Published online Aug 12, 2015. doi: 10.5501/wjv.v4.i3.295
Peer-review started: November 10, 2014
First decision: January 20, 2015
Revised: February 13, 2015
Accepted: May 5, 2015
Article in press: May 6, 2015
Published online: August 12, 2015
Processing time: 279 Days and 18.3 Hours
Foot-and-mouth disease (FMD) is a highly contagious and economically devastating disease of livestock, primarily affecting cattle, buffalo and pigs. FMD virus serotypes O, A and Asia1 are prevalent in India and systematic efforts are on to control and eventually eradicate the disease from the country. FMD epidemiology is complex due to factors like co-circulation, extinction, emergence and re-emergence of genotypes/lineages within the three serotypes, animal movement, diverse farm practices and large number of susceptible livestock in the country. Systematic vaccination, prompt diagnosis, strict biosecurity measures, and regular monitoring of vaccinal immunity and surveillance of virus circulation are indispensible features for the effective implementation of the control measures. Availability of suitable companion diagnostic tests is very important in this endeavour. In this review, the diagnostic assays developed and validated in India and their contribution in FMD control programme is presented.
Core tip: To inform scientific community, this short review summarizes existing foot-and-mouth disease diagnostics developed in the recent past and used in India. Immediate and future requirements in the diagnostics are highlighted.
- Citation: Sharma GK, Mahajan S, Matura R, Subramaniam S, Ranjan R, Biswal J, Rout M, Mohapatra JK, Dash BB, Sanyal A, Pattnaik B. Diagnostic assays developed for the control of foot-and-mouth disease in India. World J Virology 2015; 4(3): 295-302
- URL: https://www.wjgnet.com/2220-3249/full/v4/i3/295.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i3.295
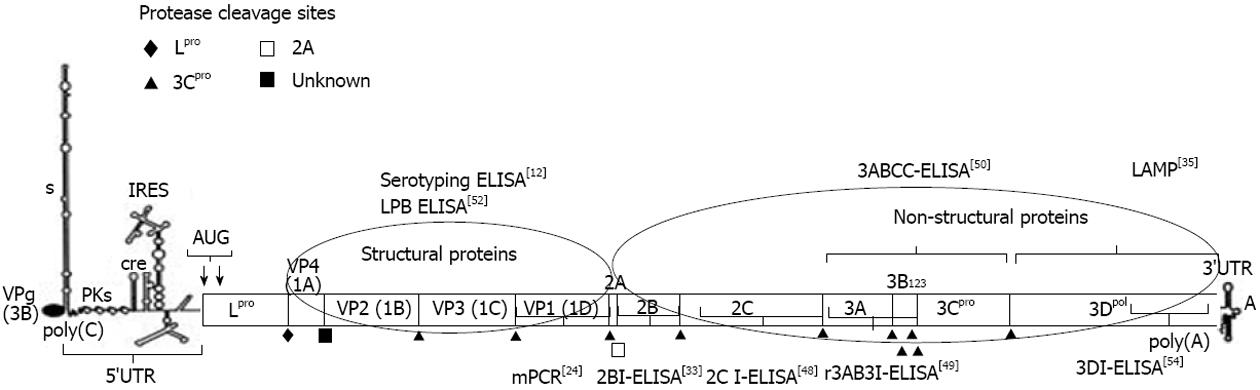
Foot and mouth disease (FMD) continues to pose threat to the livestock sector in the world. The annual direct loss due to FMD in India is estimated at United States dollar4.45 billion[1]. Economic losses due to trade barrier imposed by FMD free countries could be much more. FMD is caused by a single stranded positive sense RNA virus, belonging to the genus Aphthovirus of family Picornaviridae[2]. Seven serotypes (O, A, C, Asia1, SAT-1, SAT-2 and SAT-3) and multiple antigenic variants within the serotypes of FMD virus (FMDV) exist because of the variable antigenic nature of its structural proteins.The FMDV genome of approximately 8.5 kb is polyadenylated at 3’ terminus and carries a small protein VPg at its 5’ end[3,4]. It encodes four structural proteins (SPs) (VP1-4) and at least 8 non-structural proteins (NSPs). Structural proteins, VP1, VP2, VP3 and VP4 are formed by post-translation cleavage of a precursor coded by 1D, 1B, 1C and 1A genes, respectively[3]. Non-structural proteins of FMDV consist of L, 2A, 2B, 2C, 3A, 3B1, 3B2, 3B3, 3C, and 3D. The L gene which encodes L protein is situated at the extreme 5’ end of the coding region while all other NSPs are encoded by the P2 and P3 regions, which are situated towards the 3’ end of the viral RNA (Figure 1). The P2 region codes for 2A, 2B and 2C while P3 codes for 3A, 3B1, 3B2, 3B3, 3C and 3D.
Most of the developed countries are free from FMD, whereas the disease is present in many developing countries including India. Epidemiology of FMD in India is complex due to prevalence of many variants of FMDV serotypes (O, A, Asia1), mixed farming system, diverse landscape, animal husbandry practices and very large population (about 500 million) of susceptible livestock[5].
The disease in cattle and buffalo is characterized by high fever, depression, excessive salivation, formation of vesicles on the tongue and oral cavity, epidermis of the coronary band and inter digital space, udder and teats. Formation of vesicles in the oral cavity results in reduced food consumption, weight loss and emaciation. Vesicles may also develop in the epithelium of the pharynx, larynx, trachea, oesophagus and rumen. In young animals, it may lead to death due to myocarditis. Many times it leads to secondary bacterial infection in affected animals. While mortality is generally less than 3%, morbidity is very high and economic losses become unbearable for the farmers on account of decreased productivity and protracted convalescence in affected animals. Though, mortality is notably high in young pigs. Incubation period ranges from 2 to 14 d, but is generally shorter than a week.
FMDV can be transmitted by direct contact, aerosols, mechanical carriage by men or fomites and through animal products such as meat, offal, milk, semen or embryos. Infected pigs shed large quantities of virus in aerosols[6] and spread the virus down wind. Under favourable conditions of low temperature, high humidity and moderate winds, virus in aerosols may spread up to 250 km over sea[7] and 60 km over land[7]. Virus can remain infective on soil for 3 d in summer and for up to 28 d in winter[8].
FMD symptoms could be confused with other vesicular diseases like Swine Vesicular Disease (cattle and sheep are resistant), Vesicular Exanthema (cattle and sheep are resistant), Vesicular Stomatitis virus (sheep/goats are resistant). Availability of rapid and sensitive FMD diagnostic assays is essential in order to confirm the initial cases and prevent further spread of the disease. Infected animals may secrete the virus before clinical symptoms develop and the virus could spread rapidly in the susceptible population; hence rapid and early identification of the infected/carrier animals is critical.
Timely identification of serotype of the virus involved in the outbreak is of the utmost importance for disease control. Besides, apparently healthy animal population in endemic settings are to be regularly screened for the presence of antibodies against SPs and NSPs of FMDV and for the presence of the virus in the oro-pharynx to confirm the carrier status. Many diagnostic assays have been developed throughout the world for rapid and specific detection of FMDV and the antibodies against the FMDV proteins. Most of these assays are developed and validated considering the local requirements and prevailing virus pool, whereas some assays have been developed for use in the broad geographical areas. Now a day, molecular methods for FMD diagnosis are playing important role when compared to the conventional methods.
The episodes of FMD outbreaks are to be actively monitored, recorded and investigated in order to support the vaccination based control programme in the country. For all these activities, availability of rapid, sensitive, specific and economical diagnostic assays representing the FMDV pool in circulation is of prime importance and necessity.
A systematic vaccination based control is in operation for control and eventual eradication of FMD from India since 2003-2004 by Government of India (Department of Animal Husbandry, Dairying, and Fisheries)[5]. The total FMD susceptible livestock population in the country is about 500 million comprising of more than 300 million cattle and buffalo, 71.5 million sheep, 140.5 million goats, and 11 million pigs[9]. Availability of indigenously developed diagnostic assays is crucial and indispensible to support such a huge control programme. In this review, the role of diagnostic assays developed, validated and used over the last decade in the country (Table 1) along with their contribution in control of FMD in India is being discussed.
| FMD diagnostic assay | Specimen materials | Target region | Sensitivity | Specificity | Advantages | Disadvantages |
| Sandwich ELISA | RNA from TE, FL, TE, | VP1 protein | 80% | 100% | Easy to perform Suitable for handling large number of samples | Less sensitive, not suitable for certain type of clinical samples |
| Multiplex PCR | RNA from TE, FL, TE, Semen, Milk | 1D region | Minimum detection limit of 1 × 10-1 TCID50/mL | 100% specific for cross serotype detection | Rapid and sensitive Suitable for samples like semen and milk | High risk of generating false positives |
| Taqman real-time PCR | RNA from TE, FL, TE, Semen, Milk | 1D region | Minimum detection limit of 101.0 TCID50/mL | 100% specific for cross serotype detection | More sensitive and specific than gel based assay | high risk of generating false positives |
| Virus isolation and neutralization | Triturated material of TE, FL, TE, | -- | Gold standard assay for FMD diagnosis | Slow takes 1-4 d for confirmatory results | ||
| RNA transfection | RNA from TE, FL, TE, Semen, Milk | -- | -- | -- | FMDV can be isolated from deteriorated clinical materials | -- |
| LAMP | RNA from TE, FL, TE, Semen, Milk | 3D region | Minimum detection limit up to 1.1 × 10-4 TCID50/mL | -- | Require no specialized instruments, can be used as point-of-care diagnosis | High risk of generating false positives |
| 3AB3 I-ELISA | Serum | 3AB3 region | 96% | 99.1% -96.4% | Sensitive and Specific | Only for bovine species |
| 3ABC C-ELISA | Serum | 3ABC region | Specific assayUniversal for all species | Less sensitive than I-ELISA | ||
| 2Ct I-ELISA | Serum | 2C region | Sensitive and Specific | Only for bovine species |
FMD is primarily diagnosed by demonstrating FMDV particles or viral genome in the clinical materials viz. tongue epithelium, foot epithelium, saliva, milk and semen, etc. Detection of intact virus particles by sandwich enzyme-linked immunosorbent assay (ELISA) and virus neutralization test provides confirmatory diagnosis, whereas detection of the viral genome by polymerase chain reaction (PCR) or loop-mediated isothermal amplification (LAMP) assay is more sensitive method of diagnosis. Samples collected from the FMD suspected animals are processed and routinely analyzed by these assays. The details of the suspected clinical samples tested during the last seven years are presented in the Table 2.
| Year | Sample tested | Serotype O | Serotype A | Serotype Asia1 | Total FMD diagnosed |
| 2009-2010 | 1155 | 423 | 15 | 7 | 445 |
| 2010-2011 | 345 | 83 | 10 | 10 | 171 |
| 2011-2012 | 567 | 265 | 4 | 40 | 309 |
| 2012-2013 | 701 | 218 | 15 | 52 | 285 |
| 2013-2014 | 3130 | 1295 | 24 | 10 | 1329 |
| Total | 5898 | 2284 | 68 | 119 | 2539 |
FMD antigen detection ELISA was shown to be rapid and simpler to perform[10]. The assay is generally regarded as the primary test for FMD diagnosis especially at the regionally located FMD diagnostic laboratories in the country[11]. The suspected clinical materials are first submitted to the regionally located FMD diagnostic laboratories in the country working under ICAR-PDFMD, Mukteswar where samples are processed and tested by an in-house sandwich ELISA for identification of FMDV serotype(s). The assay is based on the detection of FMDV structural proteins (Figure 1) and utilizes the serotype specific polyclonal antibodies generated in guinea pig and rabbits[12]. This antigen-capture sandwich ELISA has 100% specificity for heterologous FMDV and 80% sensitivity for detection of complete virus particles in clinical samples[12]. The assay is easy to perform at regional FMD diagnostic laboratories and large number of samples can be processed without risk of laboratory cross contamination. The assay is being used countrywide since two decades at 23 regionally located laboratories in the country[11]. As the assay specifically detects the intact virion particles in clinical materials in a serotype specific manner, the lower sensitivity could be attributed to the improper storage and transportation of samples that leads breakdown of the virus particles. The clinical materials are then submitted to the Central Laboratory, Mukteswar for detailed virological and genome analysis. Since 2009-2010, more than 5000 clinical materials have been tested by the sandwich ELISA, both at the regional FMD diagnostic laboratories and the central laboratory.
Of the established diagnostic approaches, virus isolation (VI) in cell culture is considered as the ‘‘gold standard’’ as described in OIE Terrestrial Manual 2012[13]. This method can be highly sensitive (depending upon the cell culture system used), although it can be slow, taking between 1 and 4 d to generate the results and require a containment laboratory facility. However, virus isolation from clinical materials is indispensible for antigenic profiling of the virus and vaccine matching. Primary cells, such as bovine thyroid (BTY), are highly susceptible to a wide range of FMDV serotypes[14], but they are difficult and costly to prepare and lose FMDV susceptibility after multiple passages[15]. Primary lamb kidney (LK) cells are also very sensitive to FMDV, and unlike BTY cells, LK cells maintain their sensitivity to FMDV infection after cryopreservation[16]. Immortalized cell lines [e.g., baby hamster kidney (BHK-21) fibroblasts and porcine kidney epithelial cells], are much easier to maintain but are less susceptible to specific animal-derived FMDV serotypes[17-20]. Recently LFBK-αvβ6stable cell line has been established and was observed to be an excellent cell line for FMDV diagnostic- and research-based cell applications[21].
In India, all the clinical samples collected/submitted for FMD diagnosis are subjected to virus isolation using the cell lines (BHK21, IBRS, and LFBK cells). Virus isolates after characterization are archived in the National FMDV repository. Currently the repository contains more than 1850FMDV isolates comprising of serotype O (n = 1180), A (n = 298), Asia1 (358) and C (n = 15). The oldest isolate available in the repository is of the year 1962 (Serotype O). Such a vast pool of virus isolates aids in selection and identification of suitable vaccine candidates through vaccine matching exercise from time to time.
Isolation of virus from the clinical materials is not always possible due to several factors[22]. Under such scenarios, the transfection based virus-rescue method as described by Belsham et al[23], has been optimized in India[22]. Success rate of RNA transfection for virus isolation was observed to be 62% against 16% in conventional cell culture method that enhances the number and diversity of virus isolates being usedin vaccine matching exercise. Till date, 88 serotype O, 24 serotype Asia1 and 09 serotype A viruses have been rescued using RNA transfection method from the samples where conventional method of cell culture passage failed to isolate the virus[11].
In vitro amplification based detection of genome is more rapid and sensitive than conventional VI[24]. Initially, assays were developed targeting the conserved 3D region[25,26] and 5’ UTR region[27]. Subsequently, multiplex PCR for (mPCR) targeting VP1 region were developed for detecting FMDV and differentiating amongst the serotypes[24,28,29].
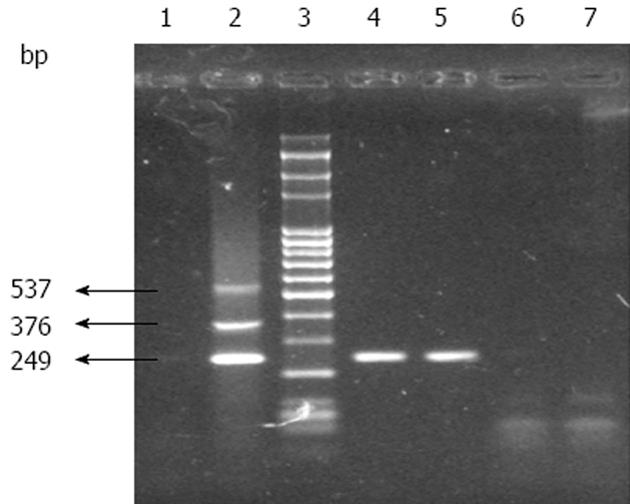
On the similar lines, mPCR was also developed in India[24] and the success rate of FMD diagnosis and serotype detection increased by 8%[30]. In this assay, the serotype-specific primers targeting 1D region and common reverse primer (NK61) targeting 2B region were used for multiplexing (Figure 1). Figure 2 indicates the mPCR based serotype identification describes the identification of serotype involved by multiplex PCR where product size of 249, 376 and 537 bp are specific for serotypes O, A, and Asia1, respectively. The minimum detection limit of the mPCR has been estimated as 1 x 10-1 TCID50/mL for serotypes O, A, and Asia1[24].
Although, the mPCR suffered from the disadvantage of generating false positives due to carry-over of PCR amplicons and thus, not considered as an ideal assay for routine testing of large numbers of samples especially at regionally located FMD diagnostic laboratories[31]. To overcome the chances of cross-contamination and make it more feasible for regional FMD laboratories, a ready-to-use thermo-stable RT-PCR mixture was developed[30]. All the components of the reaction mixture were mixed together in a vial and lyophilized (Lyodryer, United States). The lyophilized vials are to be reconstituted with nuclease free water before use and supplemented with the extracted RNA from the suspected materials followed by in-vitro amplification in a thermal-cycler. This thermostable RT-PCR mix made the assay more user friendly and clinical samples can be now diagnosed by PCR at the field level FMD diagnostic laboratories with uniformity in the results. In addition, the requirement of keeping live FMDV for positive control became obsolete. Since 2005, more than 2037 suspected clinical materials have been successfully tested by the mPCR in the country[11].
Reverse transcription-LAMP (RT-LAMP) assay is an autocycling and strand displacement DNA synthesis method[32] which has recently been employed in FMD diagnosis as point-of-care test. The RT-LAMP based targeting 3D and IRES region for detection of FMDV have been reported earlier[33,34]. In the recent past, LAMP based assay for FMDV detection and serotype differentiation (O, A and Asia 1) has been developed[35]. RT-LAMP based assay targeting 3D region has also been developed in India and is being used routinely for rapid detection of FMDV (Figure 1)[36]. LAMP assay requires only a water bath instead of a thermal-cycler as in PCR. In addition, gel documentation system is also non-essential as hydroxynaphthol blue (HNB), an azo dye is used as the indicator. The sensitivity and specificity of the RT-LAMP assay developed were estimated as 4.2 x 10-4, 2 x 10-4 and 1.1 x 10-4 TCID50/mL for FMDV serotypes O, A and Asia1 respectively. LAMP assay for FMD diagnosis was validated by simultaneous testing of the clinical samples (n = 139) by mPCR and LAMP and the results revealed higher sensitivity in case of LAMP.
Reverse-transcription real time PCR (RT-qPCR) assays have been developed and evaluated for the identification of FMDV in different parts of the world using fluorogenic dyes. Both SYBR Green and TaqMan chemistries have been widely utilised in qPCR assays for FMD, however TaqMan provide an additional advantage of multiplexing. In India, a qPCR assay targeting 1D region of FMDV was developed in multiplex format for simultaneous detection and identification of FMDV serotypes in the suspected clinical materials[37]. The sensitivity of the TaqMan based multiplex qPCR was found to be 101.7 TCID50/mL, 101.0 TCID50/mL, 101.7 TCID50/mL for serotype O, Asia1 and A respectively[37]. The qPCR assay was found to be more sensitive than gel based assay and provides an estimate through standard curve of viral load in the samples. With high sensitivity and specificity, the qPCR assay has been used as the primary tool for the detection of FMDV in persistently infected carriers among exposed ruminants which is of great importance in disease control[38].
FMDV can be actively secreted in semen of FMD infected bull before onset of clinical symptoms and up to 5-8 mo post infection[39]. It has also been reported that FMDV can survive in frozen semen straw, thus artificial insemination can possibly serve as the source of FMDV transmission to wider and farther areas. The extenders used during the production of semen straws provide the conditions conducive to survival of the virus for more than 320 d when stored at -50 °C[40]. Routinely used FMD diagnostic methods such as VI and antigen ELISA require modifications for detecting FMDV in semen samples[41,42]. Even mPCR assay was found to be far less sensitive for semen samples. The major reason behind PCR failure was the presence of PCR inhibitors in semen[28,39]. Hence, existing mPCR assay was improvised for the detection of FMDV genome in semen samples[43]. The RNA from suspected semen samples (neat or extended) was extracted by a modified method to remove the PCR inhibitors[43]. This modified mPCR has been used for screening of 980 animals for presence of FMDV genome in semen till now. It was also established that, FMDV could be detected in semen of the infected cattle bull for about 5 mo but not more than 8 mo[43].
There is co-circulation, extinction, and emergence and re-emergence of genotypes/lineages within the serotypes from time to time in India. The emergence or re-emergence of any new lineage warrants rapid and accurate detection to facilitate early warning[44,45]. Detailed nucleotide sequence of these viruses are analysed to detect emergence of any new group. A rapid multiplex PCR assay was developed for detection of the dominating VP359-deletion group of serotype A virus with 100% sensitivity and specificity[44]. Genotype differentiating RT-PCR was developed as a fast, cost-effective and user-friendly alternative to 1D region based phylogeny for detection and differentiation of genotypes VI and VII of serotype A[45]. Similarly, a simple, fast and multi-primer RT-PCR assay has been developed and validated to differentiate genetic lineages of serotype Asia1 viruses[46]. These assays have been proven as useful tools in preliminary molecular epidemiological investigation of FMD in the country.
Sero-surveillance is of prime importance in India where FMD control programme is in operation for last 10 years. As per the OIE guidelines, in regions adopting vaccination to control FMD, sero-surveillance should be performed by an assay capable of differentiating infected from vaccinated animals (DIVA)[47]. Detection of antibodies against various non-structural proteins (NSPs) of FMDV has been successfully utilized for DIVA[48,49]. Considering the complex epidemiology of the disease in the country, assays for DIVA were developed and validated taking into account the factors such as vaccine quality (in terms of level of NSP contamination in the formulation) and coverage in India. A tool box of one competitive and four indirect ELISAs utilizing 3AB3, 3ABC, and truncated 2C (2Ct) NSPs of FMDV (Figure 1) was developed in India[50-52]. The performance of these in-house DIVA assays was compared with the two commercially available kits (PrioCheck® FMDV-NS and Svanovir FMDV 3ABC-Ab ELISA kit) and indigenously developed assays were found to be equally capable in detecting infected animals among the vaccinated population[53]. However, the in-house assays performed better than the commercial kits in case of intensively vaccinated samples[53]. The r3AB3 indirect ELISA is routinely used for countrywide screening of bovines[51] and results obtained for the serum samples collected at random from the country are presented in the Table 3. The diagnostic sensitivity of this assay is 96% while the diagnostic specificity varied between the naïve and vaccinates as 99.1% and 96.4%, respectively. This assay detects antibodies to 3AB (3AB-Ab) from 10 to as late as 900 d post-infection in experimentally infected cattle. Recently 3B[54], 2B[55] and 3D[56] NSP based assays have also been developed in India and are under validation.
| Year | Total samples tested | Total positive | % animals 3AB3 reactors in India |
| 2009-2010 | 29763 | 8303 | 27.90 |
| 2010-2011 | 31042 | 8341 | 26.87 |
| 2011-2012 | 37467 | 10410 | 26.09 |
| 2012-2013 | 40934 | 10811 | 26.41 |
| 2013-2014 | 52224 | 15268 | 29.20 |
| Total | 191430 | 53133 | 27.70 |
Post vaccination sero-monitoring is critical to monitor protective antibody level in animals before and after every round of vaccination. Under the Government of India initiated vaccination based FMD control programme (FMDCP) 120 million cattle and buffaloes are routinely vaccinated at 6 mo interval to progressively build herd immunity[5]. However, vaccines against FMD only protect the animal from clinical disease and not from the super infection by other serotypes of FMDV. Additionally, the vaccine induced protection remains only for about 4-6 mo[57] and with the decline in herd immunity risk of clinical disease increases due to the creation of infection window. Therefore, quantitative estimation of protective antibody response (titer) in vaccinated animals through sero-monitoring is indispensable for devising appropriate vaccination regime and successful implementation and monitoring of the control programme[58,59]. With the current sampling policy in the country, village is considered as a herd and from each district covered under FMDCP, 10 villages are randomly selected for sampling, and from each village 20 serum samples (10 cattle and 10 buffalo) are collected at random before (0 d) and 28 d post vaccination (dpv) to have un-biased estimate of vaccination performance and the resulting level of herd immunity. Antibody titers against the serotypes O, A and Asia1 are determined by four fold dilution liquid phase blocking ELISA (LPBE)[60,61]. With the expansion of FMDCP, there is a considerable rise in the number of serum samples to be tested. Thus, a high throughput LPBE assay was developed recently to fasten the process and save time and labour (Manuscript communicated). This high throughput assay utilizes the linear regression method for extrapolation of titers of test serum samples from the known internal controls[60]. In addition, the reagents used in the assay are thermo-stable facilitating the transportation to the regional laboratories under high ambient temperature.
Considering the fact that India has a large livestock population (about 500 million) susceptible to FMD, the country requires economical companion diagnostic tests tailor-made for the suitability under Indian scenarios to run the progressive control programme for FMD. Though India is now self-sufficient to produce most of the diagnostic kits, but still a lot of improvisation is needed in the current assays. The polyclonal antibodies used in several assays could be replaced with the recombinant antibodies. Some success has been achieved in development of single-chain variable fragment (scFv)[62] but work is being continued to develop scFv and nanobodies against highly immunogenic epitopes of FMDV and assess their applicability in diagnostics.
P- Reviewer: Farzin R, Kamal SA S- Editor: Tian YL L- Editor: A E- Editor: Yan JL
| 1. | Perry BD, Sones KR, editors . Global road map for improving the tools to control foot-and-mouth disease in endemic settings. Report of a workshop held at Agra, India, 29 November-1 December 2006, and subsequent road map outputs. Nairobi, Kenya: ILRI (International Livestock Research Institute) 2007; 88. |
| 2. | Racaniello RR. Picornaviridae: the viruses and their replication. In Fields Virology. Fields BN, Knipe DM, Howley PM, editors. 4th Ed. Lippincott Williams and Wilkins, Philadelphia: Lippincott-Raven Publishers 2001; 685-722. |
| 3. | Bachrach HL. Foot-and-mouth disease. Annu Rev Microbiol. 1968;22:201-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 263] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 4. | Sangar DV. The replication of picornaviruses. J Gen Virol. 1979;45:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 73] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Pattnaik B, Subramaniam S, Sanyal A, Mohapatra JK, Dash BB, Ranjan R, Rout M. Foot-and-Mouth Disease: global status and future road map for control and prevention in India. Agric Res. 2012;1:132-147 . [DOI] [Full Text] |
| 7. | Donaldson AI, Gloster J, Harvey LD, Deans DH. Use of prediction models to forecast and analyse airborne spread during the foot-and-mouth disease outbreaks in Brittany, Jersey and the Isle of Wight in 1981. Vet Rec. 1982;110:53-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 88] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Hugh-Jones ME, Wright PB. Studies on the 1967-8 foot-and-mouth disease epidemic. The relation of weather to the spread of disease. J Hyg (Lond). 1970;68:253-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 68] [Article Influence: 1.2] [Reference Citation Analysis (1)] |
| 9. | Department of Animal Husbandry, Dairying and Fisheries. Livestock Census of India 2012. Available from: http://dahd.nic.in/dahd/WriteReadData/Livestock.pdf. |
| 10. | Ferris NP, Dawson M. Routine application of enzyme-linked immunosorbent assay in comparison with complement fixation for the diagnosis of foot-and-mouth and swine vesicular diseases. Vet Microbiol. 1988;16:201-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 161] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 11. | Annual Reports. Project Directorate on Foot and Mouth Disease. Mukteswar: 263138 Nainital (Uttaranchal). Available from: http://www.icar.org.in/files/pdfmd-vacancy-t3-2010.pdf. |
| 12. | Bhattacharya S, Pattnaik B, Venkataramanan R. Development and application of sandwich enzyme-linked immunosorbent assay (ELISA) for type identification of foot-and-mouth disease (FMD) virus in direct field materials. Ind J Animal Sci. 1996;66:1-9. |
| 13. | Available from: http://www.oie.int/wahid-prod/public.php?page=home. |
| 14. | Snowdon WA. Growth of foot-and mouth disease virus in monolayer cultures of calf thyroid cells. Nature. 1966;210:1079-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 142] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | House JA, Yedloutschnig RJ. Sensitivity of seven different types of cell cultures to three serotypes of foot-and-mouth disease virus. Can J Comp Med. 1982;46:186-189. [PubMed] |
| 16. | House C, House JA. Evaluation of techniques to demonstrate foot-and-mouth disease virus in bovine tongue epithelium: comparison of the sensitivity of cattle, mice, primary cell cultures, cryopreserved cell cultures and established cell lines. Vet Microbiol. 1989;20:99-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Swaney LM. Susceptibility of a new fetal pig kidney cell line (MVPK-1) to foot-and-mouth disease virus. Am J Vet Res. 1976;37:1319-1322. [PubMed] |
| 18. | Ferris NP, King DP, Reid SM, Hutchings GH, Shaw AE, Paton DJ, Goris N, Haas B, Hoffmann B, Brocchi E. Foot-and-mouth disease virus: a first inter-laboratory comparison trial to evaluate virus isolation and RT-PCR detection methods. Vet Microbiol. 2006;117:130-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Ferris NP, Hutchings GH, Moulsdale HJ, Golding J, Clarke JB. Sensitivity of primary cells immortalised by oncogene transfection for the detection and isolation of foot-and-mouth disease and swine vesicular disease viruses. Vet Microbiol. 2002;84:307-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | De Castro MP. Behaviour of the foot-and-mouth disease virus in cell cultures: susceptibility of the IB-RS-2 line. Arch Inst Biol. 1964;31:63-78. |
| 21. | LaRocco M, Krug PW, Kramer E, Ahmed Z, Pacheco JM, Duque H, Baxt B, Rodriguez LL. A continuous bovine kidney cell line constitutively expressing bovine αvβ6 integrin has increased susceptibility to foot-and-mouth disease virus. J Clin Microbiol. 2013;51:1714-1720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 22. | Bisht P, Mohapatra JK, Subramaniam S, Das B, Pande V, Biswal JK, Sharma GK, Rout M, Ranjan R, Dash BB. Efficient rescue of foot-and-mouth disease virus in cultured cells transfected with RNA extracted from clinical samples. J Virol Methods. 2014;196:65-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Belsham GJ, Bostock CJ. Studies on the infectivity of foot-and-mouth disease virus RNA using microinjection. J Gen Virol. 1988;69:265-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 24. | Giridharan P, Hemadri D, Tosh C, Sanyal A, Bandyopadhyay SK. Development and evaluation of a multiplex PCR for differentiation of foot-and-mouth disease virus strains native to India. J Virol Methods. 2005;126:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 25. | Meyer RF, Brown CC, House C, House JA, Molitor TW. Rapid and sensitive detection of foot-and-mouth disease virus in tissues by enzymatic RNA amplification of the polymerase gene. J Virol Methods. 1991;34:161-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Rodríguez A, Núñez JI, Nolasco G, Ponz F, Sobrino F, de Blas C. Direct PCR detection of foot-and-mouth disease virus. J Virol Methods. 1994;47:345-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 27. | Reid SM, Ferris NP, Hutchings GH, Samuel AR, Knowles NJ. Primary diagnosis of foot-and-mouth disease by reverse transcription polymerase chain reaction. J Virol Methods. 2000;89:167-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 85] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 28. | Vangrysperre W, De Clercq K. Rapid and sensitive polymerase chain reaction based detection and typing of foot-and-mouth disease virus in clinical samples and cell culture isolates, combined with a simultaneous differentiation with other genomically and/or symptomatically related viruses. Arch Virol. 1996;141:331-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 29. | Callens M, De Clercq K. Differentiation of the seven serotypes of foot-and-mouth disease virus by reverse transcriptase polymerase chain reaction. J Virol Methods. 1997;67:35-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Sharma GK, Mahajan S, Das B, Ranjan R, Kanani A, Sanyal A, Pattnaik B. Comparison of stabilisers for development of a lyophilised multiplex reverse-transcription PCR mixture for rapid detection of foot and mouth disease virus serotypes. Rev Sci Tech. 2014;33:859-867. [PubMed] |
| 31. | Hoffmann B, Beer M, Reid SM, Mertens P, Oura CA, van Rijn PA, Slomka MJ, Banks J, Brown IH, Alexander DJ. A review of RT-PCR technologies used in veterinary virology and disease control: sensitive and specific diagnosis of five livestock diseases notifiable to the World Organisation for Animal Health. Vet Microbiol. 2009;139:1-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 125] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 32. | Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5533] [Cited by in RCA: 5629] [Article Influence: 225.2] [Reference Citation Analysis (0)] |
| 33. | Dukes JP, King DP, Alexandersen S. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Arch Virol. 2006;151:1093-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 160] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 34. | Chen HT, Zhang J, Liu YS, Liu XT. Detection of foot-and-mouth disease virus RNA by reverse transcription loop-mediated isothermal amplification. Virol J. 2011;8:510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 35. | Madhanmohan M, Nagendrakumar SB, Manikumar K, Yuvaraj S, Parida S, Srinivasan VA. Development and evaluation of a real-time reverse transcription-loop-mediated isothermal amplification assay for rapid serotyping of foot-and-mouth disease virus. J Virol Methods. 2013;187:195-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 36. | Ranjan R, Kangayan M, Subramaniam S, Mohapatra JK, Biswal JK, Sharma GK, Sanyal A, Pattnaik B. Development and evaluation of a one step reverse transcription-loop mediated isothermal amplification assay (RT-LAMP) for rapid detection of foot and mouth disease virus in India. Virusdisease. 2014;25:358-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Tamil Selvan RP. Analysis of replication dynamics of mixed Foot-and-Mouth Disease virus populations using serotype differentiating multiplex QPCR. Izatnagar, India: Deemed University Indian Veterinary Research Institute 2010; . |
| 38. | Zhang Z, Alexandersen S. Detection of carrier cattle and sheep persistently infected with foot-and-mouth disease virus by a rapid real-time RT-PCR assay. J Virol Methods. 2003;111:95-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 39. | Shin J, Torrison J, Choi CS, Gonzalez SM, Crabo BG, Molitor TW. Monitoring of porcine reproductive and respiratory syndrome virus infection in boars. Vet Microbiol. 1997;55:337-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 40. | Cottral GE, Bachrach HL. Food-and-mouth disease viremia. Proc Annu Meet U S Anim Health Assoc. 1968;72:383-399. [PubMed] |
| 41. | Schultz RD, Adams LS, Letchworth G, Sheffy BE, Manning T, Bean B. A method to test large numbers of bovine semen samples for viral contamination and results of a study using this method. Theriogenology. 1982;17:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 42. | Shao JJ, Chang H, Lin T, Cong G, Du J, Guo J, Bao H, Shang Y, Yang Y, Liu X. Amplification and characterization of bull semen infected naturally with foot-and-mouth disease virus type Asia 1 by RT-PCR. Virologica Sinica. 2008;23:378-382. [RCA] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 43. | Sharma GK, Subramaniam S, De A, Das B, Dash BB, Sanyal A, Misra AK, Pattnaik B. Detection of foot-and-mouth disease virus in semen of infected cattle bulls. Ind J of AniSci. 2012;82:1472-1476. |
| 44. | Mohapatra JK, Pandey LK, Sharma GK, Barik SK, Pawar SS, Palsamy R, Pattnaik B. Multiplex PCR for rapid detection of serotype A foot-and-mouth disease virus variants with amino acid deletion at position 59 of the capsid protein VP3. J Virol Methods. 2011;171:287-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 45. | Mohapatra JK, Subramaniam S, Tosh C, Hemadri D, Sanyal A, Periyasamy TR, Rasool TJ. Genotype differentiating RT-PCR and sandwich ELISA: handy tools in epidemiological investigation of foot and mouth disease. J Virol Methods. 2007;143:117-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 46. | Mohapatra JK, Sanyal A, Hemadri D, Tosh C, Rasool TJ, Bandyopadhyay SK. A novel genetic lineage differentiating RT-PCR as a useful tool in molecular epidemiology of foot-and-mouth disease in India. Arch Virol. 2006;151:803-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 47. | Rout M, Senapati MR, Mohapatra JK, Dash BB, Sanyal A, Pattnaik B. Serosurveillance of foot-and-mouth disease in sheep and goat population of India. Prev Vet Med. 2014;113:273-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 48. | Paton DJ, de Clercq K, Greiner M, Dekker A, Brocchi E, Bergmann I, Sammin DJ, Gubbins S, Parida S. Application of non-structural protein antibody tests in substantiating freedom from foot-and-mouth disease virus infection after emergency vaccination of cattle. Vaccine. 2006;24:6503-6512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 94] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 49. | Mackay DK, Forsyth MA, Davies PR, Berlinzani A, Belsham GJ, Flint M, Ryan MD. Differentiating infection from vaccination in foot-and-mouth disease using a panel of recombinant, non-structural proteins in ELISA. Vaccine. 1998;16:446-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 113] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 50. | Mahajan S, Mohapatra JK, Pandey LK, Sharma GK, Pattnaik B. Truncated recombinant non-structural protein 2C-based indirect ELISA for FMD sero-surveillance. J Virol Methods. 2013;193:405-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 51. | Mohapatra JK, Pandey LK, Sanyal A, Pattnaik B. Recombinant non-structural polyprotein 3AB-based serodiagnostic strategy for FMD surveillance in bovines irrespective of vaccination. J Virol Methods. 2011;177:184-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 52. | Sharma GK, Mohapatra JK, Pandey LK, Mahajan S, Mathapati BS, Sanyal A, Pattnaik B. Immunodiagnosis of foot-and-mouth disease using mutated recombinant 3ABC polyprotein in a competitive ELISA. J Virol Methods. 2012;185:52-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 53. | Sharma GK, Mohapatra JK, Mahajan S, Matura R, Subramaniam S, Pattnaik B. Comparative evaluation of non-structural protein-antibody detecting ELISAs for foot-and-mouth disease sero-surveillance under intensive vaccination. J Virol Methods. 2014;207:22-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 54. | Mohapatra AK, Mohapatra JK, Pandey LK, Sanyal A, Pattnaik B. Diagnostic potential of recombinant nonstructural protein 3B to detect antibodies induced by foot-and-mouth disease virus infection in bovines. Arch Virol. 2014;159:2359-2369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 55. | Biswal JK, Jena S, Mohapatra JK, Bisht P, Pattnaik B. Detection of antibodies specific for foot-and-mouth disease virus infection using indirect ELISA based on recombinant nonstructural protein 2B. Arch Virol. 2014;159:1641-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 56. | Mahajan S, Mohapatra JK, Pandey LK, Sharma GK, Pattnaik B. Indirect ELISA using recombinant nonstructural protein 3D to detect foot and mouth disease virus infection associated antibodies. Biologicals. 2015;43:47-54. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 57. | Doel TR. Natural and vaccine-induced immunity to foot and mouth disease: the prospects for improved vaccines. Rev Sci Tech. 1996;15:883-911. [PubMed] |
| 58. | Dekker A. Why is FMD post vaccination monitoring necessary? GFRA News letter. 2012;2:2-3. |
| 59. | Rweyemamu M, Roeder P, MacKay D, Sumption K, Brownlie J, Leforban Y. Planning for the progressive control of foot-and-mouth disease worldwide. Transbound Emerg Dis. 2008;55:73-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 60. | Robiolo B, La Torre J, Duffy S, Leon E, Seki C, Torres A, Mattion N. Quantitative single serum-dilution liquid phase competitive blocking ELISA for the assessment of herd immunity and expected protection against foot-and-mouth disease virus in vaccinated cattle. J Virol Methods. 2010;166:21-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 61. | Hamblin C, Barnett IT, Crowther JR. A new enzyme-linked immunosorbent assay (ELISA) for the detection of antibodies against foot-and-mouth disease virus. II. Application. J Immunol Methods. 1986;93:123-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 86] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 62. | Sharma GK, Mahajan S, Matura R, Subramaniam S, Mohapatra JK, Pattnaik B. Production and characterization of single-chain antibody (scFv) against 3ABC non-structural protein in Escherichia coli for sero-diagnosis of Foot and Mouth Disease virus. Biologicals. 2014;42:339-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |