISPR/Cas9 gene editing technology and conditionally immortalized corneal endothelial cells (Figure 5). Further, gene editing can be used on patient derived iPS cells to develop novel corneal disease models. Gene editing can be used to treat corneal fibrosis and neovascularization by targeting pathologic genes, microRNAs, long noncoding RNAs, and/or signaling pathways driving corneal wound repair. Combat related traumatic corneal injuries present an ideal target where gene editing can be applied to maximize wound healing and tissue regeneration in corneal tissue without major adverse effects. Viral vectors and nanoparticles offer a novel platform to accomplish gene editing in corneal tissue. Real-time noninvasive intravital imaging will allow precise monitoring of gene editing success in an in vivo experimental animal model. Overall, there is tremendous potential of gene editing technology for corneal disease management as depicted in Table 1.
| Disease | Target genes for gene editing |
| Corneal fibrosis | BMP7, CTGF, Decorin, Hevin, Moesin, Smad2, Smad3, Smad4, Smad7, TGFβ1, TGFβR2, TRPA-1, Twist2, Vimentin |
| Corneal wound healing | CTGF, CNTF, EGF, EGFR1, EGFR2, Fibronectin, IGF, KGF, Laminin, Lumican, MIF, MMP-1, MMP-2, MMP-3, MMP-9, NGF, OGF, PAI-1, PAF, PDGF, rho-associated protein kinase (ROCK), TGFβ1, TGFβ2, TGFβ3, TLR4, TIMP-2, Vasohibin |
| Corneal neovascularization | Angiopoietin 1, Angiopoietin 2, Angiostatin, βFGF, Endostatin, FGFR-1, FGFR-2, FGFR-3, FGFR-4, FOXC1, HGF, IGF, IL-8, IL-1, Leptin, MMP-2, MMP-9, MMP-14, Netrin-1, Netrin-4, Neuropilin-2, NF-κB, PAI-1, PDGF, PEDF, PGF, Prox-1, ROCK, TNFα, TGFβ, TSP-1, Tie2, VCAM-1, VE-Cadherin, VEGF, VEGFR-1, VEGFR2, VEGFR-3 |
| Keratoconus | BANP-ZNF469, LOX, BNIP3, CAST, CLF1, COL4A4, COL5A1, CPT1B, CPTB1B, DOCK9, IL-1A, IL-1B, IPO5, KRT72, MPDZ-NFIB, NEFL, Noxa, PMAIP1, RAB3GAP1, SLC25A2, SLC25A4, SLC25A31, SOD2, STK24, TGFB1, TIMP1, TIMP3, UCP1, UCP3, VSX1, ZEB1 |
| Congenital hereditary endothelial dystrophy | SLC4A11 |
| Epithelial basement membrane dystrophy | TGFBI |
| Francois-neetens mouchetee fleck corneal dystrophy | PIKFYVE (PIP5K3) |
| Fuchs' endothelial corneal dystrophy | APEX1, AGBL1, COL8A2, LOXHD1, NOX4, SLC4A11, SnaI1, TCF4, TCF8, ZEB1 |
| Granular corneal dystrophy type 2 | TGFBI, TGFBIp |
| Gelatinous drop-like corneal dystrophy | TACSTD2 |
| Macular corneal dystrophy | CHST6 |
| Meesmann epithelial corneal dystrophy | KRT3, KRT12 |
| Posterior polymorphous corneal dystrophy | COL8A2, VSX1, ZEB1 |
| Reis-Bücklers' and Thiel-Behnke Corneal dystrophies | TGFBI |
| Schnyder corneal dystrophy | UBIAD1 |
The current major limitations in the field of gene editing include concerns regarding specificity, efficiency, and delivery of designer nucleases (ZFNs, TALENs and CRISPR/Cas9). The non-viral delivery systems including electroporation and protein transfection of designer nucleases have shown promising results with limited applications. The cell-specific delivery of designer nucleases such as CRISPR/Cas9 could be achieved through the recombinant viral vectors including adeno-associated virus (rAAV), integrase deficient lentivirus, baculovirus, adenovirus or nanoparticle vectors. Our laboratory has successfully identified rAAV, disabled lentivirus and nanoparticle vectors for delivering therapeutic genes into keratocytes of the mouse and rabbit corneas in vivo and human and canine corneas using ex vivo organ culture models[4,97]. The restricted cloning capacity and challenges associated with packaging of the expression cassettes limit the use of current hybrid rAAV vectors. However, recently two different promising strategies have been successfully employed to overcome the packaging limitations of rAAV. A strategy developed by a commercial vendor, proposed