Published online Feb 12, 2015. doi: 10.5501/wjv.v4.i1.25
Peer-review started: September 26, 2014
First decision: October 28, 2014
Revised: December 13, 2014
Accepted: December 29, 2014
Article in press: December 31, 2014
Published online: February 12, 2015
Processing time: 118 Days and 1.4 Hours
Hepatitis C virus (HCV) is an emerging infection worldwide and the numbers of persons infected are increasing every year. Poor blood transfusion methods along with unsafe injection practices are potential sources for the rapid spread of infection. Early detection of HCV is the need of the hour especially in high risk group population as these individuals are severely immunocompromised. Enzyme Immunoassays are the most common detection techniques but they provide no evidence of active viremia or identification of infected individuals in the antibody-negative phase and their efficacy is limited in individuals within high risk group population. Molecular virological techniques have an important role in detecting active infection with utmost specificity and sensitivity. Technologies for assessment of HCV antibody and RNA levels have improved remarkably, as well as our understanding of how to best use these tests in patient management. This review aims to give an overview of the different serological and molecular methods employed in detecting HCV infection used nowadays. Additionally, the review gives an insight in the new molecular techniques that are being developed to improve the detection techniques particularly in High Risk Group population who are severely immunocompromised.
Core tip: The review focuses on the current molecular diagnostic techniques that are being used to detect hepatitis C virus worldwide. Special emphasis is given on the detection techniques that can be used to screen the individuals with repeated blood transfusion history; particularly thalassaemic individuals, intravenous drug users and persons on hemodialysis.
- Citation: Firdaus R, Saha K, Biswas A, Sadhukhan PC. Current molecular methods for the detection of hepatitis C virus in high risk group population: A systematic review. World J Virology 2015; 4(1): 25-32
- URL: https://www.wjgnet.com/2220-3249/full/v4/i1/25.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i1.25
Hepatitis C virus (HCV) infection is a global health problem which has affected around 170 million people worldwide and is one of the major causes of deaths related to liver cirrhosis and hepatocellular carcinoma[1]. HCV can be classified to seven major genotypes and 80 subtypes[2-4]. HCV genotypes vary in patterns of geographical distribution and therapeutic response. However, the geographical and genetic diversity of this RNA virus is constantly evolving because of rapid globalization. In India, HCV infection has been reported in 0%-21% population and responsible for 14%-26% cases of chronic liver disease[5]. HCV infection is mostly transmitted through transfusion of blood or blood products. A high prevalence of HCV is found in many high-risk groups (HRG) exposed to blood or blood products like intra venous drug users (IDUs), patients with pediatric hematologic malignancies and those with thalassemia and hemophilia. India reported a higher percentage of blood donors in India (1%-1.5%) than in developed countries[6,7].
An increasing burden of HCV related liver complica-tions has been estimated particularly taking into account those who were infected before safety precautions of blood transfusions happened. A major concern is careful screening of blood and blood related products, but in developing countries like in India, the regulations for strict checking of blood and blood related products came to place only in 2001[8,9]. Recent surveys have reported that testing of blood and blood related products are poorly regulated in India[10]. In United States, data showed that death related to HCV exceeded than those by HIV. Though novel antiviral therapies are recently in the horizon with enhanced efficacy and fewer side effects but the challenge remains in detecting HCV at an early stage.
During HCV infection, though attempts are made to diagnose and differentiate acute from chronic hepatitis C infection, it is often not possible to distinguish between the two phases. The infection may be recognized only when it becomes chronic[11,12]. The serologic diagnostic tests used as first step for detecting the infection cannot distinguish between acute and chronic infection[13]. Investigations for patients with HCV infection include serological assays for antibodies to hepatitis C (anti- HCV) and molecular assays for detection of viral RNA.
The importance of low cost molecular diagnostic assays are especially important for the developing nations as they are already burdened with increasing number of hepatitis C patients who are generally economically backward. The advent of molecular diagnostic approaches has allowed for the development of nucleic acid assays that are more sensitive and specific than antibody based technologies. The linking of these assays with appropriate detection systems, therefore, makes them highly desirable for detecting HCV RNA in patient samples. Molecular techniques not only help to detect HCV RNA but confirm active state of infection, i.e., the virus is in replicating state in the patient’s body. In individuals falling in high risk diagnosis of HCV can give false negative results as these patients are already immuno-suppressed, in this scenario, molecular testing remains the best choice for detection.
