INTRODUCTION
In the current severe acute respiratory syndrome virus-2 (SARS-CoV-2) disease (COVID-19) pandemic, detailed structural knowledge of new emerging viruses in relation to developing effective treatment and interventions are very necessary[1]. Of the previous pandemics caused by emerging or re-emerging pathogenic viruses, the 1918 “Spanish Flu” pandemic had exerted a big toll on public health and world economy[2]. Viruses are complex “biomolecule capsules” with genomic nucleic acid (Deoxyribonucleic acid/Ribonucleic acid) and associated protein(s). Unlike most other microorganisms, viruses are obligate intracellular parasites that must invade live cells and hijack the host biochemical machinery to perpetuate. Though, viruses have been debated for being classified as living or non-living, they are the most populated life-forms following the prokaryotes. After the discovery of the three different ribosomes, the cellular organisms have been placed together in a universal “phylogenetic tree”. Interestingly, all viruses that do not synthesize ribosomes are re-classified into the phylogenetic tree. The International Committee on Taxonomy of Viruses however, has established a unified taxonomy for all viruses that includes 3 orders, 56 families, 9 subfamilies, and 233 genera of about 1550 species[3].
Since the early 19th century, information on the biology of viruses and their structures has remarkably advanced with experimental and computational tools and techniques. Although viruses were defined as filterable infectious agents, knowledge on their shape, size and physiochemical properties remained unknown until the isolation and characterization of tobacco mosaic virus (TMV), using a polarizing light microscope in 1953[4]. The TMV particles were further examined using X-ray diffraction concluding viruses as homogenous substances with a “protein capsid” of a definite shape and size[5]. The capsid, also called as “core”, “coat” or “nucleocapsid” protects the viral genome against a hostile environment and delivers it to the host cells. The continuation of the TMV work in different laboratories subsequently confirmed its capsid’s structural subunits[6,7]. High-resolution crystallography of the self-assembled protein subunits further improved the structural knowledge of its virions[8]. Morphologically, the assembly units of a capsid seen under electron microscope (EM) are called “capsomers” that may or may not be equal to the number of protein subunits.
Fluorescence and interferometry based microscopy are the common approaches to track the virion’s cell-surface/receptor attachment, entry, cytoplasmic motility, uncoating, genome delivery and host-protein interactions[9]. In recent decades, advancements in molecular and computational biology, high-resolution X-ray crystallography, cryo-EM and molecular dynamics simulation have elucidated atomic-level structures of several important viruses towards understanding of their virion compositions, capsid assembly or disassembly, cell-receptor interactions, antigenicity and developing antiviral strategies[9,10]. This article presents the basics of virus structures and principles underlying capsid formation as well as therapeutic implications.
VIRION ARCHITECTURE
Structurally, viruses present remarkable differences in their shapes, sizes, molecular compositions and organizations. Their geometric shapes may be spherical, polyhedral, elliptical, rod-like, and pleomorphic ranging between 20-400 nm sizes (Figure 1). While the simplest known capsids are composed of one oligomeric protein, complex capsids also contain different proteins organized in sub-structural assemblies (e.g., portals, tails, fibers or spikes etc.). A key structural basis of classification of a virus is whether the virion has a lipid “envelope” or “shell” (enveloped viruses) or not (non-enveloped viruses). Further, in some “enveloped viruses”, the shell may have further complex structures made of other glycoproteins and/or nucleoproteins. Enveloped viruses have a further level of classification by describing the morphology of their nucleocapsids- either isometric or helical. In non-enveloped viruses, capsids are defined as isometric, filamentous, and complex. An isometric virion is morphologically spherical but geometrically an icosahedron or icosadeltahedron, a universally adopted structure in human art and culture since antiquity (Figure 2). Filamentous or rod-like virions are relatively simple and helical. The complex virions are on the other hand, neither isometric nor helical, but an intricate combination of both (e.g., T2 bacteriophage)[11]. Enveloped icosahedral and helical viruses are very common in animals and rare in plants and bacteria. Comparatively, while there are relatively few purely icosahedral bacteriophages, plants almost exclusively have non-enveloped helical viruses.
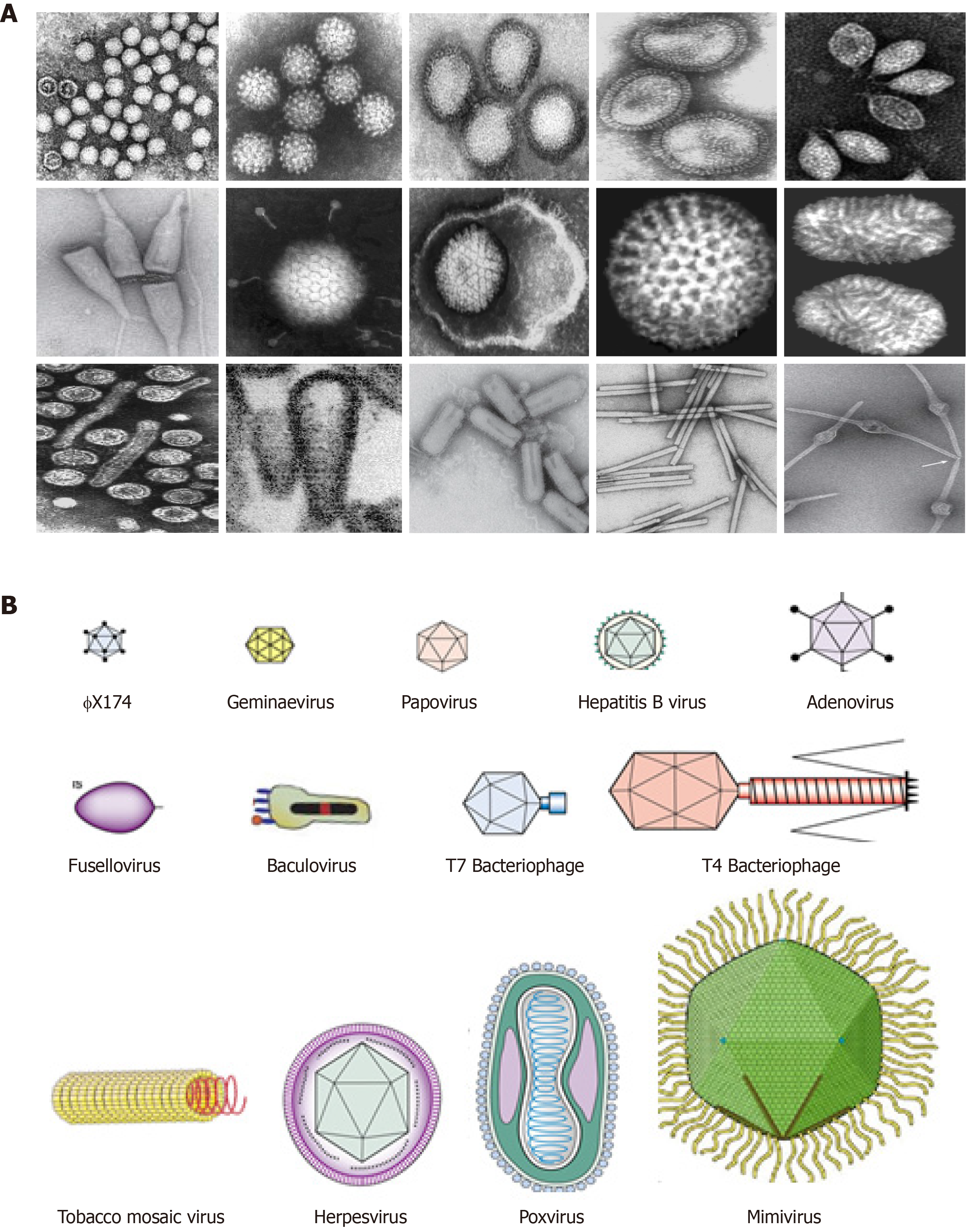
Figure 1 The structural and morphological diversity of viruses.
A: Electron micrograph of some representative animal, plant, fungal and bacterial viruses; B: Cartoon representation of virions showing their sizes (20-400 nm) and shapes (spherical, polyhedral, elliptical, rod-like etc.).
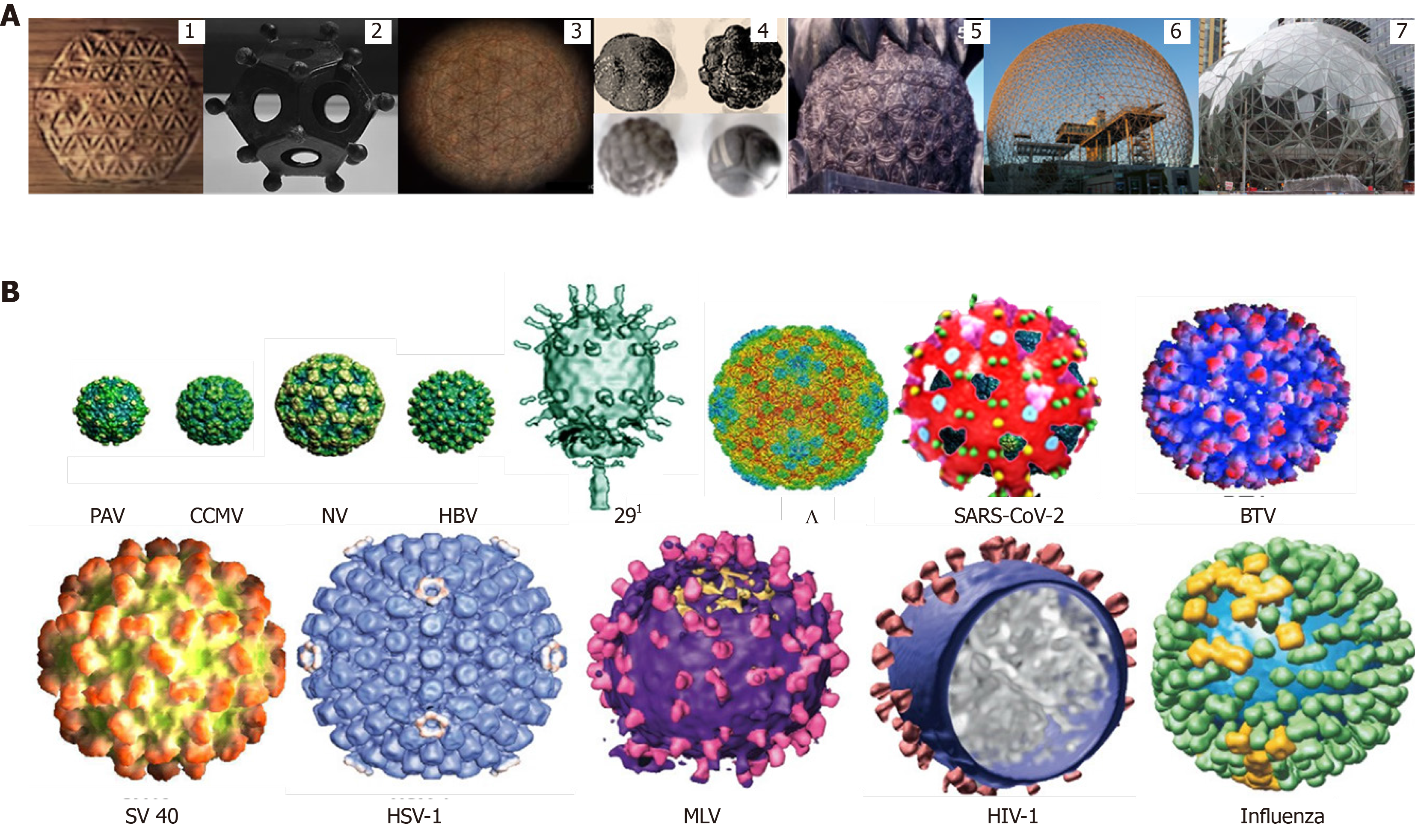
Figure 2 Morphologically spherical but geometrically polyhedral formations.
A: The virion-like universally adopted structures in human art and culture: 1. Ancient wood carving, Ephesus, Turkey; 2. Ancient Roman dodecahedron, Gallo-Roman Museum of Tongeren, Belgium; 3. Ancient petroglyph, Osirian Temple, Abydos, Egypt; 4. Neolithic stone spheres, Scotland, United Kingdom; 5. Guardian lion or Shishi, Yonghe Temple, Beijing, China; 6. Amazon Sphere, Seattle, United States; and 7. Tianjin Binhai Library, Tianjin, China; B: Spherical viruses with icosahedral capsids. PAV: Parvovirus; CMV: Cytomegalovirus; NV: Norovirus; HBV: Hepatitis B virus; 291: bacteriophage; SARS-CoV-2: Severe acute respiratory syndrome virus-2; BTV: Bluetongue virus; SV40: Simian virus 40; HSV-1: Herpes simplex virus; MLV: Murine leukemia virus; HIV-1: Human immunodeficiency virus-1; Influenza: Influenza virus.
In virion structure determination, capsid symmetry is an important factor. Inherently, a capsid’s geometric symmetry greatly contributes to its stability and balance between the packaged genome (deoxyribonucleic acid/DNA or ribonucleic acid/RNA) structure and/or the labile envelope that should melt out in a cytoplasm at a precise location and time. The physical condition for any geometrical structure’s stability is the necessity of minimum free-energy state. In view of this, the maximum number of strong interactions formed between the capsid subunits is required to attain minimum free-energy and to hold its structural integrity[12]. The MDS and Cryo-EM approaches appear to predict near experimental results on capsid stability and the structural role of packaged genome. Following the first atomic-resolution structure of TMV, a number of computational studies, such as highly coarse grained simulations and long timescale assessments on a range of capsids structures and stability have been performed[13]. As compared to non-enveloped capsids, there have been fewer simulation studies on enveloped capsids. In the relatively large sized enveloped viruses, greater structural complexity and lack of symmetry in the envelope bilayer, simulation of all components becomes relatively very complex. Nonetheless, structure of the mature enveloped human immunodeficiency virus native capsid has been recently determined using Cryo-EM and MDS[14].
CAPSID TRIANGULATION NUMBER
Viral capsid is described as empty and symmetric oligomers of one or polymers of different types of protein subunits in which viral DNA/RNA is packaged[15]. In a given capsid, the minimum number of protein subunits is determined by the symmetry of the face (i.e. triangle, square, tetrahedron etc.), and multiplying it by the number of all faces gives the total number of subunits. The triangulation number (T) is the smaller, identical equilateral-triangles that compose each triangular face, and is calculated using the law of solid geometry (T = Pf2; where P is a positive integer i.e., 1, 3 and 7; and f is face number i.e., 1, 2, 3, 4, etc.). The minimum number of subunits (n) is thus 3 for triangle, 4 for square, 12 for tetrahedron, 24 for octahedron, and 60 for dodecahedron or icosahedron. For instance, in an icosahedral capsid, the triangulation number allows to determine the number of subunits as n = 60T. A capsid volume can be increased by either increasing T value or adding equatorial capsomers. Generally, spherical viruses with capsids T > 1 tend to be of larger sizes.
CANONICAL CUBIC SYMMETRY
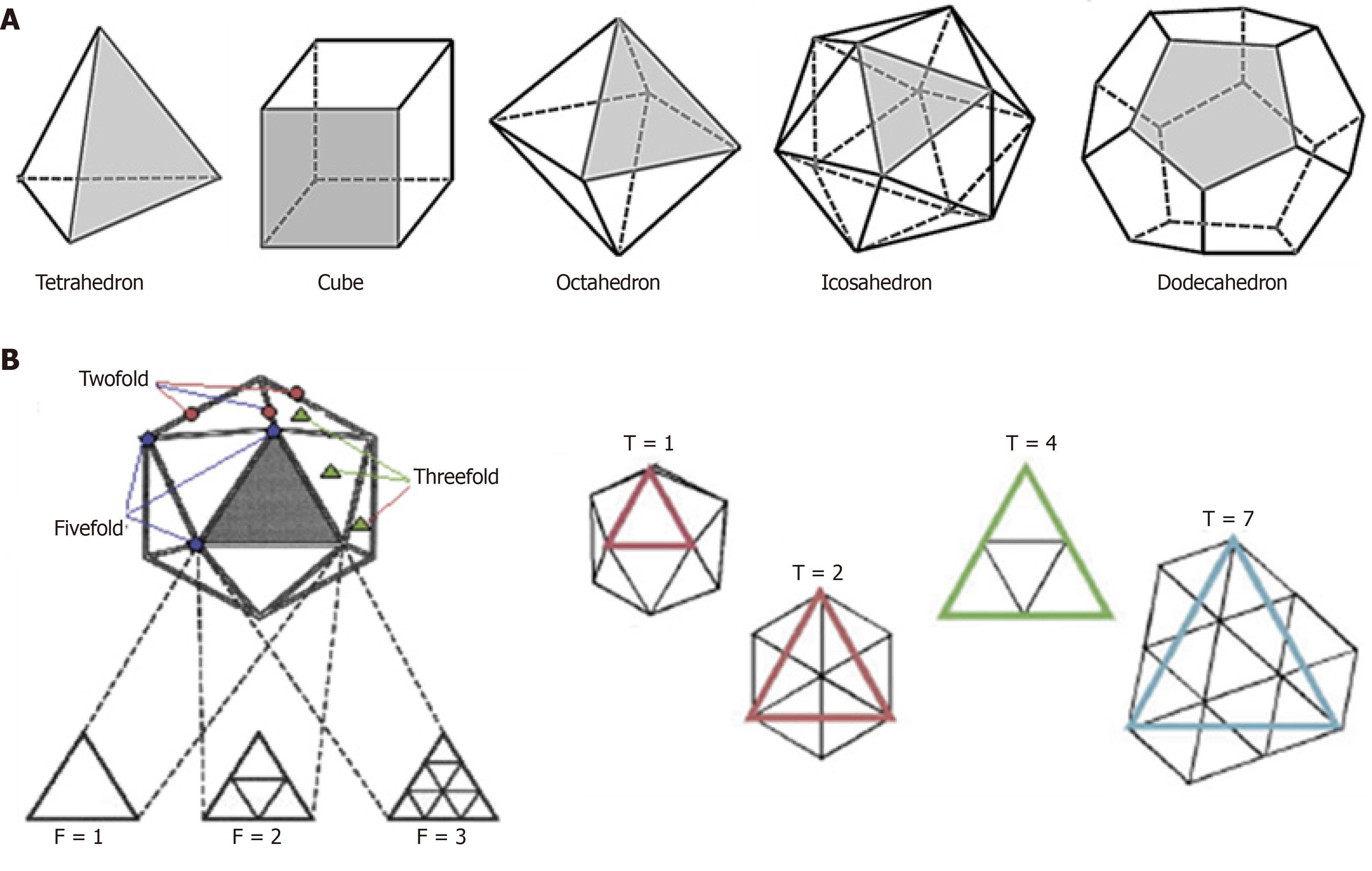
In solid geometry, the cubic symmetry is the characteristic of canonical polyhedral structures like, tetrahedron, cube, octahedron, icosahedron and dodecahedron formed of three or more identical faces, identical vertices and identical edges (Figure 3A). For example, a cube has 6 identical square faces, 8 identical vertices and 24 edges. Further, a cube has twofold rotational symmetry axes passing through the centers of the opposite faces and threefold axes along the diagonals passing through each of the vertices. The combination of these two rotational symmetry elements gives rise to additional 3 fourfold symmetry axes going through centers of the opposite faces and 6 twofold symmetry axes going through the midpoints of opposite edges. A cube with four-, three-, and twofold symmetry axes thus, allows placement of 12 identical units. In viruses, the polyhedral capsids with inherent cubic symmetry have at least 4 threefold rotational axes.
Figure 3 Cubic symmetry, the inherent characteristic of polyhedrons.
A: Tetrahedron, cube, octahedron, icosahedron and dodecahedron formed of three or more identical faces, identical vertices and identical edges; B: Icosahedral capsid (e.g., T = 1, 3, 4) with principle of axes of symmetry (e.g., 2-3-5 symmetry).
ICOSAHEDRAL SYMMETRY
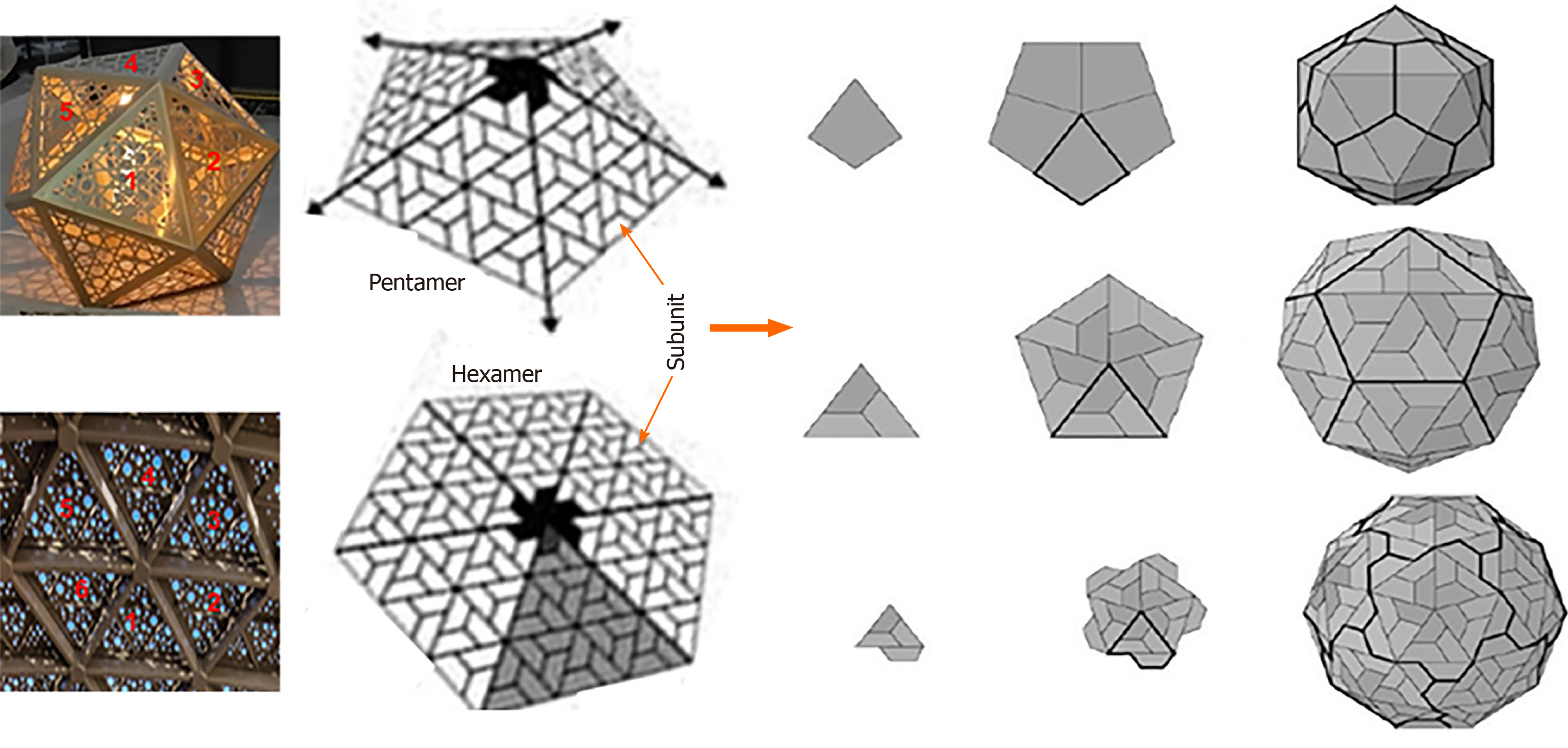
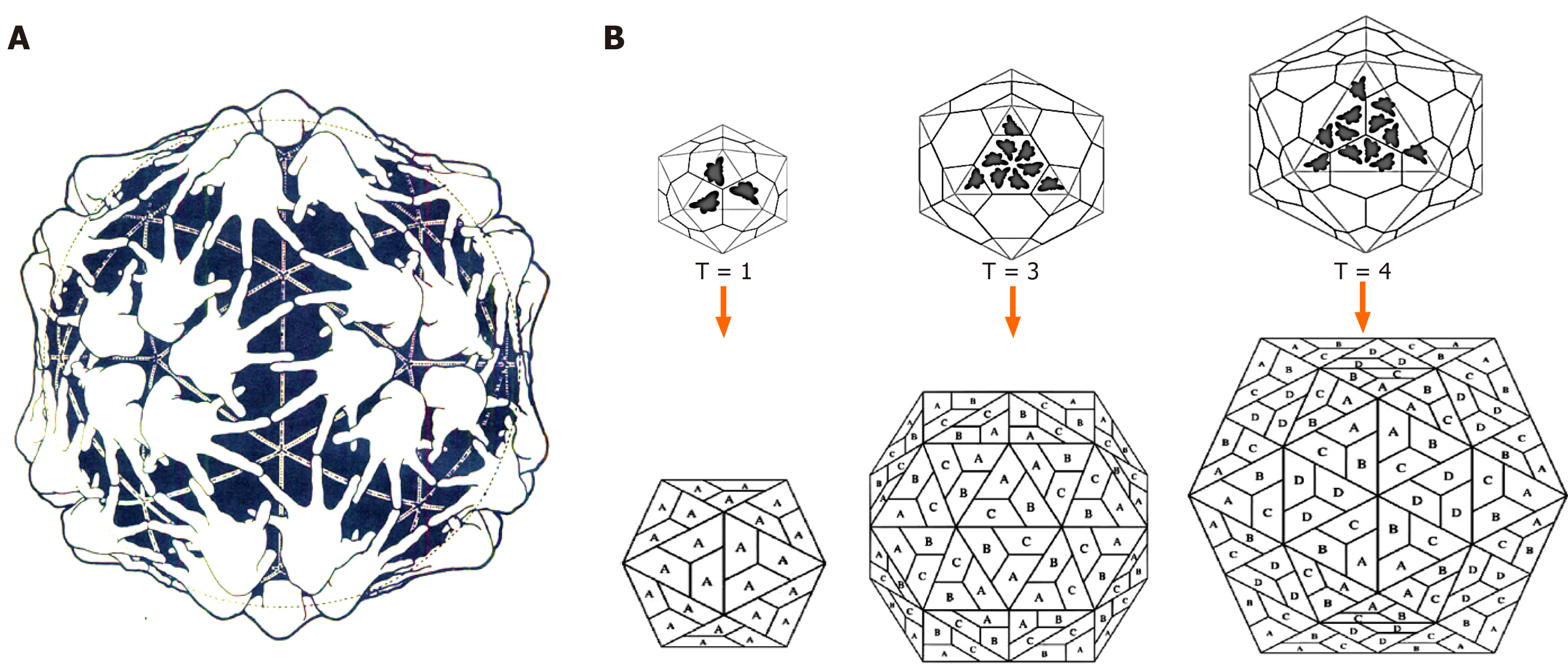
Icosahedral virions follow exclusive pathways of capsid assembly and maturation regulated by symmetry principles having three axes of symmetry: Fivefold, threefold, and twofold or 5-3-2 symmetry (Figure 3B). For example, a T = 1 icosahedron has 5-3-2 symmetry with n = 60. Notably, though most of the plant satellite viruses icosahedral capsids have n = 60, many spherical viruses have n > 60 produced by one or more genes. Moreover, an icosahedral capsid consists of f = 20 (5 on top, 5 at bottom and 10 in middle) and 12 vertices. While there are rings of five subunits (pentamers) at the vertices of each of the original faces, there are rings of six subunits (hexamers) at all the new vertices generated (Figure 4). The icosahedral capsids with pentamer and hexamer subunits are called “quasi-equivalent” that however, remains in the minimum free-energy state. Following tomato bushy stunt virus (TBSV)[16], turnip yellow mosaic virus was the second spherical virus whose icosahedral capsid was determined, using X-ray crystallography[17].
Figure 4 Icosahedral capsid formation.
Capsid formation with rings of five subunits i.e., pentamers or six subunits i.e. hexamers at the vertices of each of the faces (Left panel; Penta-/hexameric artifacts, Gallery Mall, Riyadh, Saudi Arabia).
Further, not only icosahedrally symmetric capsids have > 60 identical subunits, in several cases it is formed by subunits of different gene products. Therefore, based on their T numbers, icosahedral capsids are categorized into different classes (Figure 5).
Figure 5 Schematic presentation of icosahedral capsids.
A: A Clug model of icosahedral capsid assembly; B: Formation of icosahedral T = 1 (subunit A), T = 3 (subunits A, B and C) and T = 4 (subunits A, B, C and D) capsids.
T = 1 icosahedron
The smallest and simplest known viruses have T = 1 capsids made of a single symmetrical protein. The small plant satellite viruses, like satellite tobacco necrosis virus (STNV) icosahedron is T = 1, n = 60[18].
T = 3 icosahedron
Some virus capsids have T = 3, n = 180 structure where in each triangular face (n = 3), the subunits are asymmetrical (e.g., pentamers or hexamers). For example, in TBSV (T = 3, n = 180), each triangle is made of three identical subunits but in different conformations to accommodate the quasi-equivalent assembly[16]. In contrast, picornavirus icosahedral capsids are made of 60 copies of each of four subunits (VP1 = 60, VP2 = 60, VP3 = 60 and VP4 = 60)[19].
T = 7 icosahedron
Bacteriophage T7 icosahedron is composed of 12 pentamer and 60 hexamers with a T = 7 symmetry[8,20].
T = 13 icosahedron
The reoviruses have double-shelled isometric capsids i.e. a capsid within a capsid. The outer capsid has a T = 13, n = 780 symmetry while the inner capsid has a T = 2, n = 120 symmetry[21].
T = 16 icosahedron
The herpes simplex virus (HSV) capsid has a T = 16, n = 960 symmetry[22].
T = 25 icosahedron
In highly complex adenovirus icosahedrons, the T = 25, n = 1500 structure is made of 12 pentameric and 240 hexameric subunits, including a fiber of different proteins[23].
T = 147 icosahedron
The insect chilo iridescent virus icosahedral capsid consists of 12 pentamericand 1460 hexamericcapsomers, arranged with T = 147 symmetry[24].
T = 219 icosahedron
The marine algae Phaeocystis Pouchetii virus (PpV01) has the largest capsid diameter and T number of any icosahedral DNA virus studied[25]. PpV01 capsid consists of 2180 trimeric and 12 pentameric capsomers arranged with T = 219 quasisymmetry. Above the capsid 60 fiber-like structures project, having nearly uniform distribution on the surface.
Further, based on T numbers, three classes of icosahedron are also proposed. Of these, the two classes that follow T = Pf2, n = 60 are also referred to as P = 1 and P = 3 classes. The third one is “prolate” class of icosahedron found in T7[20] and 29[26] phages and as well as aberrant flock house virus[27]. Geometrically, prolate icosahedra are stretched along one of the axes and therefore, defined as n = 30 (T + Q) where Q is “elongation number” and Q > T. On the other hand, while an icosahedron is “obate” when Q < T and “isometric” when T = Q.
HELICAL FILAMENT OR ROD SYMMETRY
Majority of helical filamentous or rod-shaped capsid structures and assemblies belong to either plant viruses or bacteriophages[7]. In TMV, the rod-shaped capsid is made of asymmetrical subunits or capsomers in a high-aspect-ratio geometry. The subunits (n = approximately 2130) are joined in a helical circle to form symmetrical discs that are stacked on top of another, resulting in a hollow tube or rod.
HEAD-TAILED SYMMETRY
In the head-tail architecture, an isometric “icosahedral” head is attached with a “helical” tail. Though the head-tail is an inherent feature of bacteriophages e.g. T7 phage[8], many have other morphologies, too. The tails can be short, long and non-contractile or complex and contractile, and may have additional appendages such as “base-plates” and “collars”.
ENVELOPED-ISOMETRIC SYMMETRY
The Sindbis virus is the only known enveloped virus to have a geometrically symmetric virion[28]. Its single protein, isometric icosahedral core (T = 3, n = 180) is covered by an isometric glycolipid envelope (T = 4, n = 240) from which trans-membrane glycoprotein “spikes” protrude.
ENVELOPED-HELICAL SYMMETRY
In animal enveloped viruses, like retroviruses (e.g., human immunodeficiency virus, HIV), paramyxoviruses (measles and mumps viruses), orthomyxoviruses (influenza viruses), and rhabdoviruses (e.g., vesicular stomatitis virus, VSV; rabies virus, RBV) as well as plant viruses, the segmented genome is packaged in multiple compact helical/coiled filamentous nucleocapsids[29]. In influenza virus, its eight rod-shaped RNA individually encapsidated in separate cores made of matrix protein and nucleoprotein, are all contained within a spherical envelope dotted with hemagglutinin and neuraminidase spikes. In contrast, rhabdoviruses are non-isometric with bullet-shaped helical core made of nucleoprotein only. However, the structural geometry involved in the formation of rhabdovirus core still remains elusive.
ENVELOPED-SPINDLE SYMMETRY
Hyperthermophilic archaeal fuselloviruses have spindle/lemon-shaped virions with short tail-fibers attached to one end of the envelope. In sulfolobus spindle-shaped virus, for example, the capsid is made of three core proteins VP1, VP3 and VP4, including a nucleoprotein VP2[30].
VIROIDS, VIRUSOIDS OR SATELLITES
Owing to their similarities with conventional viruses, viroids, virusoids or satellites are often referred to as sub-viral particles. Viroids are the smallest phytopathogens with rod or dumb-bell shaped unencapsidated infectious RNA that however, do not synthesize any proteins[31]. Viriods do not have a capsid or outer envelope, but, as with viruses, can reproduce only within a host cell. The potato spindle tuber viroid was the first viroid discovered in 1971[32]. The hepatitis D virus is a viroid or satellite virus that requires hepatitis B virus (HBV) co-infected cells to replicate its RNA[33]. Its only synthesized core utilizes HBV envelope protein for infectious virion maturation.
CAPSID ASSEMBLY
In a capsid assembly, protein subunits are joined by maximal hydrophobic contacts and/or non-covalent interactions, and sometimes by covalent bonds. Structurally, most capsid proteins can be ascribed to a very limited number of conformational motifs i.e., “jelly-roll/antiparallel β barrel” and “HK97” leading to their perfect oligo/polymerization, stability and dynamics, formation of assembly intermediates, genome packaging and maturation[34]. While many viruses from the small STNV to the largest known Acanthamoeba polyphaga mimivirus utilize the jelly-roll motif, some mammalian DNA viruses like, HSV use HK97 motif in their capsids[35]. Evolutionarily, a capsid structure is less dynamic than the proteins of its specific motif. Also, the preference of HK97 in prokaryotic capsids and jelly-roll in eukaryotic capsids might suggest its early existence.
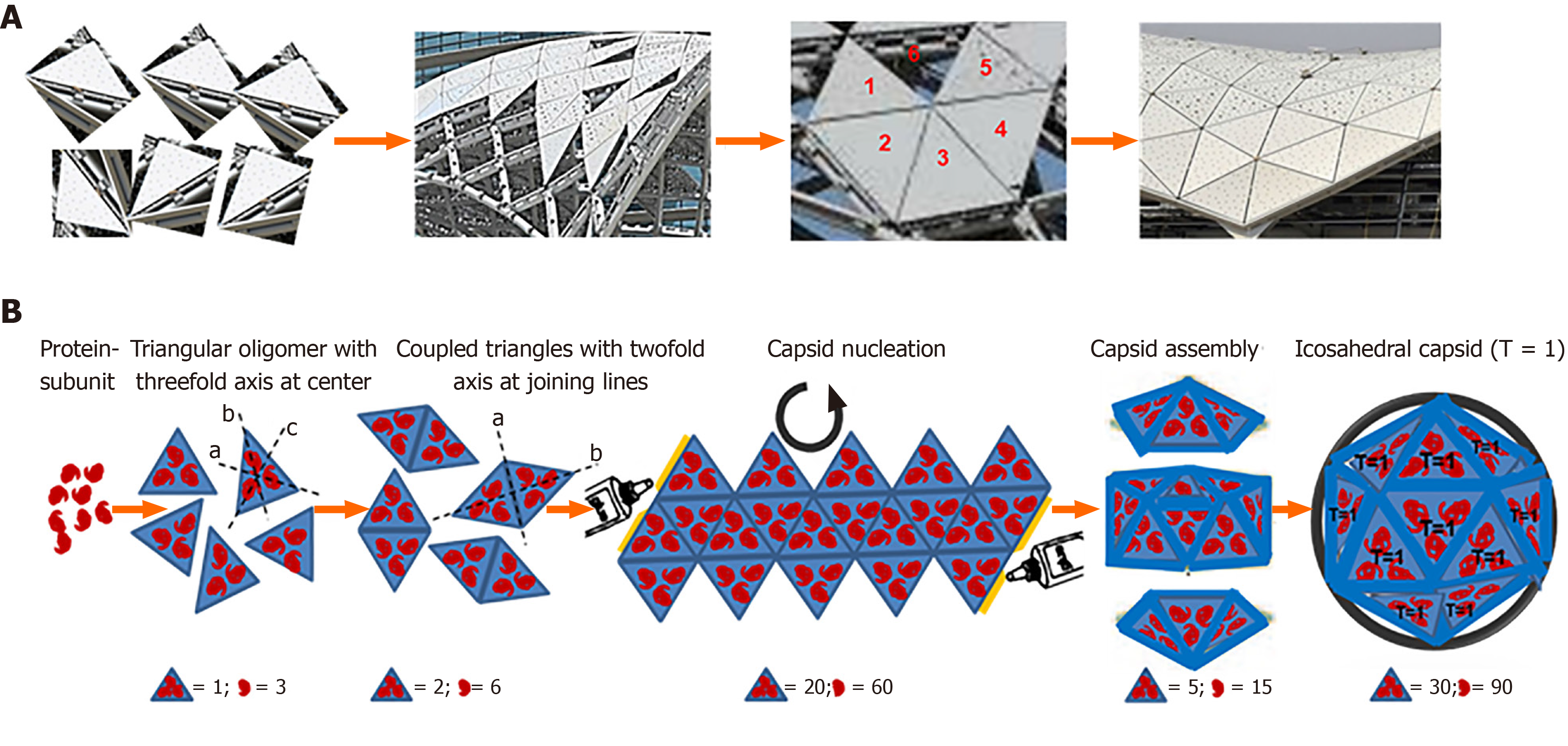
Since the capsid subunits are asymmetrical, the maximum number of inter-subunit interactions can be established only when they are arranged symmetrically. Therefore, in an ideally stable geometry, the capsid must be in a perfect symmetry and lowest possible free-energy. Watson and Crick first suggested the capsid formation by the association of multiple copies of the capsid protein(s), and the spherical viruses with cubic symmetry involving at least 4 threefold rotational symmetry axes[36]. The simplest helical capsid is assembled by first encircling the asymmetrical protein subunits to form symmetrical discs or rings, and lying one on the top of another resulting in a hollow filament or rod-like structure. In complex arrangements, the smallest number of protein subunits are placed around the vertices of a cube i.e. 12 regular pentagonal subunits to form a tetrahedron, octahedron and dodecahedron while 20 equilateral triangular subunits to form an icosahedron (Figure 6). Computationally, a non-symmetric spherical capsid can be formed using at least 12 protein subunits with substantial energy difference, favoring the symmetrical 12-mer over near/complete structure. Conversely, the relatively larger subunits i.e., 40-mer because of minor difference between their uniform sized complexes may not necessarily satisfy an icosahedral symmetry[37,38]. While there is limited knowledge on complex quaternary capsid structures, most viruses have icosahedral or helical symmetry. Notably, while nearly 50% of the virus families have icosahedral capsids, about 10% have helical capsids.
Figure 6 A cartoon presentation of an icosahedral capsid assembly.
A: Construction of a polyhedral shade with equilaterally triangular glass panes (Metro Rail Project, Riyadh, Saudi Arabia); B: Assembly of an icosahedral (T = 1) capsid of a spherical virus.
Notably, the X-ray precession image of TBSV was the first experimental evidence of icosahedral symmetry in a spherical virus[16]. Further advancements in Cryo-EM assisted structure determination of icosahedral capsids became a turning point in structural biology. The first reported MDS study on capsid self-assembly employed simple triangular shaped subunits (T = 3) with only two types of spherical virions[39]. Generally, the icosahedral capsid assembly is described by two timescales- nucleation and elongation[40]. Icosahedral reconstruction is a type of single particle three dimensional (3D) image reconstruction. Using Cryo-EM and image reconstruction, the HBV icosahedral core secondary structure has been deduced, revealing a new fold for a viral protein where in vitro expressed HBV capsid assembled to yield T = 3 as well T = 4 icosahedrons[41,42].
DEFECTIVE VIRIONS OR PARTICLES
Correct and strong interactions between protein-subunits as well as other macromolecules allow assembly of stable capsids whereas weak interactions lead to unstable or defective capsids. Formations of defective capsids are mainly reported from enveloped icosahedral alphaviruses, flaviviruses and hepadnaviruses, including irregular non-icosahedral capsids of some immature retroviruses[43]. Such defects may arise as scars at the beginning of capsid assembly to the completion where it may be incorporated stochastically during self-assembly or imposed by interactions with viral or host factors. Nonetheless, defective virions are not necessarily replication-incompetent or infectious. In HBV, for example, the newly assembled pleomorphic capsids (T = 3 and T = 4) may or may not contain viral DNA, and therefore, may or may not be infectious. In contrast, though the alphavirus capsids (T = 4) and envelope proteins appear to be well structured, a substantial fraction of Ross river virus capsid is shown to have defects[44].
Though icosahedral viruses are inherently symmetric, the imposed asymmetry can be regular, irregular or dynamic. The asymmetric or symmetric capsid modifications have both structural and biological advantages in many viruses. In regular asymmetry, the well-defined modification incapsids symmetry strengthens polymerase activities of HBV and cytoplasmic polyhedrosis virus[45,46], whereas enhances canine parvovirus and MS2 phage binding to their host cell-receptors[47,48]. Conversely, irregular asymmetry is stochastic, caused by defects trapped during capsid assemblies as observed in HBV and Ross river virus[49,50].
Biologically, identification of selective advantage of structural defects in a symmetric capsid allows the virions to better respond to their environment and exposure to internal components[4]. In both cases, such defects may facilitate capsid structural transitions, uncoating, regulated genome release, intracellular trafficking or accessibility of cellular factors. The dynamic asymmetry in capsid intermediates arises due to Brownian dynamics when internal components are exposed to the surface[49-51]. Notably, the inter-subunit hydrophobic interactions represent the primary driving force behind the thermodynamics of capsid self-assembly akin to surfactant micelle formation[52]. Analogously, while the inter-subunit electrostatic interactions can oppose hepadnavirus capsid assembly where its stability increases with ionic strength, the alphavirus capsids that appear uniformly assembled are extremely sensitivity to solution conditions[53].
VIRUS-LIKE PARTICLES
Though a nucleocapsid stability also depends on its strong interaction with the viral genome, several stable native capsidsor sub-viral particles are also formed without it, and are called virus-like particles (VLPs). The self-assembled non-infectious VLPs present the overall structure of the viral capsid or virion. For example, in hepatitis E virus (HEV), in vitro expressed capsid assembly[54], followed a high resolution Cryo-EM reconstruction for its VLPs[55]. Notably, the in vitro produced HEV-VLPs are much smaller than the authentic infectious virions, and are never detected in infected individuals. As compared to the larger native icosahedral virions (T = 3, n = 180), the HEV-VLPs display T = 1, n = 60 symmetry where T = 1 projection however, appears as spikes decorated with spherical rings akin to native virions[56]. The observed T = 1 VLPs instead of T = 3 has been suggested because of its energetically unfavorable configuration in the absence of genomic RNA. Moreover, similar to plant T = 3 capsids, the HEV-VLPs display threefold protrusions formed by P1 and twofold spikes made of P2 adopting the jelly-roll motif. Also, based on the T = 1 VLP structure, a T = 3 capsid of HEV has been modeled by using the quasi-equivalent capsid of TBSV[56].
THERAPEUTIC APPLICATIONS
A detailed 3D image of a virus particle is essential for understanding the mechanisms of capsid assembly/disassembly, antigenicity, interaction with host cell-receptors, and for designing therapeutic strategies[57,58]. The self-assembled, non-infectious VLPs mimic the real virus and present its structural immunogenic proteins as vaccine candidates. The different stages of therapeutic VLP design and development includes selection of antigenic protein or component (epitope), its expression in prokaryotic or eukaryotic system, purification and immune assays. However, to further maximize the magnitude and duration of the immunity, most of the licensed VLP-based vaccines also utilize adjuvants like, liposomes, agonists of pathogen recognition receptors, polymeric particles, emulsions, cytokines and bacterial toxins[59]. For example, some licensed prophylactic vaccines against HEV, human papilloma virus, and porcine circovirus are VLP-based vaccines. VLP technology combined with synthetic biology allows for more precise and predictable control over the composition and assembly of the capsids towards generating multivalent or cross-protective vaccines[60]. Moreover, a broad range of molecular manipulations such as encapsulation, chemical conjugation and genetic engineering further present VLPs as promising delivery agents for targeted gene therapy[61].
In addition, several viral capsid and envelope glycoproteins are exploited as drug-delivery vehicles in vitro and in vivo[62-64]. For example, brome mosaic virus (B MV) capsid assembles into different-sized therapeutic nanoparticles[64]. Recently, determination of a 1.4 Å resolution crystal structure of the novel SARS-CoV-2 nucleocapsid’s N2b domain has been determined, revealing its compact, intertwined architecture and self-assembly properties very similar to that of SARSCoV-1 and MERS-CoV[65]. Further, cryo-EM structure of its “spike” glycoprotein ectodomain-trimer has been also elucidated towards designing of vaccines and inhibitor candidates of viral entry[66]. Likewise, therapeutic virosomes are hybrid drug-delivery system that can carry genetically-modified nucleic acids, peptides, proteins and small organic molecules. A number of such prophylactic and therapeutic virosomes, especially anti-cancer products with high safety profiles are currently commercially available[67].
Compared to other enveloped icosahedral viruses, the adenovirus capsid surface has remarkable long, thin fibers primarily responsible for tethering of the viral capsid to the cell-receptor. Adenoviral capsid vectors have therefore, achieved substantial use in broad ranging therapeutic applications (e.g. hemophilia, cancer, and cystic fibrosis) in preclinical animal models and human trials[68]. Adeno-associated virus has been developed as gene therapy vector[69]. In addition, reconstituted pseudovirions of fusion-competent Sendai virus and influenza virus have been used as therapeutic gene delivery vehicles or nanoparticles[70]. Moreover, phage T4 capsid nanoparticles carrying reporter genes, vaccine candidates, enzymes, and ligands have been efficiently delivered in vitro and in vivo[71].
CONCLUSION
Viruses have remarkable differences in their geometric shapes, sizes and biomolecular compositions. Advances in molecular biology, X-ray crystallography, Cryo-EM and MDS have elucidated atomic-level understanding the structures of virions, including mechanisms of capsid assembly/disassembly, antigenicity, cell-receptor interaction, and designing therapeutic interventions. Cryo-EM combined with image analysis has provided 3D structures of icosahedral capsids that fail to form large crystals. Also, the structural details of influenza virus hemagglutinin and neuraminidase spikes, and adenovirus hexon unit are now known. These structures have further enhanced the information on antigenic surface for neutralizing antibodies, the cell-receptor site and fusion, polyprotein processing during maturation and egress as well as the interfering molecules of capsid functionality. In addition, several viral envelope and capsid proteins are exploited as targeted drug/gene-delivery vehicles. Because viral surface/envelope protein glycosylation influences antigenicity, further incorporating models of their glycan moieties would be a key to enhance full-scale virion simulations. This may further provide crucial insights into capsid assembly/ disassembly, nucleation of other components, viral genome packaging, antigenicity, interaction with cell-receptors, and therefore, exploiting for therapeutic strategies.
ACKNOWLEDGEMENTS
MKP gratefully acknowledges his mentor Dr. Shahid Jameel (Senior Scientist and CEO, Welcome Trust, United Kingdom−DBT, India Alliance) for his intellectual support and inspiration.
Manuscript source: Unsolicited manuscript
Specialty type: Virology
Country/Territory of origin: Saudi Arabia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Laassri M, Sotelo J S-Editor: Zhang L L-Editor: A P-Editor: Xing YX