that Cas9 gene could be split between pAAV-Guide-it-Up and pAAV-Guide-it-Down plasmids with 1.6 kb region of homology. In this system, sgRNA sequence against the genomic sequence of interest could be cloned into pAAV-Guide-it-Down plasmid and two separate recombinant AAVs (AAV-Up and AAV-Down) could be generated and co-transduced into target cells. Due to precise homologous recombination at the site of homology, full-length Cas9 gene driven by an upstream promoter is generated in the targeted cells leading to successful genome editing. Employing a different strategy, Ran et al[98] have recently identified six smaller Cas9 orthologs. These authors showed that Cas9 from Staphylococcus aureus (SaCas9) could edit the genome with efficiencies similar to those of Staphylococcus pyogenes (SpCas9) despite being more than 1 kilobase shorter[98]. In these studies SaCas9 and its sgRNA expression cassette were packaged into hepatocyte tropic rAAV8 to target the cholesterol regulatory gene pro-protein convertase subtilisin/kexin type 9 (Pcsk9) in the mouse liver. Following systemic delivery with rAAV, > 40% genome modification accompanied by significant reduction in serum Pcsk9 and total cholesterol levels was observed. Further, the specificity of SaCas9 was confirmed using an unbiased DSB detection method, BLESS to identify a list of candidate off-target cleavage sites. These studies highlight the potential of newer SaCas9 for AAV-mediated in vivo genome editing applications.
The possibility of undesired genetic modification is a major concern associated with current gene editing technologies. To minimize off-target activity of Cas9, Ran et al[99] have recently developed an approach that simultaneously combines a Cas9 nickase mutant with paired guide RNAs to introduce targeted DSB. Since individual nicks in the genome are repaired with high fidelity, simultaneous nicking via appropriately offset guide RNAs is required for DSB and extends the number of specifically recognized bases for target cleavage. This versatile strategy can reduce off target effects by 50- to 1500-fold in cell lines and therefore has a great potential for genome editing applications that require high fidelity as well as high specificity.
In yet another interesting study, Suzuki et al[100] have performed whole genome sequencing to evaluate the mutational load at single base resolution in individual gene-corrected hiPS cells derived from Hutchinson-Gilford progeria syndrome, sickle disease and Parkinson’s disease patients. They have reported that in single cell clones, gene correction by helper-dependent adenoviral vector (HDAdV) or TALEN exhibited few off-target effects and a low level of sequence variation. Furthermore, they have developed a TALEN-HDAdV hybrid vector, which significantly increased gene-correction efficiency in hiPS cells. Interestingly, a comparative analysis of TALENs, CRISPR/Cas9 and HDAdV revealed that HDAdVs have a clear superiority over both CRISPR/Cas9 and TALENs in gene targeting and gene correction of the HBB locus.
Utilizing a novel approach, Nihongaki et al[101] have recently developed an engineered photoactivatable Cas9 (paCas9) that enables optogenetic control of CRISPR-Cas9 genome editing by NHEJ and HDR pathways in human cells. Optogenetic paCas9 was developed by fusing the two split Cas9 fragments with photoinducible dimerization domains termed magnets. The system gets activated in response to blue light and expresses paCas9 in target cells and induces targeted genome editing which can be switched off by extinguishing the light. Development of optogenetic paCas9 will enable conditional genome editing with ultra high precision and lead to potentially innovative gene and cellular therapies for currently incurable genetic disorders.
Most recently, Zetsche et al[102] have now characterized Cpf1, a new single crRNA-guided endonuclease which lacks tracrRNA and utilizes a T rich PAM. In contrast to the well-established Cas9, which requires tracrRNA to process crRNA arrays as well as crRNA and tracrRNA to mediate interference, Cpf1 doesn’t require tracrRNA to process crRNA arrays. Furthermore, Cpf1-crRNA complexes are capable of independently cleaving target DNA molecules without any additional RNA species to generate staggered cut with a 5’ overhang unlike the blunt ends generated by Cas9. Additionally, Cpf1 has multiple advantages over Cas9 including smaller size and therefore it has a great potential to maximize high fidelity gene editing in corneal diseases.
Human germ line editing approach is currently in its infancy as its application has recently been demonstrated in China[103] and is gaining momentum in the United Kingdom. Further, CRISPR/Cas9 could be effectively used to eradicate selective group of harmful plants, animals or insects that interfere with the natural ecological balance. For example, taking a note of the fact that only female mosquitos (Aedes aegypti) which feed on blood are responsible for pathogenic transmission of dengue, yellow fever and chikungunya viruses, Hall et al[87] were able to harness the power of CRISPR/Cas9 system to knockout Nix gene leading to a population of largely feminized genetic males while induced ectopic expression of Nix resulted in genetic females with nearly complete male genitalia. This study represents a promising new approach for implementing vector-controlled strategies wherein the disease carrier female mosquitoes can be converted into harmless male mosquitoes.
Another, pressing challenge with viral vectors especially AAV and lentiviral vectors is that they have a broad tissue tropism and efficiently transduce vast majority of cell types both in vitro as well as in vivo[4]. As a result, targeted in vivo genome editing of a very specific cell type in a highly complex organ like eye is extremely challenging but not impossible. Several different approaches can be used either independently or in combination to circumnavigate and bypass this critical issue. First, a highly tissue specific promoter-enhancer combination can be used to specifically limit the expression of CRISPR-Cas9 to the desired cell type. However, tissue-specific promoters often times lack fidelity and exhibit promiscuous expression in non-targeted cells. Furthermore, transgene expression driven by tissue-specific promoters may either be inadequate for therapeutic effect or supra-physiological thereby leading to toxicity. Second approach involves either AAV capsid engineering or using a specific AAV serotype to target specific cell types. In this regard, doxycycline, rapamycin, mifepristone and tamoxifen inducible expression vectors offer an excellent choice. However, caution needs to be exercised since certain drugs like rapamycin can perturb endogenous mammalian target of rapamycin pathway. Alternatively, delivery of Cas9 vectors into the target cells using episomal expression vectors, integration deficient lentiviral vectors, adenoviral vectors and nanoparticles has a tremendous potential that needs to be explored. We believe that the development of novel hybrid genome editing vectors will lead to robust high fidelity targeted genome editing and will potentially enable futuristic gene and cellular therapies for currently incurable genetic disorders an ultimate reality.
The tremendous potential to achieve intended gene editing using ZFNs, TALENs and CRISPR/Cas9 system for the development of novel disease models and innovative therapies has been well demonstrated (Table 2). However, a theoretical risk remains that this technology can be misused and exploited for bioterrorism and may have unimaginable negative consequences. Thus, it is extremely important to develop stringent guidelines to prevent the potential misuse of CRISPR/Cas9 based innovative gene editing technology. Like any other genetic engineering technology ZFNs, TALENs, and CRISPR/Cas9 technologies can be a double-edged sword. Indeed, gene editing approach is going to play a crucial role in improving human and animal health, increasing food and biopharmaceutical production, maintaining clean environment and revolutionizing medicine.
| Target gene | Target cell | ZFN/TALEN/CRISPR | Disease | Ref. |
| α-Globin | Human iPS | ZFN | α-thalassemia | [104] |
| Tnfrsf9 | NOD mouse embryo | ZFN | Diabetes | [105] |
| HBV | Huh7 cells | ZFN | Hepatitis B | [106] |
| CCR5, CXCR4 | CD4+ T cells | ZFN | HIV | [107] |
| CCR5, IL2RG | Multiple | ZFN | HIV, X-SCID | [108] |
| TCRα, β | T cells | ZFN | Leukemia | [109] |
| HBB | Human iPS cells | ZFN | Sickle cell anemia | [110] |
| PIG-A | Human ES, iPS cells | ZFN | PNH | [111] |
| gp91(phox) | Human iPS cells | ZFN | X-CGD | [112] |
| Albumin | Mouse hepatocytes | ZFN | Hemophilia A and B | [113] |
| SCN1A | Human iPS | TALEN | Epilepsy | [114] |
| PSIP1 | HT1080, 293T, Jurkat | TALEN | HIV | [115] |
| HBB | Human iPS cells | ZFN/TALEN/CRISPR | Sickle cell anemia | [116] |
| gp91(phox) | Human iPS cells | TALEN | X-CGD | [117] |
| Ctnnb1, Apc | H2.35 | TALEN | Hepatocellular carcinoma | [118] |
| hFVIII | Human iPS cells | TALEN | Hemophilia A | [119] |
| PLN R14del | Human iPS cells | TALEN | Cardiomyopathy | [120] |
| BUB1B | HCT116 | TALEN | PCS (MVA) | [121] |
| MECP2 | Monkey zygotes | TALEN | Rett syndrome | [122] |
| Sry, Uty | Mouse blastocysts | TALEN | NA | [123] |
| Dystrophin | Myoblasts | CRISPR/Cas9 | DMD | [124] |
| FANCC | Patient fibroblasts | CRISPR/Cas9 | Fanconi anemia | [125] |
| APC, SMAD4, TP53, KRAS, PIK3CA | Human intestinal epithelial organoids | CRISPR/Cas9 | Colorectal cancer | [126] |
| FAH | Mouse liver | CRISPR/Cas9 | Tyrosinemia | [127] |
| PTEN, TP53 | Mouse liver | CRISPR/Cas9 | Liver cancer | [128] |
| DMD | Mdx mouse zygotes | CRISPR/Cas9 | DMD | [129] |
| B2M, CCR5 | CD4+ T and CD34+ HSC | CRISPR/Cas9 | NA | [130] |
| CFTR | CF intestinal organoids | CRISPR/Cas9 | Cystic Fibrosis | [131] |
| C. parvum | HCT8 | CRISPR/Cas9 | Cryptosporidiosis | [132] |
| HCV | Huh7.5 | FnCas9 | Hepatitis C | [133] |
P- Reviewer: Ramalingam S S- Editor: Wang JL L- Editor: A E- Editor: Liu SQ
| 1. | Chaurasia SS, Lim RR, Lakshminarayanan R, Mohan RR. Nanomedicine approaches for corneal diseases. J Funct Biomater. 2015;6:277-298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 2. | Burton MJ, Mabey DC. The global burden of trachoma: a review. PLoS Negl Trop Dis. 2009;3:e460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 156] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 3. | Whitcher JP, Srinivasan M, Upadhyay MP. Corneal blindness: a global perspective. Bull World Health Organ. 2001;79:214-221. [PubMed] |
| 4. | Mohan RR, Rodier JT, Sharma A. Corneal gene therapy: basic science and translational perspective. Ocul Surf. 2013;11:150-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Mohan RR, Schultz GS, Hong JW, Mohan RR, Wilson SE. Gene transfer into rabbit keratocytes using AAV and lipid-mediated plasmid DNA vectors with a lamellar flap for stromal access. Exp Eye Res. 2003;76:373-383. [PubMed] |
| 6. | Mohan RR, Sharma A, Cebulko TC, Tandon A. Vector delivery technique affects gene transfer in the cornea in vivo. Mol Vis. 2010;16:2494-2501. [PubMed] |
| 7. | Mohan RR, Sinha S, Tandon A, Gupta R, Tovey JC, Sharma A. Efficacious and safe tissue-selective controlled gene therapy approaches for the cornea. PLoS One. 2011;6:e18771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Mohan RR, Tandon A, Sharma A, Cowden JW, Tovey JC. Significant inhibition of corneal scarring in vivo with tissue-selective, targeted AAV5 decorin gene therapy. Invest Ophthalmol Vis Sci. 2011;52:4833-4841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Mohan RR, Tovey JC, Sharma A, Schultz GS, Cowden JW, Tandon A. Targeted decorin gene therapy delivered with adeno-associated virus effectively retards corneal neovascularization in vivo. PLoS One. 2011;6:e26432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Tandon A, Sharma A, Rodier JT, Klibanov AM, Rieger FG, Mohan RR. BMP7 gene transfer via gold nanoparticles into stroma inhibits corneal fibrosis in vivo. PLoS One. 2013;8:e66434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 11. | Sharma A, Tandon A, Tovey JC, Gupta R, Robertson JD, Fortune JA, Klibanov AM, Cowden JW, Rieger FG, Mohan RR. Polyethylenimine-conjugated gold nanoparticles: Gene transfer potential and low toxicity in the cornea. Nanomedicine. 2011;7:505-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 12. | Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230-234. [PubMed] |
| 13. | Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503-512. [PubMed] |
| 14. | Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA. 2001;98:8403-8410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 211] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 15. | Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1709] [Cited by in RCA: 1693] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 16. | Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J. 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 622] [Cited by in RCA: 649] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 17. | Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188-1196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 354] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 18. | Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549-555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 375] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 19. | Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1031] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 20. | Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc Natl Acad Sci USA. 1994;91:883-887. [PubMed] |
| 21. | Kim YG, Li L, Chandrasegaran S. Insertion and deletion mutants of FokI restriction endonuclease. J Biol Chem. 1994;269:31978-31982. [PubMed] |
| 22. | Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299-15304. [PubMed] |
| 23. | Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156-1160. [PubMed] |
| 24. | Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 377] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 25. | Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1482] [Cited by in RCA: 1497] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 26. | Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071-1078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4401] [Cited by in RCA: 4391] [Article Influence: 274.4] [Reference Citation Analysis (0)] |
| 27. | Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196-207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 754] [Cited by in RCA: 707] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 28. | Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2264] [Cited by in RCA: 2066] [Article Influence: 137.7] [Reference Citation Analysis (0)] |
| 29. | Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1233] [Cited by in RCA: 1214] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 30. | Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2825] [Cited by in RCA: 2794] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 31. | Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955-1958. [PubMed] |
| 32. | Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, Schneider T, Hofmann J, Kücherer C, Blau O. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1330] [Cited by in RCA: 1377] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 33. | Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao LF, Tran CA, Torres-Coronado M, Gardner A, Gonzalez N. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21:1259-1269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 34. | Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839-847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 539] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 35. | Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 842] [Cited by in RCA: 836] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 36. | Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1044] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 37. | Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 308] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 38. | Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 809] [Cited by in RCA: 778] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 39. | Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 394] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 40. | Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509-1512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2048] [Cited by in RCA: 1814] [Article Influence: 113.4] [Reference Citation Analysis (0)] |
| 41. | Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 398] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 42. | Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 314] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 43. | Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843-1846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 733] [Cited by in RCA: 683] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 44. | Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1451] [Cited by in RCA: 1372] [Article Influence: 91.5] [Reference Citation Analysis (0)] |
| 45. | Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 996] [Cited by in RCA: 958] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 46. | Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1549] [Cited by in RCA: 1527] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 47. | Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 671] [Cited by in RCA: 601] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 48. | Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460-465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 835] [Cited by in RCA: 917] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 49. | Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 519] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 50. | Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697-698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 492] [Cited by in RCA: 473] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 51. | Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 457] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 52. | Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 486] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 53. | Deml B, Kariminejad A, Borujerdi RH, Muheisen S, Reis LM, Semina EV. Mutations in MAB21L2 result in ocular Coloboma, microcornea and cataracts. PLoS Genet. 2015;11:e1005002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 54. | Park CY, Kim J, Kweon J, Son JS, Lee JS, Yoo JE, Cho SR, Kim JH, Kim JS, Kim DW. Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc Natl Acad Sci USA. 2014;111:9253-9258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 55. | Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, Chang JC, Bao G, Muench MO, Yu J. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA. 2014;111:9591-9596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 241] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 56. | Low BE, Krebs MP, Joung JK, Tsai SQ, Nishina PM, Wiles MV. Correction of the Crb1rd8 allele and retinal phenotype in C57BL/6N mice via TALEN-mediated homology-directed repair. Invest Ophthalmol Vis Sci. 2014;55:387-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 57. | Panda SK, Wefers B, Ortiz O, Floss T, Schmid B, Haass C, Wurst W, Kühn R. Highly efficient targeted mutagenesis in mice using TALENs. Genetics. 2013;195:703-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 58. | Reddy P, Ocampo A, Suzuki K, Luo J, Bacman SR, Williams SL, Sugawara A, Okamura D, Tsunekawa Y, Wu J. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell. 2015;161:459-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 222] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 59. | Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med. 2013;19:1111-1113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 315] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 60. | Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 391] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 61. | Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 387] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 62. | Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31:251-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 284] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 63. | Menon T, Firth AL, Scripture-Adams DD, Galic Z, Qualls SJ, Gilmore WB, Ke E, Singer O, Anderson LS, Bornzin AR. Lymphoid regeneration from gene-corrected SCID-X1 subject-derived iPSCs. Cell Stem Cell. 2015;16:367-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 64. | Hu Z, Ding W, Zhu D, Yu L, Jiang X, Wang X, Zhang C, Wang L, Ji T, Liu D. TALEN-mediated targeting of HPV oncogenes ameliorates HPV-related cervical malignancy. J Clin Invest. 2015;125:425-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 84] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 65. | Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10336] [Cited by in RCA: 11174] [Article Influence: 931.2] [Reference Citation Analysis (0)] |
| 66. | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11979] [Cited by in RCA: 11019] [Article Influence: 847.6] [Reference Citation Analysis (0)] |
| 67. | Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833-838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1434] [Cited by in RCA: 1373] [Article Influence: 114.4] [Reference Citation Analysis (0)] |
| 68. | Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 952] [Cited by in RCA: 898] [Article Influence: 74.8] [Reference Citation Analysis (0)] |
| 69. | Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6555] [Cited by in RCA: 7009] [Article Influence: 584.1] [Reference Citation Analysis (0)] |
| 70. | Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62-67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1254] [Cited by in RCA: 1350] [Article Influence: 122.7] [Reference Citation Analysis (0)] |
| 71. | Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 300] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 72. | Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428-431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 318] [Cited by in RCA: 315] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 73. | Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33:102-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 599] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 74. | Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2015] [Cited by in RCA: 2115] [Article Influence: 176.3] [Reference Citation Analysis (0)] |
| 75. | Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2799] [Cited by in RCA: 2791] [Article Influence: 232.6] [Reference Citation Analysis (0)] |
| 76. | Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 547] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 77. | Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3572] [Cited by in RCA: 3868] [Article Influence: 351.6] [Reference Citation Analysis (0)] |
| 78. | Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1502] [Cited by in RCA: 1475] [Article Influence: 134.1] [Reference Citation Analysis (0)] |
| 79. | Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 273] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 80. | Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741-743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 749] [Cited by in RCA: 686] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 81. | Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028-1034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 681] [Cited by in RCA: 713] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 82. | Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1480] [Cited by in RCA: 1462] [Article Influence: 121.8] [Reference Citation Analysis (0)] |
| 83. | Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2349] [Cited by in RCA: 2255] [Article Influence: 187.9] [Reference Citation Analysis (0)] |
| 84. | Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31:681-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 508] [Article Influence: 46.2] [Reference Citation Analysis (0)] |
| 85. | Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31:684-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 344] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 86. | Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1203] [Cited by in RCA: 1243] [Article Influence: 103.6] [Reference Citation Analysis (0)] |
| 87. | Hall AB, Basu S, Jiang X, Qi Y, Timoshevskiy VA, Biedler JK, Sharakhova MV, Elahi R, Anderson MA, Chen XG. SEX DETERMINATION. A male-determining factor in the mosquito Aedes aegypti. Science. 2015;348:1268-1270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 215] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 88. | Kistler KE, Vosshall LB, Matthews BJ. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11:51-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 270] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 89. | Dong S, Lin J, Held NL, Clem RJ, Passarelli AL, Franz AW. Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito, Aedes aegypti. PLoS One. 2015;10:e0122353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 90. | Basu S, Aryan A, Overcash JM, Samuel GH, Anderson MA, Dahlem TJ, Myles KM, Adelman ZN. Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc Natl Acad Sci USA. 2015;112:4038-4043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 91. | Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579-E2586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1687] [Cited by in RCA: 1891] [Article Influence: 145.5] [Reference Citation Analysis (0)] |
| 92. | Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275-9282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 556] [Cited by in RCA: 552] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 93. | Chen Y, Cao J, Xiong M, Petersen AJ, Dong Y, Tao Y, Huang CT, Du Z, Zhang SC. Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 128] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 94. | Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 443] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 95. | Courtney DG, Moore JE, Atkinson SD, Maurizi E, Allen EH, Pedrioli DM, McLean WH, Nesbit MA, Moore CB. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 100] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 96. | Zhong H, Chen Y, Li Y, Chen R, Mardon G. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci Rep. 2015;5:8366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 97. | Mohan RR, Sharma A, Netto MV, Sinha S, Wilson SE. Gene therapy in the cornea. Prog Retin Eye Res. 2005;24:537-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 98. | Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1785] [Cited by in RCA: 2014] [Article Influence: 201.4] [Reference Citation Analysis (0)] |
| 99. | Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2581] [Cited by in RCA: 2530] [Article Influence: 210.8] [Reference Citation Analysis (0)] |
| 100. | Suzuki K, Yu C, Qu J, Li M, Yao X, Yuan T, Goebl A, Tang S, Ren R, Aizawa E. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell. 2014;15:31-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 101. | Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33:755-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 460] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 102. | Zetsche B, Gootenberg Jonathan S, Abudayyeh Omar O, Slaymaker Ian M, Makarova Kira S, Essletzbichler P, Volz Sara E, Joung J, van der Oost J, Regev A. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759-771. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3250] [Cited by in RCA: 3136] [Article Influence: 313.6] [Reference Citation Analysis (0)] |
| 103. | Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363-372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 715] [Cited by in RCA: 650] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 104. | Chang CJ, Bouhassira EE. Zinc-finger nuclease-mediated correction of α-thalassemia in iPS cells. Blood. 2012;120:3906-3914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 105. | Chen YG, Forsberg MH, Khaja S, Ciecko AE, Hessner MJ, Geurts AM. Gene targeting in NOD mouse embryos using zinc-finger nucleases. Diabetes. 2014;63:68-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 106. | Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Mol Ther. 2010;18:947-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 107. | Didigu CA, Wilen CB, Wang J, Duong J, Secreto AJ, Danet-Desnoyers GA, Riley JL, Gregory PD, June CH, Holmes MC. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 108. | Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 715] [Cited by in RCA: 655] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 109. | Provasi E, Genovese P, Lombardo A, Magnani Z, Liu PQ, Reik A, Chu V, Paschon DE, Zhang L, Kuball J. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat Med. 2012;18:807-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 365] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 110. | Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LF. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717-1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 246] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 111. | Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 438] [Cited by in RCA: 416] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 112. | Zou J, Sweeney CL, Chou BK, Choi U, Pan J, Wang H, Dowey SN, Cheng L, Malech HL. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117:5561-5572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 194] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 113. | Sharma R, Anguela XM, Doyon Y, Wechsler T, DeKelver RC, Sproul S, Paschon DE, Miller JC, Davidson RJ, Shivak D. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777-1784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 236] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 114. | Chen W, Liu J, Zhang L, Xu H, Guo X, Deng S, Liu L, Yu D, Chen Y, Li Z. Generation of the SCN1A epilepsy mutation in hiPS cells using the TALEN technique. Sci Rep. 2014;4:5404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 115. | Fadel HJ, Morrison JH, Saenz DT, Fuchs JR, Kvaratskhelia M, Ekker SC, Poeschla EM. TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol. 2014;88:9704-9717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 116. | Huang X, Wang Y, Yan W, Smith C, Ye Z, Wang J, Gao Y, Mendelsohn L, Cheng L. Production of Gene-Corrected Adult Beta Globin Protein in Human Erythrocytes Differentiated from Patient iPSCs After Genome Editing of the Sickle Point Mutation. Stem Cells. 2015;33:1470-1479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 143] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 117. | Dreyer AK, Hoffmann D, Lachmann N, Ackermann M, Steinemann D, Timm B, Siler U, Reichenbach J, Grez M, Moritz T. TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells. Biomaterials. 2015;69:191-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 118. | Zhang S, Li L, Kendrick SL, Gerard RD, Zhu H. TALEN-mediated somatic mutagenesis in murine models of cancer. Cancer Res. 2014;74:5311-5321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 119. | Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS. Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell Stem Cell. 2015;17:213-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 226] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 120. | Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, Kong CW, Rushing S, Hansen J, Ceholski D. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 121. | Ochiai H, Miyamoto T, Kanai A, Hosoba K, Sakuma T, Kudo Y, Asami K, Ogawa A, Watanabe A, Kajii T. TALEN-mediated single-base-pair editing identification of an intergenic mutation upstream of BUB1B as causative of PCS (MVA) syndrome. Proc Natl Acad Sci USA. 2014;111:1461-1466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 122. | Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell. 2014;14:323-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 155] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 123. | Wang H, Hu YC, Markoulaki S, Welstead GG, Cheng AW, Shivalila CS, Pyntikova T, Dadon DB, Voytas DF, Bogdanove AJ. TALEN-mediated editing of the mouse Y chromosome. Nat Biotechnol. 2013;31:530-532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 124. | Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 360] [Cited by in RCA: 340] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 125. | Osborn MJ, Gabriel R, Webber BR, DeFeo AP, McElroy AN, Jarjour J, Starker CG, Wagner JE, Joung JK, Voytas DF. Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum Gene Ther. 2015;26:114-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 126. | Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 838] [Article Influence: 83.8] [Reference Citation Analysis (0)] |
| 127. | Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 710] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 128. | Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380-384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 627] [Cited by in RCA: 588] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 129. | Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 520] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 130. | Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, Vrbanac V, Garrison BS, Stortchevoi A, Bryder D. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 362] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 131. | Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 990] [Article Influence: 90.0] [Reference Citation Analysis (0)] |
| 132. | Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, Cipriano MJ, Striepen B. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature. 2015;523:477-480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 266] [Cited by in RCA: 254] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 133. | Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci USA. 2015;112:6164-6169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 173] [Article Influence: 17.3] [Reference Citation Analysis (0)] |