This review aims to give an overview of the different serological and viral genome based laboratory tests which has become instrumental in the management of HCV infection to diagnose viral infection, and more importantly guide treatment decisions which could be of enormous help to clinicians.
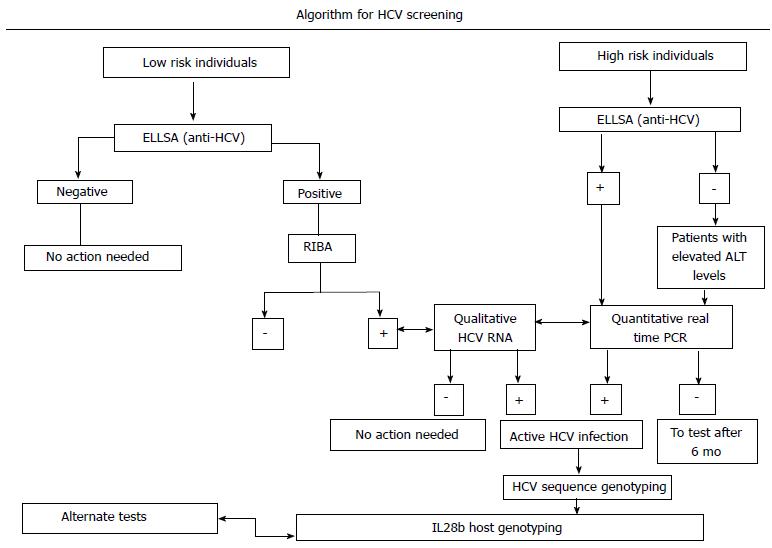
The investigation of HCV diagnosis starts with serological assays for detecting antibodies to HCV followed by molecular assays for detecting HCV RNA (Figure 1). Initial diagnosis of HCV infection is classically done by serologic methods either by determining anti HCV antibody by EIAs or by immunoblot assays and by determining the presence of HCV RNA. The advent of simple rapid immunoassays has significantly reduced the risk of HCV transmission, but concern remains for patients in high risk groups[13-16]. Studies have shown that false negative results in rapid tests might arise in patients who are severely immunocompromised such as those co-infected with HIV[17], in patients on hemodialysis, IDUs, thalassaemic. In these patient groups molecular detection by reverse transcription polymerase chain reaction (RT-PCR) remains the best method for detection.
Rapid immunoassay tests are based on the principle to detect HCV antigens from core, NS3, NS4 and NS5 regions of the virus. In western countries, these tests are used besides nucleic acid testing, and used only as point of care tests, but in developing countries these tests are solely relied in commercial places for detection of HCV[18]. Commercial kits like OraQuick rapid HCV antibody test use device that delivers HCV antibody test results in 20 min using a single drop of whole blood. The kit was approved for laboratory use in United States from June 2010. The OraQuick is very accurate, with sensitivity and specificity performance that meets the standards for FDA approval[19]. Though rapid kits have been extensively used for surveillance purposes, they are not well suited for high risk groups and immunocompromised patients[20,21]. In developing countries like India, WHO has recommended certain kits for rapid testing for surveillance purposes.
Enzyme immunoassays are the most common screening test for HCV [enzyme immunoassay (EIA), microparticle EIA, chemiluminescence immunoassay (CIA)] that detects anti-HCV antibodies in plasma or serum. These assays are relatively easy to use, does not require expert technicians, automation is simple, have a low variability and are inexpensive.
Three generations of EIA antibody testing have been developed since 1989. In the first generation of EIA developed in 1992, c100-3 epitope from the non-structural NS4 regions was incorporated. A newer and better second generation EIA-2 was developed next which contained HCV antigens from core, NS3, NS4 regions[22-24]. The third generation of EIA developed contained modified antigens form core, NS3, and a slightly modified antigen from NS5 region. The incorporation of these antigens increased the overall sensitivity to 97%, which was better than the second generation assays. The mean time to seroconversion in the improved third generation kits has gone down to 2-3 wk as compared to 4-6 wk in the second generation kits. EIA methods have several advantages as the kits are relatively inexpensive and highly sensitive too, but one of the disadvantages is that it may give false positive results in routine blood donors and asymptomatic adults. For this reason, Centres for Disease control and prevention has recommended the supplementary tests like RIBA or PCR based methods to confirm positive ELISA tests unless the signal-to-cut-off ratio is above a predetermined threshold[25].
False-negative EIA especially occur in patients with major immunosuppression (advanced HIV infection or organ transplantation recipients), patients with chronic renal failure on long-term hemodialysis, and patients with acute or early HCV infection have been reported in HCV EIA[26].
In developing countries like India, WHO recommends the use of 3rd generation HCV EIA kits. Several commercial kits which employ the structural and non-structural antigens i.e., core, E1, E2, NS3, NS4 and NS5 are in use. The 3rd generation kits are better than the previous versions with improved specificity and sensitivity. The use of EIA kits in India is limited as it requires expertise in handling and in developing countries like India where the health care resources are already burdened; it becomes very difficult to reach the people[27].
In the recombinant immunoblot assay (RIBA) assays, multiple HCV antigens are individually displayed on a nitrocellulose strips as bands. In positive HCV infection, RIBA results show two reactive bands, in intermediate infection show one positive band. Since positive cases in RIBA, show two bands they are considered more sensitive than EIA. However, they are not considered as independent gold standard as two tests contain similar antigens to detect HCV antibody[28-30].
HCV core antigen testing was developed as an alternative to nucleic acid testing (NAT assays). The HCV core antigen detects viral antibodies within the sero-conversion period and can be used to utilize to monitor antiviral therapy. But till now is not commercially used as detection system[31]. The Architect HCV Ag assay (Abbott) which is a quantitative core antigen based assay, is commercially available in Europe. The assay is chemiluminesce based immunoassay in an automated platform[32]. In these assay, micro particles are coated with a monoclonal antibody against HCV core antigen. Studies have shown that HCV core antigen could be detected within first two weeks of acute infection. The core antigen based testing has a sensitivity ranging from 80% to 99% and a specificity ranging from 96% to 100%. The core antigen based assay could be an important detection technique in individuals who are in High Risk Group, because the core protein is the most conserved viral antigen amongst HCV types and, was therefore, a likely antigenic probe. Also the core protein is one the first protein which is synthesized and therefore could be an attractive target for molecular detection in high risk group patients who are immunocompromised. Studies have demonstrated two major B-cell epitopes at the N-terminus of core: amino acids (aa) 5-23 and 39-74 using peptide reactivity to sera from patients with chronic HCV can be used as antigenic determinants[33,34].
Molecular diagnostic assays represent are an integral part in the management of HCV patients. Both qualitative and quantitative HCV molecular assays are used in the diagnosis of acute and chronic infection. The principle of qualitative HCV assays includes viral RNA isolation, complementary DNA (cDNA) synthesis, PCR amplification and detection of PCR amplicons. Qualitative HCV RNA test detects the presence of HCV circulating in the blood and is among the most sensitive tests available. Since HCV is a RNA virus, reverse transcription PCR is used to detect viral RNA[35-37]. The viral genome is 9.6kb long, contains a single open reading frame that is translated to produce a single protein product, which is then further processed to produce functional proteins for viral replication and propagation[36]. At the 5’ and 3’ ends of the viral RNA are the untranslated region (UTR) that are not translated into proteins but are important to translation and replication of the viral RNA. Most of the commercial and in-house PCR amplification strategies are targeted against the 5’ UTR region as there is more than 90% sequence identity among different HCV genotypes, with some segments nearly identical among different strains[37-39]. The secondary and tertiary structures of this region are also largely conserved and this one of the first regions which is transcribed.
Other than the 5’ UTR region, the core and the 3’ UTR region are also targeted for PCR based detection of HCV[40-44]. A recent study showed that detection based on the sequence of the core region could reliably identify subtypes as well as major genotypes since the sequence divergence was greater than the divergence of the 5’UTR sequence. Though there are other regions like the E1, E2, NS2, which can be used as detection targets for PCR amplification but they are not in much use as there is a lack of conservation in the primer binding sites[45-50].
Detection of viral RNA is useful in diagnosing HCV infection prior to sero-conversion, distinguishing active from resolved infection, and diagnosing chronic hepatitis carriers who are HCV antibody negative, especially among HRGs. Nucleic acid testing is recommended: (1) for confirmation of HCV RNA in cases where patients are HCV seropositive; (2) to confirm the presence of HCV viremia in patients who are seronegative but immunocompromised such as HIV infected individuals; (3) in babies who are born to HCV positive mothers- as antibody testing in babies can give false positive results upto 18 mo of age; and (4) for determining the baseline value before starting the anti-viral therapy. Molecular detection of HCV includes both qualitative and quantitative assays. The qualitative HCV RNA testing is very popular due to its higher sensitivity, but a major disadvantage of the qualitative assays is that it only determines the presence or absence of HCV RNA. On the other hand, quantitative HCV RNA determines the HCV RNA level and thus provides prognostic information for treatment. Nowadays, there are several widely used commercial tests which are used to detect the presence of HCV RNA in patient’s serum. One of the commercial assays used is the Cobas Amplicor HCV version 2.0 (Roche Molecular Diagnostics, Pleasonton, CA, United States) based on a standard RT-PCR is available for the qualitative measurement of HCV RNA. The lowest detection limit is 50 IU/mL whatever the HCV genotype[51,52]. Another assay commercially used is versant HCV qualitative assay (Siemens Healthcare Diagnostics, Deerfield, IL, United States) which is based on transcription mediated amplification technique. In this assay, first viral RNA is isolated from the patient’s serum and then amplified by utilizing two enzymes (reverse transcriptase and T7 RNA polymerase). These amplicons are further detected via hybridization protection assay (HPA) in which only hybridized probes remain chemiluminescent and are detected in a luminometer. Analytical sensitivity is 10 IU/mL for most genotypes and 5.3 IU/mL for genotype 1[53].
HCV quantitative assay is used to determine the number of international units of HCV RNA per millimeter of serum or plasma (IU/mL) in known HCV positive patients.
Recently, real time PCR based detection systems have become widely available and are considered as the detection method of choice by many clinicians. The advantages of this technique are that they have a very low limit of detection, have a broad dynamic range. Several companies now market the real time PCR assays: the COBASs Ampliprep/Cobas TaqMan assay (CAP/CTM, Roche Molecular Diagnostics) and the real-time HCV assay (also named AccuGenes HCV, Abbott Molecular Inc., Des Plaines, IL, United States). These assays have the advantage of having a broad dynamic range of amplification, thus improving the limits of detection (LOD) to 10 IU/mL, and linear quantification up to 107-108 IU/mL[54,55].
The quantitation of HCV viral RNA in Cobas Amplicor is performed using the HCV Quantitation Standard. The HCV quantitation standard is a non-infectious armoured RNA construct consists of HCV sequences with identical primer binding sites as the HCV RNA target and a unique probe binding region that allows HCV Quantitation Standard amplicon to be distinguished from HCV target amplicon. The HCV Quantitation Standard is pipetted into each individual sample and control at a known copy number and is then HCV amplification by PCR is carried out. The COBAS TaqMan HCV Test, v2.0 uses reverse transcription and PCR amplification primers against the highly conserved 5’ untranslated region of the HCV genome[56].
The Versant HCV quantitative test (Siemens Healthcare Diagnostics) which is HCV RNA assay based on signal amplification by branched DNA (bDNA). In this assay, single stranded DNA molecules are present; which acts as probe DNA molecules. Next an extender DNA molecule is added. Once the capture and extender molecules are in their proper place they are hybridized and the sample is added. The bDNA assay version 3.0 is has been reported to have a lower detection limit of 615 IU/mL to 8 million IU/mL whatever the HCV genotype[57].
The advantage of RT-PCR is that it allows continuous monitoring of amplicon kinetics during the exponential phase before the amplification reaches its plateau. This allows for a good correlation between the initial numbers of template copies whereas in qualitative assays based on PCR, amplicon detection was at the end[56,58]. Thus the use quantitation techniques have greatly enhanced the sensitivity and reliability in detection techniques.
There are at least seven genotypes and over 80 subtypes of HCV. Different assays are used to determine genotype such as sequencing and hybridization[2]. Most genotype assays use amplification of specific region of viral genome by PCR followed by direct DNA sequencing. While a variety of techniques are used, the gold standard for HCV genotyping is nucleotide sequencing, which can be done by using core (C), envelope (E1), or the non-structural (NS5B) regions which can be amplified by reverse transcription followed by polymerase chain reaction[59-63]. Most diagnostic assays commonly target the 5’ UTR but in research settings core and or NS5B region is usually sequenced as this region is more conserved amongst all genotypes. Genotypes are very useful for determining the duration of treatment regimens and predicting treatment response[64-68].
One of the emerging diagnostic assays is nanoparticle based diagnostic assay. Quantum dot and gold based nanoparticle based diagnostic assay[69-71]. Quantum dots are nanoparticles made of semiconductors that emit light at different spectra; the emission is dependent on the size which greatly increases the ability to multiplex[72-74]. Another novel technique being developed recently is the use of aptamers as capture molecules. Aptamers are short, single stranded oligonucleotide that can fold into specific 3-dimensional structures and recognize target molecules such as small chemicals, proteins, and even cells[75]. These techniques have been used for various diagnostic applications because of their ability to bind their targets with high affinity and specificity.
Molecular diagnostic testing for HCV has provided a crucial tool for addressing significant controversies in HCV management. NATs for detecting HCV RNA remain the mainstay for detecting HCV infection in individuals in high risk group population. Nucleic acid test not only helps to detect HCV RNA but confirms active state of viral infection, i.e., the virus is in replicating state in the patient’s body. However, in developing countries due to financial constraints and lack of technical expertise in clinical settings, these tests are difficult to perform and time consuming. In these settings, the most widely employed screening tests are the HCV rapid immunoassays. However, it is the need of the hour to effectively design strategies to detect HCV infection even in sero-conversion period.
P- Reviewer: Bare P S- Editor: Ji FF L- Editor: A E- Editor: Wu HL
| 1. | WHO. Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J Viral Hepat. 1999;6:35-47. [PubMed] |
| 2. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 982] [Article Influence: 89.3] [Reference Citation Analysis (1)] |
| 3. | Kuiken C, Simmonds P. Nomenclature and numbering of the hepatitis C virus. Methods Mol Biol. 2009;510:33-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 4. | Kato N. Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation. Microb Comp Genomics. 2000;5:129-151. [PubMed] |
| 5. | Mehta SK, Singh V, Bhasin DK, Kumar YR, Kochhar R. Hepatitis C virus in patients with acute and chronic liver disease. Indian J Gastroenterol. 1992;11:146. [PubMed] |
| 6. | Jindal N, Arora U, Singh K. Prevalence of human immunodeficiency virus (HIV), hepatitis B virus, and hepatitis C virus in three groups of populations at high risk of HIV infection in Amritsar (Punjab), Northern India. Jpn J Infect Dis. 2008;61:79-81. [PubMed] |
| 7. | Irshad M, Acharya SK, Joshi YK. Prevalence of hepatitis C virus antibodies in the general population & amp; in selected groups of patients in Delhi. Indian J Med Res. 1995;102:162-164. [PubMed] |
| 8. | Agarwal N, Chatterjee K, Coshic P, Borgohain M. Nucleic acid testing for blood banks: an experience from a tertiary care centre in New Delhi, India. Transfus Apher Sci. 2013;49:482-484. [PubMed] |
| 9. | Pahuja S, Sharma M, Baitha B, Jain M. Prevalence and trends of markers of hepatitis C virus, hepatitis B virus and human immunodeficiency virus in Delhi blood donors: a hospital based study. Jpn J Infect Dis. 2007;60:389-391. [PubMed] |
| 10. | Chandrashekar S. Half a decade of mini-pool nucleic acid testing: Cost-effective way for improving blood safety in India. Asian J Transfus Sci. 2014;8:35-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Morishima C, Gretch DR. Clinical use of hepatitis C virus tests for diagnosis and monitoring during therapy. Clin Liver Dis. 1999;3:717-740. [PubMed] |
| 12. | Richter SS. Laboratory assays for diagnosis and management of hepatitis C virus infection. J Clin Microbiol. 2002;40:4407-4412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Clemens JM, Taskar S, Chau K, Vallari D, Shih JW, Alter HJ, Schleicher JB, Mimms LT. IgM antibody response in acute hepatitis C viral infection. Blood. 1992;79:169-172. [PubMed] |
| 14. | Somi MH, Etemadi J, Ghojazadeh M, Farhang S, Faramarzi M, Foroutan S, Soleimanpour M. Risk factors of HCV seroconversion in hemodialysis patients in tabriz, iran. Hepat Mon. 2014;14:e17417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Nafishah A, Asiah MN, Syimah AT, Mohd Zahari TH, Yasmin A, Normi M, Anza E, Shahnaz M, Narazah MY. Rate of seroconversion in repeat blood donors at the national blood centre, kuala lumpur. Indian J Hematol Blood Transfus. 2014;30:105-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 16. | Atrah HI, Hutchinson F, Gough D, Ala FA, Ahmed MM. Hepatitis C virus seroconversion rate in established blood donors. J Med Virol. 1995;46:329-333. [PubMed] |
| 17. | van der Helm J, Geskus R, Sabin C, Meyer L, Del Amo J, Chêne G, Dorrucci M, Muga R, Porter K, Prins M. Effect of HCV infection on cause-specific mortality after HIV seroconversion, before and after 1997. Gastroenterology. 2013;144:751-760.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Available from: http://jmitra.co.in/download/Procedure/Manual-HCV-Tri-Dot.pdf. |
| 19. | OraSure Technologies. Step-by-Step Instructions: For OraQuick® HCV Rapid Antibody Test. Available from: http://hcvadvocate.org/Hepatitis/factsheets_pdf/OraQuick_HCV_Rapid_Antibody_Test.pdf. |
| 20. | WHO. List of prequalified in vitro diagnostic products (updated 2014 December 16). Available from: http: //www.who.int/diagnostics_laboratory/evaluations/PQ_list/en/. |
| 21. | Martinot-Peignoux M, Marcellin P, Xu LZ, Bernuau J, Erlinger S, Benhamou JP, Larzul D. Reactivity to c33c antigen as a marker of hepatitis C virus multiplication. J Infect Dis. 1992;165:595-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 22. | Filice G, Patruno S, Campisi D, Chiesa A, Orsolini P, Debiaggi M, Bruno R, Tinelli M. Specificity and sensitivity of 3rd generation EIA for detection of HCV antibodies among intravenous drug-users. New Microbiol. 1993;16:35-42. [PubMed] |
| 23. | Hart-Malloy R, Carrascal A, Dirienzo AG, Flanigan C, McClamroch K, Smith L. Estimating HCV prevalence at the state level: a call to increase and strengthen current surveillance systems. Am J Public Health. 2013;103:1402-1405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | François M, Dubois F, Brand D, Bacq Y, Guerois C, Mouchet C, Tichet J, Goudeau A, Barin F. Prevalence and significance of hepatitis C virus (HCV) viremia in HCV antibody-positive subjects from various populations. J Clin Microbiol. 1993;31:1189-1193. [PubMed] |
| 25. | Chevaliez S, Pawlotsky JM. Diagnosis and management of chronic viral hepatitis: antigens, antibodies and viral genomes. Best Pract Res Clin Gastroenterol. 2008;22:1031-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Kamili S, Drobeniuc J, Araujo AC, Hayden TM. Laboratory diagnostics for hepatitis C virus infection. Clin Infect Dis. 2012;55 Suppl 1:S43-S48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 27. | Thakur V, Gupta RC, Arankale V, Sarin SK. Low specificity of the third generation ELISA for HCV detection in voluntary blood donors in India. J Int Fed Clin Chem. 1999;14:1. |
| 28. | Damen M, Zaaijer HL, Cuypers HT, Vrielink H, van der Poel CL, Reesink HW, Lelie PN. Reliability of the third-generation recombinant immunoblot assay for hepatitis C virus. Transfusion. 1995;35:745-749. [PubMed] |
| 29. | Tobler LH, Stramer SL, Kleinman SH, Brodsky JP, Todd DS, Busch MP. Misclassification of HCV-viremic blood donors as indeterminate by RIBA 3.0 because of human superoxide dismutase reactivity. Transfusion. 2001;41:1625-1626. [PubMed] |
| 30. | Pawtosky JM. Significance of indeterminate second generation RIBA and resolution by third generation RIBA. Hepatitis C virus: New Diagnostic Tools. Paris: John Libbey Eurotext 1994; 177-188. |
| 31. | Chevaliez S, Soulier A, Poiteau L, Bouvier-Alias M, Pawlotsky JM. Clinical utility of hepatitis C virus core antigen quantification in patients with chronic hepatitis C. J Clin Virol. 2014;61:145-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 32. | Ross RS, Viazov S, Salloum S, Hilgard P, Gerken G, Roggendorf M. Analytical performance characteristics and clinical utility of a novel assay for total hepatitis C virus core antigen quantification. J Clin Microbiol. 2010;48:1161-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 33. | Song D, Kang JE, Kim SY, Hwang SH, Kim HH, Lee EY, Son HC. Evaluation of ARCHITECT HCV core antigen assay. Korean J Lab Med. 2010;30:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 34. | Medici MC, Furlini G, Rodella A, Fuertes A, Monachetti A, Calderaro A, Galli S, Terlenghi L, Olivares M, Bagnarelli P. Hepatitis C virus core antigen: analytical performances, correlation with viremia and potential applications of a quantitative, automated immunoassay. J Clin Virol. 2011;51:264-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 35. | Moradpour D, Penin F. Hepatitis C virus proteins: from structure to function. Curr Top Microbiol Immunol. 2013;369:113-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 36. | Bukh J, Purcell RH, Miller RH. Sequence analysis of the 5’ noncoding region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:4942-4946. [PubMed] |
| 37. | Yanagi M, St Claire M, Emerson SU, Purcell RH, Bukh J. In vivo analysis of the 3’ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci USA. 1999;96:2291-2295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 165] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 38. | Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451-2455. [PubMed] |
| 39. | Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570-583. [PubMed] |
| 40. | Bartenschlager R, Cosset FL, Lohmann V. Hepatitis C virus replication cycle. J Hepatol. 2010;53:583-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 41. | McLauchlan J. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J Viral Hepat. 2000;7:2-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 230] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 42. | Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi SI, Ichikawa M, Kajita T, Moradpour D, Wands JR, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048-6055. [PubMed] |
| 43. | Kunkel M, Watowich SJ. Conformational changes accompanying self-assembly of the hepatitis C virus core protein. Virology. 2002;294:239-245. [PubMed] |
| 44. | Goffard A, Dubuisson J. Glycosylation of hepatitis C virus envelope proteins. Biochimie. 2003;85:295-301. [PubMed] |
| 45. | Keck ZY, Op De Beeck A, Hadlock KG, Xia J, Li TK, Dubuisson J, Foung SK. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J Virol. 2004;78:9224-9232. [PubMed] |
| 46. | Han JH, Houghton M. Group specific sequences and conserved secondary structures at the 3’ end of HCV genome and its implication for viral replication. Nucleic Acids Res. 1992;20:3520. [PubMed] |
| 47. | Blight KJ, Rice CM. Secondary structure determination of the conserved 98-base sequence at the 3’ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345-7352. [PubMed] |
| 48. | Tanaka T, Kato N, Cho MJ, Shimotohno K. A novel sequence found at the 3’ terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215:744-749. [PubMed] |
| 49. | Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsutsumi M, Shimomura H, Tsuji T, Date T. Genetic organization and diversity of the 3’ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255-261. [PubMed] |
| 50. | Vernelen K, Claeys H, Verhaert H, Volckaerts A, Vermylen C. Significance of NS3 and NS5 antigens in screening for HCV antibody. Lancet. 1994;343:853. [PubMed] |
| 51. | Albadalejo J, Alonso R, Antinozzi R, Bogard M, Bourgault AM, Colucci G, Fenner T, Petersen H, Sala E, Vincelette J. Multicenter evaluation of the COBAS AMPLICOR HCV assay, an integrated PCR system for rapid detection of hepatitis C virus RNA in the diagnostic laboratory. J Clin Microbiol. 1998;36:862-865. [PubMed] |
| 52. | DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy ZG, Rosenstraus M. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chem. 1996;42:1915-1923. [PubMed] |
| 53. | Colucci G, Gutekunst K. Development of a quantitative PCR assay for monitoring HCV viraemia levels in patients with chronic hepatitis C. J Viral Hepat. 1997;4 Suppl 1:75-78. [PubMed] |
| 54. | Hawkins A, Davidson F, Simmonds P. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by quantiplex HCV RNA assay versions 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J Clin Microbiol. 1997;35:187-192. [PubMed] |
| 55. | Beld M, Sentjens R, Rebers S, Weegink C, Weel J, Sol C, Boom R. Performance of the New Bayer VERSANT HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche COBAS Amplicor HCV Monitor, Version 2.0, assay. J Clin Microbiol. 2002;40:788-793. [PubMed] |
| 56. | Lee SC, Antony A, Lee N, Leibow J, Yang JQ, Soviero S, Gutekunst K, Rosenstraus M. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J Clin Microbiol. 2000;38:4171-4179. [PubMed] |
| 57. | Desombere I, Van Vlierberghe H, Couvent S, Clinckspoor F, Leroux-Roels G. Comparison of qualitative (COBAS AMPLICOR HCV 2.0 versus VERSANT HCV RNA) and quantitative (COBAS AMPLICOR HCV monitor 2.0 versus VERSANT HCV RNA 3.0) assays for hepatitis C virus (HCV) RNA detection and quantification: impact on diagnosis and treatment of HCV infections. J Clin Microbiol. 2005;43:2590-2597. [PubMed] |
| 58. | Zeuzem S, Lee JH, Franke A, Rüster B, Prümmer O, Herrmann G, Roth WK. Quantification of the initial decline of serum hepatitis C virus RNA and response to interferon alfa. Hepatology. 1998;27:1149-1156. [PubMed] |
| 59. | Davidson F, Simmonds P, Ferguson JC, Jarvis LM, Dow BC, Follett EA, Seed CR, Krusius T, Lin C, Medgyesi GA. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5’ non-coding region. J Gen Virol. 1995;76:1197-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 292] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 60. | Murphy DG, Willems B, Deschênes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5’ untranslated region sequences. J Clin Microbiol. 2007;45:1102-1112. [PubMed] |
| 61. | Casanova YS, Boeira Tda R, Sisti E, Celmer Á, Fonseca AS, Ikuta N, Simon D, Lunge VR. A complete molecular biology assay for hepatitis C virus detection, quantification and genotyping. Rev Soc Bras Med Trop. 2014;47:287-294. [PubMed] |
| 62. | Furione M, Simoncini L, Gatti M, Baldanti F, Grazia Revello M, Gerna G. HCV genotyping by three methods: analysis of discordant results based on sequencing. J Clin Virol. 1999;13:121-130. [PubMed] |
| 63. | Bouchardeau F, Cantaloube JF, Chevaliez S, Portal C, Razer A, Lefrère JJ, Pawlotsky JM, De Micco P, Laperche S. Improvement of hepatitis C virus (HCV) genotype determination with the new version of the INNO-LiPA HCV assay. J Clin Microbiol. 2007;45:1140-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 88] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 64. | Saha K, Firdaus R, Biswas A, Mukherjee A, Sadhukhan PC. A novel nested reverse-transcriptase polymerase chain reaction method for rapid hepatitis C virus detection and genotyping. Indian J Med Microbiol. 2014;32:130-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 65. | Cantaloube JF, Laperche S, Gallian P, Bouchardeau F, de Lamballerie X, de Micco P. Analysis of the 5’ noncoding region versus the NS5b region in genotyping hepatitis C virus isolates from blood donors in France. J Clin Microbiol. 2006;44:2051-2056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 66. | Saha K, Firdaus R, Biswas A, Mukherjee A, Sarkar K, Chakrabarti S, Sadhukhan PC. Transmission dynamics of hepatitis C virus among intra venous drug users in the border state of Manipur, India. Infect Genet Evol. 2014;24:57-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 67. | Lole KS, Jha JA, Shrotri SP, Tandon BN, Prasad VG, Arankalle VA. Comparison of hepatitis C virus genotyping by 5’ noncoding region- and core-based reverse transcriptase PCR assay with sequencing and use of the assay for determining subtype distribution in India. J Clin Microbiol. 2003;41:5240-5244. [PubMed] |
| 68. | Shawky SM, Guirgis BS, Azzazy HM. Detection of unamplified HCV RNA in serum using a novel two metallic nanoparticle platform. Clin Chem Lab Med. 2014;52:565-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 69. | Liu J, Zhang GX. [A protein array based on quantum dots (QDs) encoded microbeads for detection of hepatitis C virus]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2013;27:67-69. [PubMed] |
| 70. | Roh C, Lee HY, Kim SE, Jo SK. A highly sensitive and selective viral protein detection method based on RNA oligonucleotide nanoparticle. Int J Nanomedicine. 2010;5:323-329. [PubMed] |
| 71. | Biju V, Itoh T, Anas A, Sujith A, Ishikawa M. Semiconductor quantum dots and metal nanoparticles: syntheses, optical properties, and biological applications. Anal Bioanal Chem. 2008;391:2469-2495. [PubMed] |
| 72. | Ghasemi Y, Peymani P, Afifi S. Quantum dot: magic nanoparticle for imaging, detection and targeting. Acta Biomed. 2009;80:156-165. [PubMed] |
| 73. | Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818-822. [PubMed] |
| 74. | Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505-510. [PubMed] |
| 75. | Yang D, Meng X, Yu Q, Xu L, Long Y, Liu B, Fang X, Zhu H. Inhibition of hepatitis C virus infection by DNA aptamer against envelope protein. Antimicrob Agents Chemother. 2013;57:4937-4944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |