Published online Nov 12, 2015. doi: 10.5501/wjv.v4.i4.313
Peer-review started: July 10, 2015
First decision: July 31, 2015
Revised: October 14, 2015
Accepted: October 23, 2015
Article in press: October 27, 2015
Published online: November 12, 2015
Processing time: 135 Days and 22.9 Hours
Hepatitis delta virus (HDV) is the etiologic agent of the most severe form of virus hepatitis in humans. Sharing some structural and functional properties with plant viroids, the HDV RNA contains a single open reading frame coding for the only virus protein, the Delta antigen. A number of unique features, including ribozyme activity, RNA editing, rolling-circle RNA replication, and redirection for a RNA template of host DNA-dependent RNA polymerase II, make this small pathogen an excellent model to study virus-cell interactions and RNA biology. Treatment options for chronic hepatitis Delta are scarce and ineffective. The disease burden is perhaps largely underestimated making the search for new, specific drugs, targets, and treatment strategies an important public health challenge. In this review we address the main features of virus structure, replication, and interaction with the host. Virus pathogenicity and current treatment options are discussed in the light of recent developments.
Core tip: Hepatitis delta virus (HDV) is the etiologic agent of probably the most severe form of virus hepatitis. HDV replication and spread depends on the presence of hepatitis B virus which provides the envelope proteins coded exclusively by its own genome. About 20 million people are currently chronically infected with HDV and no specific therapy is still available. Here, we review the current knowledge on HDV biology, epidemiology, pathogenesis, and treatment. Future trends and perspectives are discussed in the light of recent developments on HDV biology and its interaction with the host.
- Citation: Cunha C, Tavanez JP, Gudima S. Hepatitis delta virus: A fascinating and neglected pathogen. World J Virology 2015; 4(4): 313-322
- URL: https://www.wjgnet.com/2220-3249/full/v4/i4/313.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i4.313
Over 35 years have passed since Rizzetto et al[1] reported the discovery of what has been called Delta antigen in a patient with diagnosis of severe hepatitis B infection. Subsequent research on the nature of this antigen led to the identification, in 1980, of a new hepatotropic virus, hepatitis delta virus (HDV)[2,3]. This new infectious agent was later found to be a sub-viral agent dependent on the presence, in infected cells, of hepatitis B virus (HBV) to accomplish the replication cycle[3,4]. In nature, both viruses, HBV and HDV, share the same envelope proteins coded exclusively by the HBV genome[5,6].
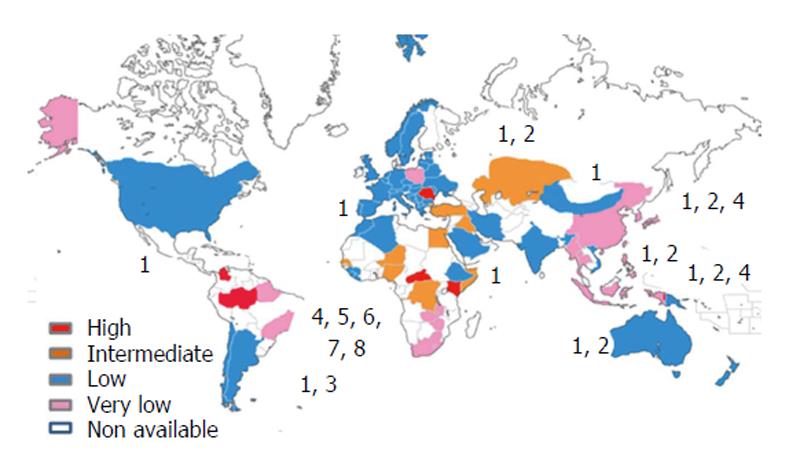
Today, the World Health Organization estimates that about 400 million people are chronically infected with HBV worldwide[7,8], of which approximately 20 million are co-infected with HDV[9,10]. The Amazon basin and some central African and east European countries are among the regions with higher prevalence. However, there is still a considerable lack of information concerning a significant number of countries mostly situated in Africa, Asia, and Latin America (Figure 1). The geographic distribution of the so far identified eight HDV clades is also far from being uniform. Clade 1 may be found worldwide, in contrast with clade 3 which seems to be confined to the Amazon region (Figure 1). The most frequent outcome of the acute co-infection with HDV is virus clearance and patient’s recovery. However, in up to 5% of the infected individuals a chronic form of HDV infection will develop[11]. In the case of super-infection, when a chronic HBV carrier gets super-infected with HDV, the outcome is distinct. About 70%-90% of super-infected individuals will become chronic carriers for both viruses, HBV and HDV[12].
As compared to the individuals that are chronic carriers of HBV alone, HDV additionally increases the risk of hepatocellular carcinoma (HCC) and mortality threefold and twofold, respectively, in HDV/HBV carriers[13,14]. Currently, in clinical practice, there are no drugs used that directly and specifically target HDV. None of the currently approved anti-HBV drugs efficiently blocks HDV infection[7,9,14-17].
All of the above, given additional HDV-inflicted liver pathogenesis, and inability to efficiently circumvent HDV infection by anti-HBV drugs, makes HDV a very serious pathogen, and it does call for additional attention to HDV and development of specific anti-HDV interventions.
HDV is mostly endemic in low income countries in which the budget for new, potentially expensive drugs is, of course, not the first priority. Accordingly, development of new treatment options based on specific drugs has not only proved to be difficult (the virus apparently does not code for any specific enzymatic activity that could be targeted) but may also represent an uninteresting option for pharmaceutical companies, speaking from a strictly financial point of view.
Nevertheless, this small human pathogen bears a set of features that make it a formidable model to study fundamental aspects of host-pathogen interactions and RNA biology including mechanisms of transcription, replication, and genome evolution[18,19]. The small size and structure of the genome bearing only one open reading frame (ORF), which is edited by host enzymes, its ribozyme activity and still largely undeciphered mechanism of RNA-directed RNA replication, are prominent examples of the uniqueness of this human pathogen[19].
In this review, we will address the specific features of HDV structure and replication, its interaction with host cells and HBV. Future perspectives of research based on recent important developments will be discussed.
The virus: HDV is an enveloped spherical subviral agent about 36 nm in diameter[19]. The virus particle contains a ribonucleoprotein (RNP) core consisting of one copy of the RNA genome and approximately 200 copies of the only virus encoded protein, the Delta antigen (HDAg)[20]. The HDV envelope contains hepatitis B virus surface antigens (HBsAg), provided solely by HBV. In accordance, the two viruses share virtually indistinguishable envelopes[6].
The virus genome is a circular single-stranded RNA molecule of around 1.7 kb and negative polarity[21,22]. A significant degree of internal base-pairing (about 70% of all nucleotides) is an important feature, with potential not yet unveiled functional implications, observed in this molecule[23,24]. This structure is similar to that described for plant viroids, albeit the latters have a smaller size and do not code for any protein (Table 1). On the contrary, the HDV genome displays one ORF which codes for the only viral protein, the Delta antigen[25-27]. This protein can be found in virions under two distinct forms: Small (S-HDAg, 195 aa) and large (L-HDAg; 213 or 214 aa, depending on the genotype). L-HDAg is synthesized mainly later in the replication cycle[28,29] as a consequence of an editing mechanism that takes place in the so-called anti-genome, an exact copy of the genome that arises as a replicative intermediate during RNA replication. The editing reaction is catalyzed by cellular adenosine deaminase 1 which converts an amber stop codon into a tryptophan codon (UGG) allowing a 57 nucleotide and consequently 19 aa extension of the ORF[30,31].
| HDV (1700 nt) | Pospiviroidae (200-400 nt) | Avsunviroidae (200-400 nt) |
| Circular ssRNA | Circular ssRNA | Circular ssRNA |
| Extensive intramolecular base pairing | Extensive intramolecular base pairing | Extensive intramolecular base pairing |
| A DNA-directed RNA polymerase makes both plus and minus strands | A DNA-directed RNA polymerase makes both plus and minus strands | A DNA-directed RNA polymerase makes both plus and minus strands |
| Encodes for protein | No proteins encoded | No proteins encoded |
| Virion maturation depends on a helper virus | Replication does not depend on the presence of a helper virus | Replication does not depend on the presence of a helper virus |
| Symmetric rolling circle RNA replication | Asymmetric rolling circle RNA replication | Symmetric rolling circle RNA replication |
| Replicates in the nucleus | Replicates in the nucleus | Replicates in chloroplasts |
| Ribozyme activity | No ribozyme activity | Ribozyme activity |
Both L-HDAg and S-HDAg share the same functional domains with the exception of the L-HDAg-specific C-terminal extension, which bears an isoprenylation signal present in cysteine residue 211[32]. Farnesylation of this residue is reported to be crucial albeit not sufficient for interaction with HBsAg and subsequent virion packaging and release from the cells[33,34]. The common functional motifs are a nuclear localization signal (NLS; aa 66-75), a coiled-coil domain (aa 12-60), and a bipartite arginine-rich RNA binding domain (aa 97-107 and 136-146; ARM1 and ARM2, respectively)[35-37]. More recently, however, it was shown that mutation in the core arginines of both ARM1 and ARM2 did not impair the RNA-binding ability of a C-terminal HDAg-160 truncated form of HDAg[38]. The authors suggested that HDAg establishes numerous contacts with HDV RNA to assemble ribonucleoprotein complexes.
Delta antigens: Several properties have been assigned to S-HDAg but none related to any known enzymatic activity. Among the reported putative and observed functions are the promotion of nuclear import of HDV RNPs[39], regulation of HDV RNA editing[40], facilitation of ribozyme cleavage (chaperone)[41,42], and facilitation of accumulation of processed RNA transcripts[43,44]. Both Delta antigens are post-translationally modified by host enzymes. Several post-translational modifications (PTM) have been described in HDAg and these include phosphorylation, methylation, acetylation, and sumoylation[45-48]. Phosphorylation occurs at multiple sites and can be mediated by different host kinases, dsRNA-activated protein kinase R, protein kinase C, and ERK1/2[49-51]. All these modifications may have distinct functional significance but it seems consensual that they are all involved in promoting virus RNA replication[52].
Methylation of Arg 13 on S-HDAg by arginine methyltransferase I was reported and proposed to be important to enhance both genomic RNA and mRNA synthesis[46]. Additionally, cellular p300 acetyltransferase was found to acetylate Lys72 on the NLS of S-HDAg[53]. Although speculative, this modification may have impact on the efficiency of nuclear import.
Finally, sumoylation was the most recent PTM to be reported on S-HDAg. It occurs at multiple lysine residues and is catalyzed by host small ubiquitin-related modifier isoform 1. Sumoylation was proposed to be important to promote genomic RNA and mRNA synthesis[48].
Undoubtedly, these observations represent only a tiny part of the whole picture drawn by HDAgs inside the cell. In fact, Delta antigens can also be found as peptides of different smaller sizes in the nucleus of HDV replicating cells[54]. Do these additional smaller forms correspond to distinct functional features? The answer is still far from being clear as no evidence supporting this point of view have been reported. In addition, it has been shown that S-HDAg can form multimers in HDV replicating cells[20,55,56]. These multimers may play an important role in virus replication by facilitating the accumulation of virus RNAs. Moreover, it is known that HDAgs are basic proteins with an estimated overall + 12 charge[57]. Thus, it is not surprising that, at least in vitro, the protein can bind nonspecifically to several types of nucleic acids including dsDNA and several distinct RNAs[58].
Furthermore, S-HDAg may also be involved in sequestering and manipulating host cell components to facilitate HDV replication. In this context, it is not surprising that the search for S-HDAg interacting proteins unveiled a considerable number of potential partners. First Cao et al[59] used an immunoprecipitation followed by mass spectrometry approach being able to identify more than 100 host proteins in the assay. Later, Gowans et al[60] performed a yeast two-hybrid screen using a human liver cDNA library and identified 30 host candidate proteins capable of specifically interacting with S-HDAg. Making use of RNA silencing strategies some of these candidate interactions were found to be of potential functional significance. However, the above mentioned strong positive charge of HDAgs compels one to be careful when analyzing the specificity and role of these interactions in the HDV replication cycle.
S-HDAg is predicted to be an intrinsically disordered protein, a property already assigned to several other virus and cellular proteins[61]. This feature may be responsible for the lack of success in all, to our knowledge at least in three different laboratories, attempts to crystalize and solve the 3D structure of the Delta antigen. These properties of the Delta antigen make the study of HDV biology much more complex than perhaps initially believed. However, as we shall discuss below, they are not the only most important ones.
HDV replication: HDV replication takes place in the nucleus of infected cells[60,62,63]. The study of the HDV replication has long been difficult due to the lack of an appropriate cell culture system capable of supporting all steps of the virus life cycle, from attachment to release from the cells. Primary human hepatocytes have been long the only cells known to support the complete life cycle of HDV[64]. These are expensive and not easy to cultivate. Thus, other approaches needed to be developed and a number of alternatives arose with time. Among them are the Hepa RG cell line and the stably transfected HEK-293 cells expressing S-HDAg under the control of a tetracycline inducible promoter[65,66]. Although not representing ideal models, they became important tools for HDV research. The recent identification of the sodium-taurocholate co-transporting polypeptide (NTCP, encoded by SLC10A1) as the bona fide receptor for HBV and HDV culminated a long run that included a number of tested hypothesis and putative isolations[67,68]. It represented an important breakthrough since it allowed engineering cell lines overexpressing it and consequently also supporting the initial steps of virus attachment and entry. So far, these human NTCP-expressing cell lines include human HepG2 and Huh7 as well as mouse Hepa1-6, AML-12, and primary mouse hepatocytes[69].
After the uncoating of virus particles, HDV RNPs are transported to the nucleus, where RNA replication takes place[70]. The existing data indicates replication of the virus genome involves a double rolling-circle mechanism with formation of multimeric anti-genomic and genomic molecules[71]. These RNA multimers are cleaved at precise monomeric intervals by a rybozime activity present in both genomic and antigenomic molecules[72,73]. The presence of ribozymes in HDV RNAs is a feature shared with the viroid family of Avsunviroidae[74] (Table 1).
Although it is well established that the presence of S-HDAg stimulates virus RNA accumulation, the precise role of this virus antigen in the mechanism of HDV RNA replication remains elusive. Controversy on which host polymerase or polymerases are involved in synthesis of genomes and antigenomes lasted, for a long time. Some groups claimed that both RNA pol I and pol II are involved in genome and antigenome synthesis, respectively[75]. Mainly, these evidences were obtained in in vitro assays using different inhibitory concentrations of α-amanitin and on reports showing the presence of virus RNA in the nucleolus[75,76]. By contrast, other groups, using different types of transcription inhibitors, actinomycin D, 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole, α-amanitin, provided data suggesting the involvement of solely RNA pol II[77]. Furthermore, the presence of virus RNA in the nucleolus could not be observed in the absence of Delta antigen suggesting that this presence lacks functional relevance[62,63]. In recent years, the use of immunoprecipitation and proteomic approaches, among others, led to the identification of several pol I, pol II, and pol III subunits as binding partners for HDV RNA[59]. These results need to be interpreted with care since the observed binding to HDV RNA could be a result of indirect interaction through other non-identified partners. However, independently of the host polymerase(s) involved in replication of virus RNAs a striking question is still hanging in the air: How does the virus redirect a host DNA-dependent RNA polymerase to use an RNA template? Here, the eventual participation of the S-HDAg, which as mentioned before displays a net positive charge and intrinsic disorder, may play a crucial role allowing the virus to overcome obstacles posed by the host environment for its replication.
The search for promoter sequences in virus RNA has also been followed by a few groups with inconclusive results. Yet, there is evidence from in vivo models supporting that mRNA synthesis initiates at nt 1630[78,79]. It may additionally be possible that multiple binding sequences for host RNA polymerases are present both in the virus genome and antigenome. This “nonspecific” binding could be a consequence of the RNA secondary structure bearing an extensive base-pairing with a number of predicted internal loops. Additionally, S-HDAg could also play an important role since it can bind nonspecifically to several nucleic acids, from dsDNA to ssRNA. It could be possible that S-HDAg plays a role as mediator between the virus RNA and a host RNA polymerase promoting its binding to several sequences in the genome and antigenome. Alternatively, S-HDAg could simply act as a chaperone, stabilizing RNA molecules and making them available for transcription. Assembly of HDV virions takes place in the cytoplasm. In this cellular compartment HBV-derived HBsAgs interact with HDV RNPs that are exported from the nucleus[80,81]. This interaction was shown to be mediated by L-HDAgs[82,83]. Tavanez et al[81] used heterokaryon assays to show that HDV RNPs shuttle between the nucleus and the cytoplasm. The authors claimed that nuclear import is mediated by an NLS located in Delta antigens (aa 66-75) and provided evidence that export to the cytoplasm is mediated by a cis-acting sequence in virus RNA[35]. However, Lee et al[84] (2001) have shown a year before that aa 198-210 in L-HDAg were able to promote the export of a reporter protein. More recently, Freitas and Cunha used a well-established CAT reporter system to investigate a possible presence of nuclear export elements (NEEs) in HDV RNAs[85]. The authors showed that NEEs may be present in both genomic and antigenomic molecules and that nuclear export is, at least in part, sensitive to leptomycin B, an inhibitor of the host CRM1-mediated export pathway. Whether a NES present in L-HDAg or a NEE in virus RNA are responsible for promoting HDV RNP export may be considered still controversial. Consequently, further research is mandatory to unequivocally answer this question.
Clinical manifestations and therapy: It is widely and for a longtime known that HDV infection is associated with a broad range of clinical manifestations, from asymptomatic to fulminant hepatitis. In the latter cases, mortality often reaches 80% of the affected individuals[86,87].
Concomitant infection of HBV and HDV usually displays more severe symptoms when compared with a single HBV infection. Nevertheless, the most frequent outcome is virus clearance, a situation reported in about 95% of the cases[88]. In contrast, HDV super-infection of chronic HBV patients results in progression to chronicity in up to 80% of patients. Moreover, about 60%-70% of these patients will develop cirrhosis[89]. These patients usually progress more rapidly to cirrhosis, show increased liver decompensation, and eventually death when compared with those chronically infected with HBV alone[90,91].
The factors influencing the distinct clinical course in coinfected and superinfected patients are still poorly understood. In both cases the organism produces a strong anti-HDAg antibody response which is, unfortunately, unable to modulate the course of infection[92-94]. The majority of superinfected patients progresses to chronic disease independent of the presence of high titers of anti-HDV antibodies. Despite the limited number of studies there are evidences supporting a role of cytotoxic T cells in HDV infection including the destruction of infected hepatocytes[95]. In any case, immunology of HDV infection is perhaps one of the most poorly understood aspects of the disease.
From the histologic point of view there are no detectable differences between anomalies observed in the liver of HDV-infected patients and patients with other acute or chronic virus liver disease[96,97]. These anomalies mostly consist of hepatocellular necrosis and inflammation and may represent, at least in part, a consequence of the immune response of the host. Proteomic and systems biology approaches have more recently been used to investigate changes in protein expression patterns and metabolic pathways altered during HDV replication. Although the model systems used can hardly be considered ideal, the obtained results provided consistent evidence that HDV replication results in significant alterations in pyruvate and glycolysis metabolism[98-100]. Of note, these studies have shown that cancer was the most likely disease associated with HDV replication and provided evidence that the G2/M cell cycle checkpoint is altered as a consequence of the presence of the virus[100]. Definitely, these observations, of which a significant number of arise from proteomic experiments and analysis, need to be interpreted and handled with care. In any case, it seems uncontroversial that further research on liver biopsies of infected patients may possibly help confirming these findings.
There is no efficient therapy for chronic HBV/HDV infection. Pegylated interferon-α (PEG-IFN-α) is perhaps the most popular therapy and the one that has shown some antiviral activity against HDV[15,101]. However, the efficacy is limited - a temporary reduction in virus titers is usually observed in 15%-40% patients - and the need for prolonged administration often results in severe adverse effects[101,102]. These effects include fatigue, weight loss, and psychiatric disturbances. Ribavirine, lamivudine and other nucleotide analogues have also been tested but have shown a very limited, if any, efficacy[103-106]. The Hep-Net International hepatitis D intervention trial included 77 patients from Germany, Greece, and Turkey. In this study a PEG-IFN-α2a therapy was compared with adefovir and a combination of PEG-IFN-α2a and adefovir[107]. Adefovir showed a very limited efficacy and the combination therapy based on PEG-IFN-α2a and adefovir was only superior in reducing HBsAg levels but not in HDV RNA[17]. In any case, HDV RNA relapses were often observed in a long-term follow-up (median time 4.5 years). The nucleoside analog entecavir, which showed antiviral efficacy in the woodchuck model of hepatitis B, was assayed in thirteen chronic hepatitis D patients for one year also proving to be ineffective[17]. It thus seems evident that current anti-HBV drugs are unable to efficiently circumvent HDV infection.
Today, it is usually recommended to treat chronic hepatitis D with PEG-IFN-α for at least one year if the patient tolerates the eventual adverse effects. However, in patients with advanced liver disease, liver transplantation may represent the only available option[108]. It is thus clear that current therapeutic options are unsatisfactory and there is an urgent need for more effective and specific anti-HDV drugs that will directly target HDV. Prenylation inhibitors may become an interesting and effective option and have been shown to be safe when used to treat neoplasias[109,110]. As discussed before, prenylation of L-HDAg is essential for interaction with HBsAg and virion assembly, and thus may be regarded as a potential target for therapeutic intervention.
Most recently, and as a consequence of the identification of NCTP as the host cell HDV receptor, inhibitors of viral entry have been tested and proposed as potential anti-viral drugs. Namely, Myrcludex B, a synthetic N-acylated preS1 lipopeptide and cyclosporine A were shown to inhibit virus entry by interfering with the receptor functions of NCTP, however, currently there is no data available regarding the performance of this drug in actual HDV-infected individuals[111,112].
However, it is clear that a higher investment in research of fundamental aspects of HDV biology as well as of anti-HDV specific compounds is crucial in order to improve the quality of life and life expectancy of chronic HBV/HDV carriers.
In the past few years a number of interesting developments have occurred in the field of HDV research and its interaction with HBV.
Using super-infection with WHV-enveloped HDV of the woodchucks that were chronic carriers of WHV and already developed HCCs, it was found that HDV was able to infect fractions of the cells of WHV-induced HCCs. These results suggest that at least a certain percentage of HCC cells in vivo express functional WHV receptors and support the attachment, entry, trafficking, and complete replication cycle of HDV[113]. The data also opens new avenues of research that will further address the mechanisms of the relationship between established HCCs and ongoing virus infection.
A second study compared several types of HDV that differed only by the envelope proteins of HBV that coated the virions[114]. Twenty five different types of HBV envelope proteins that belonged to twenty five different HBV variants of nine genotypes A-I were analyzed. It was found that all nine HBV genotypes tested were able to support the production of infectious HDV virions that contained HDV genome of genotype I. Significant differences in infectivity were found for the envelope proteins of different HBV variants. The data generated strongly suggest that HBV envelope proteins facilitate not only attachment and entry, but also at least one additional immediate post-entry step of the HDV life cycle. In addition, testing of infectivity suggested that it cannot be concluded that the envelope proteins of HBV produced during chronic stage of HBV infection are mainly responsible for assembly of the virions with diminished infectivity. The study also suggested that correctly regulated disassembly of HDV RNP from the HBV envelope proteins after entry is critical for the overall infectivity of HDV particles[114].
Finally, a third recent study demonstrated that infectious HDV virions can be assembled by the envelope proteins derived from the naturally integrated HBV DNA in the absence of ongoing HBV replication[115]. These findings suggest that HDV can possibly persist in vivo in the absence of HBV replication (or when HBV replication is suppressed by a drug), when functional HBV envelope proteins are supplied from integrated HBV DNA. Such a mechanism of HDV persistence was not explored previously. The results obtained explain, at least in part, inability of anti-HBV drugs to efficiently block HDV infection in vivo. Additionally, they also suggest that HDV can be actually a more independent and more significant pathogen than it is currently assumed[116].
As discussed earlier, HDV bears a number of characteristics similar to those found in plant viroids (Table 1).
These similarities may allow speculation on a possible HDV origin from the plant world. According to this hypothesis, HDV could have evolved to encode the Delta antigen thus providing an explanation for its larger genome when compared with viroids[117]. However, a deeper analysis of this homology was evaluated as non-significant and this hypothesis seems to be, at least for the time being, ruled out.
One of the key features of HDV genomic and antigenomic RNA molecules is their ribozyme activity. Ribozymes are considered to be characteristic of viroids. However, the two HDV ribozymes are not only structurally different from those of Avsunviroidae but also display similarities to several HDV-like ribozymes found in eukaryotes[74,116]. This finding rather supports the hypothesis of a human transcriptome origin of HDV.
We can thus conclude that the plant or animal origins of HDV are still questionable and highly speculative. But this is one of the many fascinating questions that still remain to be unveiled for this awkward and awesome virus.
Almost 40 years after its discovery, HDV remains a challenge for clinicians and researchers. It is disconcerting simplicity, with a small RNA genome and a single protein, the Delta antigen, make it an excellent model not only for virologists but also for those interested in RNA and cell biology. The virus bears a number of unique features including a RNA-directed RNA replication mechanism of the genome catalyzed by host RNA polymerase II. Enzymatic activities were not identified in Delta antigens thus making difficult the identification of potential targets for specific and effective therapies. Development of such therapies is crucial to reduce the number of chronic patients progressing to cirrhosis and hepatocellular carcinoma. The burden of disease caused by HDV is most probably underestimated since there is a considerable lack of epidemiologic data from several countries where HBV is highly prevalent.
In conclusion, despite considerable progress made in HDV research a significant number of questions remain to be answered concerning fundamental aspects of its biology, pathogenesis, and interaction with the host. The next few years will hopefully bring to light new answers but also new exciting questions, helping understand this fascinating pathogen, and contributing to reducing morbidity and mortality among infected individuals.
João Paulo Tavanez is a recipient of a Fundação para a Ciência e Tecnologia post-doctoral fellowship.
P- Reviewer: Marzuillo P, Mishra PK, Russell RS S- Editor: Qiu S L- Editor: A E- Editor: Liu SQ
| 1. | Rizzetto M, Canese MG, Aricò S, Crivelli O, Trepo C, Bonino F, Verme G. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977;18:997-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 642] [Article Influence: 13.4] [Reference Citation Analysis (1)] |
| 2. | Rizzetto M, Canese MG, Gerin JL, London WT, Sly DL, Purcell RH. Transmission of the hepatitis B virus-associated delta antigen to chimpanzees. J Infect Dis. 1980;141:590-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 343] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 3. | Rizzetto M, Hoyer B, Canese MG, Shih JW, Purcell RH, Gerin JL. delta Agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc Natl Acad Sci USA. 1980;77:6124-6128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 314] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 4. | Ponzetto A, Negro F, Popper H, Bonino F, Engle R, Rizzetto M, Purcell RH, Gerin JL. Serial passage of hepatitis delta virus in chronic hepatitis B virus carrier chimpanzees. Hepatology. 1988;8:1655-1661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Smedile A, Rizzetto M, Gerin JL. Advances in hepatitis D virus biology and disease. Prog Liver Dis. 1994;12:157-175. [PubMed] |
| 6. | Sureau C. The role of the HBV envelope proteins in the HDV replication cycle. Curr Top Microbiol Immunol. 2006;307:113-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Gish RG. Current treatment and future directions in the management of chronic hepatitis B viral infection. Clin Liver Dis. 2005;9:541-565, v. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Akbar F, Yoshida O, Abe M, Hiasa Y, Onji M. Engineering immune therapy against hepatitis B virus. Hepatol Res. 2007;37 Suppl 3:S351-S356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Heidrich B, Manns MP, Wedemeyer H. Treatment options for hepatitis delta virus infection. Curr Infect Dis Rep. 2013;15:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 10. | Reinheimer C, Doerr HW, Berger A. Hepatitis delta: on soft paws across Germany. Infection. 2012;40:621-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Hadziyannis SJ. Review: hepatitis delta. J Gastroenterol Hepatol. 1997;12:289-298. [PubMed] |
| 12. | Lau DT, Kleiner DE, Park Y, Di Bisceglie AM, Hoofnagle JH. Resolution of chronic delta hepatitis after 12 years of interferon alfa therapy. Gastroenterology. 1999;117:1229-1233. [PubMed] |
| 13. | Rizzetto M, Verme G, Recchia S, Bonino F, Farci P, Aricò S, Calzia R, Picciotto A, Colombo M, Popper H. Chronic hepatitis in carriers of hepatitis B surface antigen, with intrahepatic expression of the delta antigen. An active and progressive disease unresponsive to immunosuppressive treatment. Ann Intern Med. 1983;98:437-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 262] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 14. | Fattovich G, Giustina G, Christensen E, Pantalena M, Zagni I, Realdi G, Schalm SW. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. The European Concerted Action on Viral Hepatitis (Eurohep). Gut. 2000;46:420-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 362] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 15. | Yurdaydin C. Treatment of chronic delta hepatitis. Semin Liver Dis. 2012;32:237-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 16. | Heidrich B, Yurdaydın C, Kabaçam G, Ratsch BA, Zachou K, Bremer B, Dalekos GN, Erhardt A, Tabak F, Yalcin K. Late HDV RNA relapse after peginterferon alpha-based therapy of chronic hepatitis delta. Hepatology. 2014;60:87-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 221] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 17. | Kabaçam G, Onder FO, Yakut M, Seven G, Karatayli SC, Karatayli E, Savas B, Idilman R, Bozdayi AM, Yurdaydin C. Entecavir treatment of chronic hepatitis D. Clin Infect Dis. 2012;55:645-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Taylor JM. Hepatitis delta virus. Virology. 2006;344:71-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 141] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 19. | Alves C, Branco C, Cunha C. Hepatitis delta virus: a peculiar virus. Adv Virol. 2013;2013:560105. [PubMed] |
| 20. | Gudima S, Chang J, Moraleda G, Azvolinsky A, Taylor J. Parameters of human hepatitis delta virus genome replication: the quantity, quality, and intracellular distribution of viral proteins and RNA. J Virol. 2002;76:3709-3719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Saldanha JA, Thomas HC, Monjardino JP. Cloning and sequencing of RNA of hepatitis delta virus isolated from human serum. J Gen Virol. 1990;71:1603-1606. [PubMed] |
| 22. | Makino S, Chang MF, Shieh CK, Kamahora T, Vannier DM, Govindarajan S, Lai MM. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature. 1987;329:343-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 245] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 23. | Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 526] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 24. | Chen PJ, Kalpana G, Goldberg J, Mason W, Werner B, Gerin J, Taylor J. Structure and replication of the genome of the hepatitis delta virus. Proc Natl Acad Sci USA. 1986;83:8774-8778. [PubMed] |
| 25. | Bergmann KF, Gerin JL. Antigens of hepatitis delta virus in the liver and serum of humans and animals. J Infect Dis. 1986;154:702-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 118] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Bonino F, Heermann KH, Rizzetto M, Gerlich WH. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J Virol. 1986;58:945-950. [PubMed] |
| 27. | Weiner AJ, Choo QL, Wang KS, Govindarajan S, Redeker AG, Gerin JL, Houghton M. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J Virol. 1988;62:594-599. [PubMed] |
| 28. | Luo GX, Chao M, Hsieh SY, Sureau C, Nishikura K, Taylor J. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol. 1990;64:1021-1027. [PubMed] |
| 29. | Chao M, Hsieh SY, Taylor J. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990;64:5066-5069. [PubMed] |
| 30. | Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 252] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 31. | Wong SK, Lazinski DW. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci USA. 2002;99:15118-15123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 150] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 32. | Glenn JS, Watson JA, Havel CM, White JM. Identification of a prenylation site in delta virus large antigen. Science. 1992;256:1331-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 248] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 33. | Lee CZ, Chen PJ, Lai MM, Chen DS. Isoprenylation of large hepatitis delta antigen is necessary but not sufficient for hepatitis delta virus assembly. Virology. 1994;199:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 34. | Otto JC, Casey PJ. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J Biol Chem. 1996;271:4569-4572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Alves C, Freitas N, Cunha C. Characterization of the nuclear localization signal of the hepatitis delta virus antigen. Virology. 2008;370:12-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | Lee CZ, Lin JH, Chao M, McKnight K, Lai MM. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J Virol. 1993;67:2221-2227. [PubMed] |
| 37. | Zuccola HJ, Rozzelle JE, Lemon SM, Erickson BW, Hogle JM. Structural basis of the oligomerization of hepatitis delta antigen. Structure. 1998;6:821-830. [PubMed] |
| 38. | Daigh LH, Griffin BL, Soroush A, Mamedov MR, Casey JL. Arginine-rich motifs are not required for hepatitis delta virus RNA binding activity of the hepatitis delta antigen. J Virol. 2013;87:8665-8674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Xia YP, Yeh CT, Ou JH, Lai MM. Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J Virol. 1992;66:914-921. [PubMed] |
| 40. | Cheng Q, Jayan GC, Casey JL. Differential inhibition of RNA editing in hepatitis delta virus genotype III by the short and long forms of hepatitis delta antigen. J Virol. 2003;77:7786-7795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 41. | Huang ZS, Wu HN. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J Biol Chem. 1998;273:26455-26461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Wang CC, Chang TC, Lin CW, Tsui HL, Chu PB, Chen BS, Huang ZS, Wu HN. Nucleic acid binding properties of the nucleic acid chaperone domain of hepatitis delta antigen. Nucleic Acids Res. 2003;31:6481-6492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 43. | Lazinski DW, Taylor JM. Relating structure to function in the hepatitis delta virus antigen. J Virol. 1993;67:2672-2680. [PubMed] |
| 44. | Wu TT, Netter HJ, Lazinski DW, Taylor JM. Effects of nucleotide changes on the ability of hepatitis delta virus to transcribe, process, and accumulate unit-length, circular RNA. J Virol. 1997;71:5408-5414. [PubMed] |
| 45. | Hong SY, Chen PJ. Phosphorylation of serine 177 of the small hepatitis delta antigen regulates viral antigenomic RNA replication by interacting with the processive RNA polymerase II. J Virol. 2010;84:1430-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 46. | Li YJ, Stallcup MR, Lai MM. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J Virol. 2004;78:13325-13334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 47. | Mu JJ, Tsay YG, Juan LJ, Fu TF, Huang WH, Chen DS, Chen PJ. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology. 2004;319:60-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 48. | Tseng CH, Cheng TS, Shu CY, Jeng KS, Lai MM. Modification of small hepatitis delta virus antigen by SUMO protein. J Virol. 2010;84:918-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 49. | Chen CW, Tsay YG, Wu HL, Lee CH, Chen DS, Chen PJ. The double-stranded RNA-activated kinase, PKR, can phosphorylate hepatitis D virus small delta antigen at functional serine and threonine residues. J Biol Chem. 2002;277:33058-33067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 50. | Yeh TS, Lo SJ, Chen PJ, Lee YH. Casein kinase II and protein kinase C modulate hepatitis delta virus RNA replication but not empty viral particle assembly. J Virol. 1996;70:6190-6198. [PubMed] |
| 51. | Chen YS, Huang WH, Hong SY, Tsay YG, Chen PJ. ERK1/2-mediated phosphorylation of small hepatitis delta antigen at serine 177 enhances hepatitis delta virus antigenomic RNA replication. J Virol. 2008;82:9345-9358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 52. | Huang WH, Chen CW, Wu HL, Chen PJ. Post-translational modification of delta antigen of hepatitis D virus. Curr Top Microbiol Immunol. 2006;307:91-112. [PubMed] |
| 53. | Huang WH, Mai RT, Lee YH. Transcription factor YY1 and its associated acetyltransferases CBP and p300 interact with hepatitis delta antigens and modulate hepatitis delta virus RNA replication. J Virol. 2008;82:7313-7324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 54. | Wang JG, Lemon SM. Hepatitis delta virus antigen forms dimers and multimeric complexes in vivo. J Virol. 1993;67:446-454. [PubMed] |
| 55. | Cornillez-Ty CT, Lazinski DW. Determination of the multimerization state of the hepatitis delta virus antigens in vivo. J Virol. 2003;77:10314-10326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 56. | Lin BC, Defenbaugh DA, Casey JL. Multimerization of hepatitis delta antigen is a critical determinant of RNA binding specificity. J Virol. 2010;84:1406-1413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 57. | Kuo MY, Goldberg J, Coates L, Mason W, Gerin J, Taylor J. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J Virol. 1988;62:1855-1861. [PubMed] |
| 58. | Alves C, Cheng H, Roder H, Taylor J. Intrinsic disorder and oligomerization of the hepatitis delta virus antigen. Virology. 2010;407:333-340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 59. | Cao D, Haussecker D, Huang Y, Kay MA. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication. RNA. 2009;15:1971-1979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 60. | Gowans EJ, Baroudy BM, Negro F, Ponzetto A, Purcell RH, Gerin JL. Evidence for replication of hepatitis delta virus RNA in hepatocyte nuclei after in vivo infection. Virology. 1988;167:274-278. [PubMed] |
| 61. | Casaca A, Fardilha M, da Cruz e Silva E, Cunha C. The heterogeneous ribonuclear protein C interacts with the hepatitis delta virus small antigen. Virol J. 2011;8:358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 62. | Cunha C, Monjardino J, Cheng D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus RNA in the nucleus of human cells. RNA. 1998;4:680-693. [PubMed] |
| 63. | Han Z, Alves C, Gudima S, Taylor J. Intracellular localization of hepatitis delta virus proteins in the presence and absence of viral RNA accumulation. J Virol. 2009;83:6457-6463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 64. | Sureau C, Jacob JR, Eichberg JW, Lanford RE. Tissue culture system for infection with human hepatitis delta virus. J Virol. 1991;65:3443-3450. [PubMed] |
| 65. | Sureau C. The use of hepatocytes to investigate HDV infection: the HDV/HepaRG model. Methods Mol Biol. 2010;640:463-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 66. | Chang J, Gudima SO, Tarn C, Nie X, Taylor JM. Development of a novel system to study hepatitis delta virus genome replication. J Virol. 2005;79:8182-8188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 67. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1602] [Article Influence: 123.2] [Reference Citation Analysis (1)] |
| 68. | Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 623] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 69. | Li H, Zhuang Q, Wang Y, Zhang T, Zhao J, Zhang Y, Zhang J, Lin Y, Yuan Q, Xia N. HBV life cycle is restricted in mouse hepatocytes expressing human NTCP. Cell Mol Immunol. 2014;11:175-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 70. | Chou HC, Hsieh TY, Sheu GT, Lai MM. Hepatitis delta antigen mediates the nuclear import of hepatitis delta virus RNA. J Virol. 1998;72:3684-3690. [PubMed] |
| 71. | Modahl LE, Lai MM. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J Virol. 1998;72:5449-5456. [PubMed] |
| 72. | Wadkins TS, Been MD. Core-associated non-duplex sequences distinguishing the genomic and antigenomic self-cleaving RNAs of hepatitis delta virus. Nucleic Acids Res. 1997;25:4085-4092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 73. | Wang KS, Choo QL, Weiner AJ, Ou JH, Najarian RC, Thayer RM, Mullenbach GT, Denniston KJ, Gerin JL, Houghton M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature. 1986;323:508-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 537] [Cited by in RCA: 531] [Article Influence: 13.6] [Reference Citation Analysis (1)] |
| 74. | Flores R, Grubb D, Elleuch A, Nohales MÁ, Delgado S, Gago S. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 2011;8:200-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 89] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 75. | Macnaughton TB, Shi ST, Modahl LE, Lai MM. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J Virol. 2002;76:3920-3927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 99] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 76. | Li YJ, Macnaughton T, Gao L, Lai MM. RNA-templated replication of hepatitis delta virus: genomic and antigenomic RNAs associate with different nuclear bodies. J Virol. 2006;80:6478-6486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 77. | Chang J, Nie X, Gudima S, Taylor J. Action of inhibitors on accumulation of processed hepatitis delta virus RNAs. J Virol. 2006;80:3205-3214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 78. | Gudima S, Dingle K, Wu TT, Moraleda G, Taylor J. Characterization of the 5’ ends for polyadenylated RNAs synthesized during the replication of hepatitis delta virus. J Virol. 1999;73:6533-6539. [PubMed] |
| 79. | Gudima S, Wu SY, Chiang CM, Moraleda G, Taylor J. Origin of hepatitis delta virus mRNA. J Virol. 2000;74:7204-7210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 80. | Jenna S, Sureau C. Effect of mutations in the small envelope protein of hepatitis B virus on assembly and secretion of hepatitis delta virus. Virology. 1998;251:176-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 81. | Tavanez JP, Cunha C, Silva MC, David E, Monjardino J, Carmo-Fonseca M. Hepatitis delta virus ribonucleoproteins shuttle between the nucleus and the cytoplasm. RNA. 2002;8:637-646. [PubMed] |
| 82. | Chang FL, Chen PJ, Tu SJ, Wang CJ, Chen DS. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc Natl Acad Sci USA. 1991;88:8490-8494. [PubMed] |
| 83. | Ryu WS, Bayer M, Taylor J. Assembly of hepatitis delta virus particles. J Virol. 1992;66:2310-2315. [PubMed] |
| 84. | Lee CH, Chang SC, Wu CH, Chang MF. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J Biol Chem. 2001;276:8142-8148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 85. | Freitas N, Cunha C. Searching for nuclear export elements in hepatitis D virus RNA. World J Virol. 2013;2:123-135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 86. | Farci P, Niro GA. Clinical features of hepatitis D. Semin Liver Dis. 2012;32:228-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 87. | Lee WM. Acute liver failure. N Engl J Med. 1993;329:1862-1872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 441] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 88. | Caredda F, Rossi E, d’Arminio Monforte A, Zampini L, Re T, Meroni B, Moroni M. Hepatitis B virus-associated coinfection and superinfection with delta agent: indistinguishable disease with different outcome. J Infect Dis. 1985;151:925-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 89. | Fattovich G, Boscaro S, Noventa F, Pornaro E, Stenico D, Alberti A, Ruol A, Realdi G. Influence of hepatitis delta virus infection on progression to cirrhosis in chronic hepatitis type B. J Infect Dis. 1987;155:931-935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 141] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 90. | Smedile A, Farci P, Verme G, Caredda F, Cargnel A, Caporaso N, Dentico P, Trepo C, Opolon P, Gimson A. Influence of delta infection on severity of hepatitis B. Lancet. 1982;2:945-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 277] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 91. | Buti M, Homs M, Rodriguez-Frias F, Funalleras G, Jardí R, Sauleda S, Tabernero D, Schaper M, Esteban R. Clinical outcome of acute and chronic hepatitis delta over time: a long-term follow-up study. J Viral Hepat. 2011;18:434-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 92. | Purcell RH, Rizzetto M, Gerin JL. Hepatitis delta virus infection of the liver. Semin Liver Dis. 1984;4:340-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 93. | DeCock KM, Govindarajan S, Redeker AG. Serological response to hepatitis delta virus in hepatitis D. Lancet. 1987;1:1438. [PubMed] |
| 94. | Fiedler M, Roggendorf M. Immunology of HDV infection. Curr Top Microbiol Immunol. 2006;307:187-209. [PubMed] |
| 95. | Huang YH, Tao MH, Hu CP, Syu WJ, Wu JC. Identification of novel HLA-A*0201-restricted CD8+ T-cell epitopes on hepatitis delta virus. J Gen Virol. 2004;85:3089-3098. [PubMed] |
| 96. | Colombari R, Dhillon AP, Piazzola E, Tomezzoli AA, Angelini GP, Capra F, Tomba A, Scheuer PJ. Chronic hepatitis in multiple virus infection: histopathological evaluation. Histopathology. 1993;22:319-325. [PubMed] |
| 97. | Mathurin P, Thibault V, Kadidja K, Ganne-Carrié N, Moussalli J, El Younsi M, Di Martino V, Lunel F, Charlotte F, Vidaud M. Replication status and histological features of patients with triple (B, C, D) and dual (B, C) hepatic infections. J Viral Hepat. 2000;7:15-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 98. | Mota S, Mendes M, Penque D, Coelho AV, Cunha C. Changes in the proteome of Huh7 cells induced by transient expression of hepatitis D virus RNA and antigens. J Proteomics. 2008;71:71-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 99. | Mota S, Mendes M, Freitas N, Penque D, Coelho AV, Cunha C. Proteome analysis of a human liver carcinoma cell line stably expressing hepatitis delta virus ribonucleoproteins. J Proteomics. 2009;72:616-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 100. | Mendes M, Pérez-Hernandez D, Vázquez J, Coelho AV, Cunha C. Proteomic changes in HEK-293 cells induced by hepatitis delta virus replication. J Proteomics. 2013;89:24-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 101. | Goyal A, Murray JM. Effect of interferon-alpha therapy on hepatitis D virus. Hepatology. 2015;61:2117-2118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 102. | Heller T, Rotman Y, Koh C, Clark S, Haynes-Williams V, Chang R, McBurney R, Schmid P, Albrecht J, Kleiner DE. Long-term therapy of chronic delta hepatitis with peginterferon alfa. Aliment Pharmacol Ther. 2014;40:93-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 103. | Triantos C, Kalafateli M, Nikolopoulou V, Burroughs A. Meta-analysis: antiviral treatment for hepatitis D. Aliment Pharmacol Ther. 2012;35:663-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 104. | Gunsar F, Akarca US, Ersoz G, Kobak AC, Karasu Z, Yuce G, Ilter T, Batur Y. Two-year interferon therapy with or without ribavirin in chronic delta hepatitis. Antivir Ther. 2005;10:721-726. [PubMed] |
| 105. | Yurdaydin C, Bozkaya H, Onder FO, Sentürk H, Karaaslan H, Akdoğan M, Cetinkaya H, Erden E, Erkan-Esin O, Yalçin K. Treatment of chronic delta hepatitis with lamivudine vs lamivudine + interferon vs interferon. J Viral Hepat. 2008;15:314-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 99] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 106. | Wedemeyer H, Yurdaydìn C, Dalekos GN, Erhardt A, Çakaloğlu Y, Değertekin H, Gürel S, Zeuzem S, Zachou K, Bozkaya H. Peginterferon plus adefovir versus either drug alone for hepatitis delta. N Engl J Med. 2011;364:322-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 359] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 107. | Nattermann J, Nitschmann S. [Therapy of hepatitis delta. The Hep-Net International Delta Hepatitis Intervention Trial]. Internist (Berl). 2011;52:1365-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 108. | Roche B, Samuel D. Liver transplantation in delta virus infection. Semin Liver Dis. 2012;32:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 109. | Bordier BB, Ohkanda J, Liu P, Lee SY, Salazar FH, Marion PL, Ohashi K, Meuse L, Kay MA, Casey JL. In vivo antiviral efficacy of prenylation inhibitors against hepatitis delta virus. J Clin Invest. 2003;112:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 110. | Koh C, Yurdaydin C, Cooper S, Cory D, Dahari H, Haynes-Williams V, Winters MA, Bys M, Choong IC, R Idilman R. Prenylation inhibition with lonafarnib decreases hepatitis D levels in humans. Hepatology. 2014;60:1092A. |
| 111. | Volz T, Giersch K, Allweiss L, Bhadra OD, Petersen J, Lohse AW, Lütgehetmann M, Urban S, Dandri M. Myrcludex-B inhibits establishment of HDV super-infection in HBV infected mice and reduces HDV viremia in stably HBV/HDV co-infected mice. J Hepatol. 2015;62, S514. |
| 112. | Nkongolo S, Ni Y, Lempp FA, Kaufman C, Lindner T, Esser-Nobis K, Lohmann V, Mier W, Mehrle S, Urban S. Cyclosporin A inhibits hepatitis B and hepatitis D virus entry by cyclophilin-independent interference with the NTCP receptor. J Hepatol. 2014;60:723-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 197] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 113. | Freitas N, Salisse J, Cunha C, Toshkov I, Menne S, Gudima SO. Hepatitis delta virus infects the cells of hepadnavirus-induced hepatocellular carcinoma in woodchucks. Hepatology. 2012;56:76-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 114. | Freitas N, Abe K, Cunha C, Menne S, Gudima SO. Support of the infectivity of hepatitis delta virus particles by the envelope proteins of different genotypes of hepatitis B virus. J Virol. 2014;88:6255-6267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 115. | Freitas N, Cunha C, Menne S, Gudima SO. Envelope proteins derived from naturally integrated hepatitis B virus DNA support assembly and release of infectious hepatitis delta virus particles. J Virol. 2014;88:5742-5754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 116. | Flores R, Ruiz-Ruiz S, Serra P. Viroids and hepatitis delta virus. Semin Liver Dis. 2012;32:201-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 117. | Taylor J, Pelchat M. Origin of hepatitis delta virus. Future Microbiol. 2010;5:393-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |