Published online Nov 12, 2013. doi: 10.5501/wjv.v2.i4.152
Revised: September 20, 2013
Accepted: October 15, 2013
Published online: November 12, 2013
Processing time: 133 Days and 15.9 Hours
AIM: To probe the organizational structure of the adsorption apparatus of bacteriophage epsilon 15 (E15) using genetic and biochemical methodology
METHODS: Hydroxylamine was used to create nonsense mutants of bacteriophage E15. The mutants were then screened for defects in their adsorption apparatus proteins, initially by measuring the concentrations of free tail spike proteins in lysates of cells that had been infected by the phage mutants under non-permissive growth conditions. Phage strains whose infected cell lysates contained above-average levels of free tail spike protein under non-permissive growth conditions were assumed to contain nonsense mutations in genes coding for adsorption apparatus proteins. These mutants were characterized by classical genetic mapping methods as well as automated sequencing of several of their genes. Finally, sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography were used to examine the protein compositions of the radioactive particles produced when the various mutants were grown on a non-permissive host cell in the presence of 35S-methionine and co-purified along with E15wt phage on CsCl block gradients.
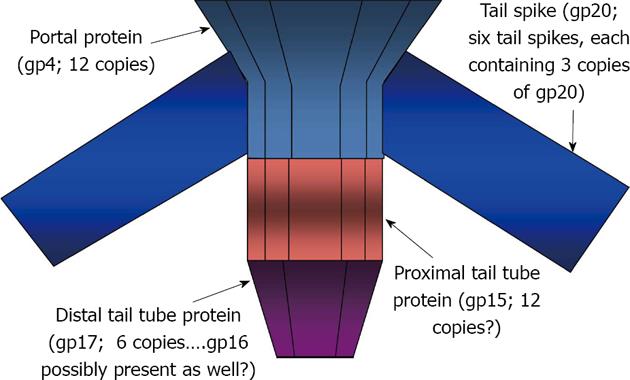
RESULTS: Our results are consistent with gp4 forming the portal ring structure of E15. In addition, they show that proteins gp15 and gp17 likely comprise the central tube portion of the E15 adsorption apparatus, with gp17 being more distally positioned than gp15 and dependent upon both gp15 and gp16 for its attachment. Finally, our data indicates that tail spike proteins comprised of gp20 can assemble onto nascent virions that contain gp7, gp10, gp4 and packaged DNA, but which lack both gp15 and gp17, thereby forming particles that are of sufficient stability to survive CsCl buoyant density centrifugation.
CONCLUSION: The portal ring (gp4) of E15 is bound to tail spikes (gp20) and the tail tube (gp15 and gp17); gp17’s attachment requires both gp15 and gp16.
Core tip: Epsilon 15 (E15) is a temperate, serotype-converting bacteriophage that specifically infects group E1 Salmonellae bacteria. This paper presents genetic and biochemical evidence regarding the identities and positional relationships of the proteins that comprise the tail tube structure of E15. As such, it makes a small contribution towards what may someday be a fuller understanding, not only of how E15 stabilizes its packaged DNA, but also, how it triggers release of its DNA when the phage encounters a susceptible Salmonella host cell.
- Citation: Guichard JA, Middleton PC, McConnell MR. Genetic analysis of structural proteins in the adsorption apparatus of bacteriophage epsilon 15. World J Virol 2013; 2(4): 152-159
- URL: https://www.wjgnet.com/2220-3249/full/v2/i4/152.htm
- DOI: https://dx.doi.org/10.5501/wjv.v2.i4.152
Salmonella bacteria are enteric organisms that constitute a serious source of gastro-intestinal infection in humans and agriculturally important animals[1]. Bacteriophages provide an important mechanism of genetic variation and gene exchange among Salmonella bacteria (and thus, the potential for enhanced pathogenicity) through their ability to promote lateral transfer of host cell genes. Understanding the structural features of phage DNA packaging and adsorption/DNA ejection apparati is an important step in being able to fully assess how phage contribute to genetic variation within their Salmonella hosts.
Bacteriophage epsilon15 (E15) is a temperate, Group E1 Salmonella-specific phage that belongs to the Order “Caudovirales” and the Family “Podoviridae”[2]. At the genomic level[3], it closest relatives are the Salmonella-specific viruses, SPN1S (NCBI Accession number JN391180.1) and SPN9TCW (NCBI Accession number JQ691610.1) but it also shares 36 related genes in common with the E. coli O1H57-specific phage, V10 (NCBI Accession number DQ126339.2). E15 was among the first Salmonella-specific phages to be discovered and was a popular experimental model for Japanese and US investigators in the 50’s, 60’s and 70’s, both because of its ability to cause serotype conversion and because of its enzymatically active tail spikes, which display endorhamnosidase activity towards the host cell O-polysaccharide structure[4-9]. The publication of the E15 genome sequence by our laboratory in 2002 (NCBI Accession number AY150271.1) stimulated renewed interest in E15, this time as a model system for investigating virion structure by cryo-electron microscopy (cryo-EM), matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry and other methods[3,10-14]. These studies, combined with earlier genetic and biochemical investigations[6], have revealed the following: (1) gp7 and gp10 together comprise the capsid of E15; (2) E15’s enzymatically active tail spikes are homotrimers of gp20; and (3) other major proteins in E15 virions include gp4, gp15 and gp17. Circumstantial evidence, including size, relative abundance within virion particles and the position of its gene just downstream of those coding for the small and large terminase subunits in the late transcript are all consistent with gp4 being the portal protein of E15[3].
In addition to being a powerful tool for elucidating virion capsid structures, cryo-EM can also be used effectively to decipher the structure of a phage adsorption apparatus, especially if the adsorption apparatus can be detached intact from the virion capsid and prepared in purified form. Such was the case for the Group B Salmonella-specific phage, P22, and the resulting structure that was determined by cryo-EM analysis of these P22 adsorption apparati (termed “tail machines”) is, in a word, spectacular[15,16]. To date, no one has reported having successfully purified the intact adsorption apparatus of phage E15.
In this paper, we present genetic and biochemical data that is consistent with gp4 forming the portal ring structure of E15; in addition, our data indicates that the centrally-positioned tail tube portion of the adsorption apparatus is likely comprised of gp15 and gp17, with gp17 being more distally positioned than gp15 and dependent upon both gp15and gp16 for its attachment. Finally, our data indicates that tail spike proteins comprised of gp20 can form stable associations with nascent virus particles that contain gp7, gp10, gp4 and packaged dsDNA, but which lack both gp15 and gp17. This implies that tail spikes bind directly to the portal ring during the assembly process that leads to the formation of mature virions.
Parental phages E15 and E15vir (a clear plaque mutant with a missense mutation in gp38, the major repressor protein) as well as bacterial host strains Salmonella enterica subsp. enterica serovar Anatum A1 and Salmonella enterica subsp. enterica serovar Anatum 37A2Su+ all came originally from the laboratory of Dr. Andrew Wright (Tufts University, Boston, MA). E15 (am2) is a nonsense mutant of E15 that is unable to produce tail spike proteins[6]. Propagation of bacteria and phage was in trypticase soy broth, unless otherwise indicated.
Isolation of phage nonsense mutants with adsorption apparatus defects
Nonsense mutants of E15vir were generated by hydroxylamine mutagenesis[17] and were detected initially by an anaerobic, double layer plating method that dramatically increases plaque size[18]. Hydroxylamine-treated phage were mixed with an amber suppressor strain (Salmonella anatum 37A2Su+) in the bottom LB soft agar layer, then overlaid with a second soft agar layer containing the non-suppressing parental strain Salmonella anatum A1. Turbid-looking plaques were cloned and re-screened to verify their inability to form plaques on Salmonella anatum A1.
Phage nonsense mutants isolated by the method described above were subsequently screened individually for potential defects in adsorption apparatus proteins other than the tail spike by measuring the level of free tail spike protein in lysates of non-permissively infected cells. The tail spike assay was based on a method developed earlier in an investigation involving phage P22 tailspikes[19]; It involved UV-irradiating 10000RPM (10K) supernatant fractions obtained from lysates of Salmonella anatum A1 cells infected by E15vir nonsense mutants, then incubating the irradiated 10K supernatants with E15 “heads” obtained by infecting Salmonella anatum A1 with E15 (am2), an E15 nonsense mutant that is unable to produce tail spike protein. Following incubation, reaction mixes were plated at varying dilutions on the permissive host strain, Salmonella anatum 37A2Su+, in order to titer the number of E15 (am2) “heads” that were made infectious by the binding of tail spike proteins in vitro.
Genetic mapping and sequencing of Epsilon15 nonsense mutations: E15vir nonsense mutants isolated and screened as described above were characterized (along with the known tailspike nonsense mutant, am2) using classical in vivo complementation and two-factor recombination assay procedures that have been previously described[6]. These genetic mapping studies revealed the number of complementation groups (i.e., genes) defined by the nonsense mutants and also allowed for an approximation of their locations relative to the E15 tail spike gene. Shortly after the mapping of the nonsense mutations using classical methods, the genomic sequence of E15 was completed by our lab. Gene 20 was then shown by sequencing analysis to contain the am2 nonsense mutation (i.e., gp20 is the tailspike protein) and in addition, was observed to be the distal-most gene in the late mRNA transcript of E15[3].
Each E15vir mutant believed to be defective in an adsorption apparatus protein was subjected to DNA sequence analyses for genes 15, 16 and 17, in an effort to assign a gene identity for its nonsense mutation. The bracketing, Frwrd and Rvrse primer pairs used for initial PCR amplification of the three genes are shown below, with underlined bases representing modifications made in order to facilitate cloning of the PCR products into plasmids. Gene 15: E15.Orf15.Frwrd, AGGGATCCAAATGCCAGTTGTACCTACAG, E15.Orf15.Rvrse, ATACATAAGCTTTTATTCAACCCTCACG; Gene 16: E15.Orf16.Frwrd, TGGATCCATGGCTGATGTATTTTCACT, E15.Orf16.Rvrse, ACACATGCCTGCAGCATTATGGATTCCT; Gene 17: E15.Orf17.Frwrd, GAGGGATCCATAATGAAACAGGCATGTGT, E15.Orf17.Rvrse, GTTAAGGGTACCATCATTGTCCTA.
Because of their large sizes (ranging from 1928 to 2782 basepairs), the resulting PCR products were sequenced not only with the same Frwrd and Rvrse primers that had been used to produce them, but also with several additional primers known to bind internally within each PCR product. The internal sequencing primers were as follows: Gene 15: E15.g15.W12689: GGCGCTGCTCATGGCTGGAGTCATGAACAG, E15.g15.W13264: CGCGGCTATCGGTCTTTCTCAGTTACCTAC, E15g15.W13879: GGAGGCGGCTGCGCTGTCTGAACAGGTAC; Gene 16: E15.g16.W15213: CGGCAGGCATGGCCCTTCCTGCTGCTGTTG, E15.g16:W15689:TAGCGAACAGCCAGCGCATCCTGGATAAC; Gene 17: E15.g17.W17092: GCGGCAAAGTCTGCACAGTTCCAGATCCTG, E15.g17.W17717: GACCTGACGCTGCGCGAAACTTTTCCCTTG, E15.g17.W18214: GCGGCGTTCGGGCTGTTGATGTACAAAAAC.
Taq polymerase is somewhat error-prone[20], so in order to generate PCR products suitable for accurate DNA sequencing, PCR reaction mixes were prepared on a large scale (250 μL), then separated into five 50 μL aliquots prior to commencing the thermocycling reaction. Upon completion of PCR, the five aliquots were recombined into a single 250 μL sample and the DNA product was purified using a QIAGEN PCR purification column. Automated DNA sequencing reactions were performed by the Microchemical Core Facility at San Diego State University.
Preparation and analysis of 35S-methionine labeled, virion-like particles produced by phage nonsense mutants under non-permissive conditions: Preparations of 35S-methionine labeled, wild type E15vir phage particles and non-infectious, virion-like particles produced by the nonsense mutants were obtained by incubating mid-log phase Salmonella anatum A1 cells grown in low sulfate medium with phage (multiplicity of infection of 10) for ten minutes at 0 °C, then adding 35S-methionine to a final concentration of 10 uCi/mL and shifting the incubation temperature to 37 °C. At T = 90 min, cell cultures were lysed with chloroform, then centrifuged for 10 min at 10000 RPM in order to remove cellular debris. The resulting 10K supernatant fractions were loaded onto CsCl block gradients and centrifuged for 30 min at 38000 RPM on a Beckman L8-80M ultracentrifuge (an excess of cold E15wt phage was included in each sample as a carrier). Particles displaying virion-like densities (i.e., the ability to pass readily through a 1.375 g/cm3 CsCl layer and settle onto a 1.6 g/cm3 CsCl layer along with non-radioactive E15wt carrier phage) were dialyzed, normalized for cpm and electrophoresed on 12% sodium dodecyl sulfate-protective antigen (SDS-PA) gels. The gels were subsequently dried on Whatman 3M paper and the paper was exposed to Kodak X-Omat X-ray film in order to detect radioactive proteins by autoradiography.
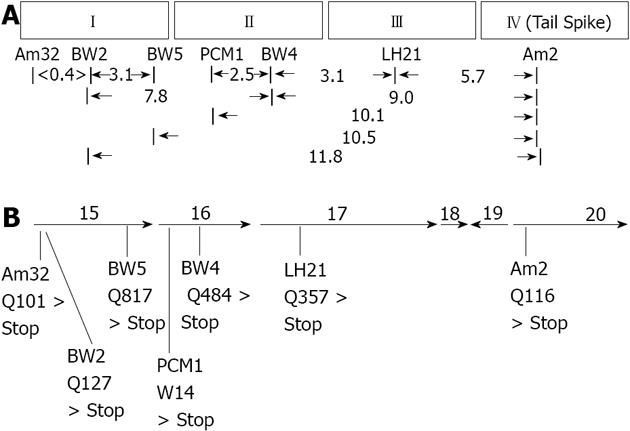
We reasoned that cell lysates produced by infection of Salmonella anatum A1 with E15vir phage containing nonsense mutations in genes coding for adsorption apparatus proteins other than the tail spike should contain higher than normal levels of free tail spike protein. Cell lysates produced by infection with different E15 nonsense mutants were therefore screened for their ability to provide tail spike proteins to E15 (am2) “heads”in vitro, thereby rendering the heads infectious. Six E15vir nonsense mutants whose lysates had tail spike levels surpassing that of an E15vir lysate were identified, then further analyzed using classical genetic mapping methods. The six mutants were shown to define three complementation groups (i.e., genes), which mapped in close proximity to each other as well as to the tail spike gene, defined by nonsense mutation am2 (Figure 1A). After confirming by DNA sequencing that the am2 mutation lay within gene 20 (the last gene in E15’s “late” mRNA transcript), PCR primers were used to amplify and sequence three genes for each of the six mutants; namely 15, 16 and 17. Genes 15 and 17 were chosen for sequence analysis because the pI values, overall sizes, and tryptic digestion fragment sizes of their inferred polypeptide products closely matched those of E15 virion proteins shown by SDS-PA/autoradiography to be missing in virion-like particles formed by the various nonsense mutants under non-permissive conditions[3]. Gene 16 was included for sequence analysis as well because the genetic mapping data showed that the collection of six nonsense mutations with potential adsorption apparatus defects defined three different genes. Other neighboring genes (i.e., 13, 14, 18 and 19) all coded for inferred proteins that were either very small or strongly hydrophobic, and were therefore not included in the sequencing analysis.
The DNA sequencing data (Figure 1B) revealed the presence of unique amber nonsense mutations in gene 15 for the three non-complementing phage mutants am32, BW2 and BW5. Non-complementing mutants pericentriolar material 1 (PCM1) and BW4 both contained unique amber nonsense mutations in gene 16, while mutant luteinizing hormone 21 (LH21), which the classical mapping data showed to be in a complementation group of its own, was found to contain a unique amber nonsense mutation in gene 17. The positions of the nonsense mutations determined by DNA sequencing correlated nicely with the linear map order that had been established for them previously by recombination analysis. In every case, the nonsense mutation had resulted from a hydroxylamine-induced C > T transition (either CAG > TAG, or TGG > TAG).
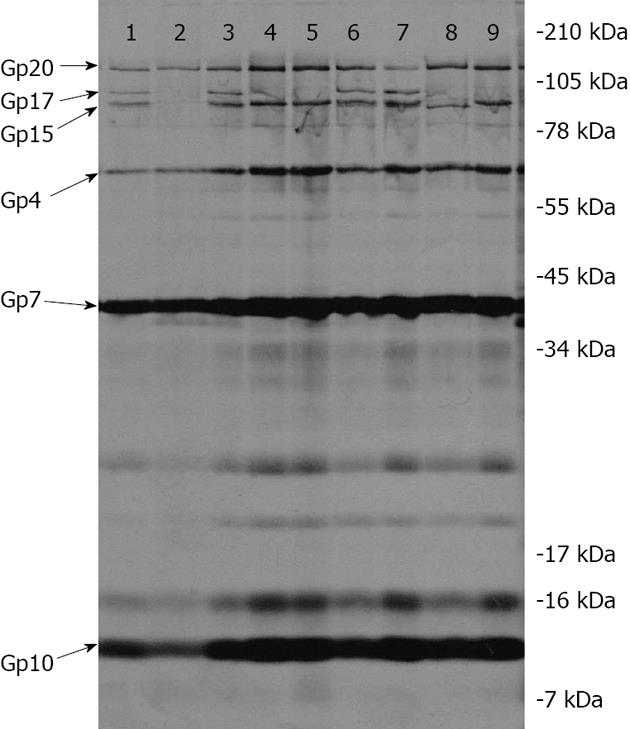
MALDI-TOF mass spectrometry analyses of trypsin-digestion products obtained from purified E15 virion proteins[10] indicate that after the tail spike protein, gp20 (1070 amino acids, 115676 daltons), the next two largest proteins contained in E15 virions are gp17 (918 amino acids, 100841 daltons) and gp15 (842 amino acids, 91012 daltons). When 35S-methionine-labeled particles produced by the various nonsense mutants under non-permissive conditions were co-purified with non-radioactive, “carrier” E15wt phage on CsCl block gradients, then analyzed by SDS-PAGE and autoradiography, it was observed that the two gene 16 mutants (PCM1 and BW4) and the gene 17 mutant (LH21) all produced good yields of radioactive particles relative to E15wt (118%, 154% and 100%, respectively, with a mean of 124 ± 28% SD) and that these particles all lacked gp17 (Figure 2, Lanes 4, 5 and 9). The three gene 15 mutants (am32, BW2 and BW5) all produced lower quantities of radioactive particles than E15wt (17%, 23% and 44%, respectively, with a mean of 28 ± 14% SD). The am32 and BW2 mutants, whose nonsense mutations mapped at codons 101 and 127, respectively, of gene 15 (845 codons), produced particles that lacked both gp15 and gp17 (Figure 2, Lane 2). Mutant BW5, whose nonsense mutation maps at codon 817 of gene 15, produced particles lacking gp17 but containing a novel protein with a slightly faster mobility than that of gp15; a protein most likely comprised of amino acids 1 through 816 of gp15 (Figure 2, Lane 8). The quantity of the slightly truncated gp15 protein in BW5 particles is reduced, relative to the quantity of gp15 observed in E15vir and the various gp17-deficient mutants (see Lane 9, for example), thus indicating that its ability to assemble onto nascent virion particles has been diminished by the loss of 29 C-terminal amino acids, but not entirely eliminated. The 10K supernatant fractions obtained from cells infected by the three gene 15 mutants (am32, BW2 and BW5) were also analyzed by SDS-PAGE and autoradiography. All three supernatants contained a protein that co-migrated with the gp17 protein of E15wt (data not shown).
The two gene 16 nonsense mutants analyzed in this study (PCM1 and BW4) both produced good yields (118% and 154%, respectively, relative to wt E15) of non-infectious, virion-like particles that are missing gp17 (Figure 2, Lanes 4 and 5). As was the case for the three gene 15 mutants, a protein with gp17-like mobility was present in the 10K supernatant fractions of cells infected by PCM1 and BW4 (data not shown).
Every nonsense mutant that was studied produced radioactive particles that contained DNA, as judged by their ability to co-sediment with E15wt virions through CsCl at 1.375 g/mL and layer onto the 1.6 g/mL solution. In addition, all of the mutants, whether gp17-deficient or both gp15- and gp17-deficient, displayed normal quantities of the two known capsid proteins, gp7 and gp10, as well as gp4. Yields of the radioactive particles that lacked both gp15 and gp17 were significantly lower than those of particles that lacked gp17 only, suggesting that maximum stability of packaged DNA is achieved when both gp4 and gp15 are present. All of the mutant phage particles contained sufficient gp20 tail spike protein for easy detection by autoradiography (see lanes 2, 4, 5, 8, 9 of Figure 2).
The complete absence of both gp15 and gp17 in high-density particles produced by mutants am32 and BW2, whose nonsense mutations both map near the beginning of gene 15, combined with the gp17-only deficiency observed in high density particles produced by the gene 17 nonsense mutant (LH21), argues for a model in which gp15 and gp17 occupy penultimate and terminal positions, respectively, within a peripheral E15 virion structure that we hypothesize is the tail tube. The missing 29 amino acids at the C-terminal end of the gp15-like protein that is produced by BW5 phage under non-permissive conditions must be critical for gp17 binding since no gp17 protein was detected in these particles.
We currently do not know why gp16 is required for gp17’s assembly onto nascent virions. The gp16 protein is inferred to have 634 amino acids and our two gene 16 nonsense mutations, PCM1 and BW4, are positioned at codons 14 and 484, respectively. The predicted mass for gp16 is 67364 daltons and its inferred overall methionine content (2.4%) falls within the range of methionine contents inferred for the other known virion proteins (from as low as 1.3% for gp20 to as high as 5.2% for gp4). In other words, if gp16 is present in E15 virions in appreciable quantities, then it should contain sufficient 35S-methionine to show up in our autoradiogram. Faint protein bands were observed above the 78 kDa marker and above and below the 55 kDa marker on the gel (Figure 2), but none of these three proteins appeared to be diminished in quantity in the gene 16 mutants, relative to the other mutants or to E15vir. It is conceivable that gp16 is a virion protein that was not detected in our experiment because it co-migrated with gp4 protein (the inferred mass for gp4 is 61657 daltons). If that is true, though, one can argue that the quantity of gp16 in virions must be quite small, since the intensities of the gp4 bands in the two gene 16 mutants do not appear to be diminished, relative to those of E15vir and the other nonsense mutants that were analyzed. It should be noted that both our lab and at least one other have detected gp16 tryptic fragments in purified E15 virions using MALDI-TOF analysis[10]; the other lab has more recently hypothesized that gp16 is a tail tube protein[21]. While the data in this paper does not support that hypothesis, we remain open to the possibility and are continuing to explore the role played by gp16 in E15 virion assembly. It has also been hypothesized that gp17 functions as a pilot (or ejection) protein for E15[21]; this seems highly unlikely since ejection proteins, as the name implies, exit the capsid along with the DNA during the infection process[22,23]. Our results clearly show that E15 particles lacking gp17 retain stably packaged DNA within their capsids, as evidenced by their ability to co-purify in high yields with E15wt carrier phage on CsCl block gradients; furthermore, the same holds true, albeit to a lesser degree, for particles that are lacking both gp15 and gp17.
Frankly, we were surprised that tail spikes were present in all of the particles produced by our nonsense mutants. The initial screening procedure used to identify nonsense mutants for this study was based on the assumption that mutations resulting in adsorption apparatus defects would hinder tail spike assembly onto the virion, thereby resulting in higher than normal levels of free tail spike protein in the infected cell lysates, as well as the production of phage particles lacking tail spike proteins. Our current explanation is that gp4 forms the portal ring structure and perhaps, with help from immediately adjacent capsid proteins, provides a significant part of the binding surface(s) to which gp20 tail spikes normally attach during virion assembly. Interestingly, in their first cryo-EM paper dealing with E15, Jiang et al[10] reported that two of E15’s six tail spikes occupy positions around the tail tube that place them in very close contact with the capsid. If these two tailspikes are more firmly bound in gp17- and gp15-/gp17-deficient particles than the other four, then that might explain both the presence of gp20 in the mutant particles as well as the enhanced levels of tail spike protein in their infected cell lysates.
Figure 3 sums up our current model for the structure of the E15 adsorption apparatus: (1) gp4 forms the portal ring structure and perhaps, with help from neighboring capsid proteins, provides a binding surface that is sufficient for attachment of tail spikes (gp20); (2) gp15 and gp17 form the central tail tube, with gp17 occupying the more distal position and interacting with gp15 by 4o interactions that cannot occur if the C-terminal 29 amino acids of gp15 are missing. The association of gp17 with gp15 is also gp16-dependent but we do not know yet whether or not gp16 forms part of the tail tube. We are currently continuing our study of E15 adsorption apparatus structure and function by conducting phenotypic suppression experiments with an E15 mutant in our collection that under non-permissive conditions, adsorbs to cells and degrades O-polysaccharide normally, but fails to eject its DNA[6].
The best understood Salmonella-specific phage in the Podoviridae family is P22 and recent X-ray crystallography and cryo-EM studies have revealed features of the proteins that comprise its capsid, portal, tail tube, needle and tail spikes in exquisite detail[15,16,24,25]. The dodecameric, ring-shaped portal structure of P22 is comprised of gp1; below the portal ring is the tail tube, comprised of twelve copies of gp4 (bound directly to the portal) and six copies of gp10, which are bound to gp4. Attached to the distal portion of gp10 is P22’s “needle” structure, which is comprised of three copies of gp26. The six laterally-positioned, homo-trimeric tail spikes of P22 are comprised of gp9 and are thought to be associated with a binding surface generated cooperatively by proteins gp4 and gp10 at their point of junction on the sides of the tail tube[15].
Gene homology studies indicate that of the three Podoviridae phages known to infect Group E Salmonellae, namely E15, Epsilon34 (E34) and g341, two (E34 and g341) likely have adsorption apparatus protein compositions and organizations that are similar to that of P22[26,27]. Phage E15, on the other hand, has clearly taken a different path; Its tail spike protein is gp20, which at 1070 amino acids (aa) is about 63% larger, on average, than those of E34 (606 aa), g341 (705 aa) and P22 (667 aa) and is homologous with them only in a short stretch of amino acids at the N-terminal end that are thought to be critical for assembly onto the virion. Although they appear to occupy similar positions in the tail tube, there is no apparent structural homology between the proximal tail tube proteins of E15 and P22 (gp15 and gp4, respectively) or between their distal tail tube proteins (gp17 and gp10, respectively). There are stoichiometric similarities, though, in that densitometry measurements of Coomassie Blue-stained proteins of wild type E15 virions, followed by normalization for size differences, indicate that tail spikes (gp20), proximal tail tube proteins (gp15) and distal tail tube proteins (gp17) are present in E15 virions at approximately a 3/2/1 ratio, which matches the well-established 18/12/6 ratios of tail spike (gp9), proximal tail tube (gp4) and distal tail tube (gp10) proteins known to be present in P22 virions. No homolog of the P22 “needle” protein (gp26) is present among inferred bacteriophage E15 proteins, but that is not surprising since the tail tubes of negatively-stained E15 virions do not display the “needle-like” protuberance that is seen in electron micrographs of P22[6]. The “needle” is thought to play a role in the movement of the P22’s genome across the bacterial cell envelope during an infection[28]. How E15 compensates for its lack of a “needle” protein remains to be determined.
The authors thank Jonathon King (MIT) for reading the manuscript and for providing useful suggestions and encouragement. We’re also grateful to Michael McClelland (Vaccine Research Institute of San Diego) and Jack Johnson (The Scripps Research Institute, La Jolla, CA), both of whom provided space in their laboratories for the principle author to perform experiments and write during his sabbatical.
In April, 2010 (http://www.hhs.gov/asl/testify/2010/04/t20100428b.html), the Director of the Center for Disease Control reported to the House Subcommittee on Human Health that approximately 1.4 million Americans are infected annually with foodborne strains of Salmonellae bacteria. He further stated that the incidence of antibiotic resistance among isolates of Salmonella strains obtained in hospitals, stock animals and the food supply were all on the rise. Generalized transduction by bacteriophages is a major method for the horizontal transfer of genes between Salmonella organisms and thus, likely plays a role in their evolving pathogenicity.
The ability of a bacteriophage to infect a bacterium is governed by the nature of its adsorption apparatus. The adsorption apparatus is a collection of proteins that cooperate together to maintain the stability of the phage’s packaged genome until the moment when a susceptible host cell is encountered. At that point, the same sets of proteins interact with each other in an entirely different manner to trigger ejection of the phage genome and facilitation of its transport into the host cell cytoplasm.
Recent cryo-electron microscopy studies on virions of the Group E1 Salmonella-specific bacteriophage, epsilon 15 (E15) have yielded highly detailed information on the composition and structure of the phage’s capsid. Those same investigators have also produced the first close-up view of the adsorption apparatus of E15. This paper presents data regarding the identities of the proteins that comprise E15’s adsorption apparatus; in addition, the data presented herein provides some insight into the ways these proteins interact with each other in order to form the adsorption apparatus.
Compared with other salmonellae-specific members of the podoviridae family, bacteriophage E15 appears to be unique when it comes to the collection of proteins that comprise its adsorption apparatus. Perhaps, in addition to the uniqueness of their physical characteristics, the manner in which these proteins interact with each other to control the stability of packaged DNA as well as its release in response to the proper environmental cue will also prove to be novel, and thus, worthy of further study.
Adsorption apparatus pertains to those proteins that are stably associated with the mature virion, either through direct binding interactions with the portal ring or else, by virtue of their association with other proteins that are bound to the portal ring.
The authors used genetic and biochemical methods to examine compositional and organizational aspects of the adsorption apparatus of bacteriophage E15. Although preliminary, the results are sufficient for establishing a simple model that should be possible to refine with further experimentation.
P- Reviewers: Datta S, Ghiasi SM, Jaime GL, Kamal SA, Menendez-Arias L, Piergiuseppe De B S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 1. | Hoelzer K, Isabel A, Switt M, Wiedmann M. Animal contact as a source of human non-typhoidal salmonellosis. Veterinary Research. 2011;34. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 285] [Cited by in RCA: 306] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 2. | Ackermann HW. Tailed bacteriophages: the order caudovirales. Adv Virus Res. 1998;51:135-201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 283] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 3. | Kropinski AM, Kovalyova IV, Billington SJ, Patrick AN, Butts BD, Guichard JA, Pitcher TJ, Guthrie CC, Sydlaske AD, Barnhill LM. The genome of epsilon15, a serotype-converting, Group E1 Salmonella enterica-specific bacteriophage. Virology. 2007;369:234-244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 49] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 4. | Kanegasaki S, Wright A. Studies on the mechanism of phage adsorption: interaction between phage epsilon15 and its cellular receptor. Virology. 1973;52:160-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Losick R, Robbins PW. Mechanism of epsilon-15 conversion studies with a bacterial mutant. J Mol Biol. 1967;30:445-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 45] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | McConnell M, Reznick A, Wright A. Studies on the initial interactions of bacteriophage epsilon15 with its host cell, Salmonella anatum. Virology. 1979;94:10-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 7. | Robbins PW, Keller JM, Wright A, Bernstein RL. Enzymatic and kinetic studies on the mechanism of O-antigen conversion by bacteriophage epsilon-15. J Biol Chem. 1965;240:384-390. [PubMed] |
| 8. | Takeda K, Uetake H. In vitro interaction between phage and receptor lipopolysaccharide: a novel glycosidase associated with Salmonella phage epsilon15. Virology. 1973;52:148-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Uetake H, Luria SE, Burrous JW. Conversion of somatic antigens in Salmonella by phage infection leading to lysis or lysogeny. Virology. 1958;5:68-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 92] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 248] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 11. | Petrov AS, Lim-Hing K, Harvey SC. Packaging of DNA by bacteriophage epsilon15: structure, forces, and thermodynamics. Structure. 2007;15:807-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 172] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 13. | Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, Jakana J, Marsh MP, Booth CR, Shinkawa T, Nakata M. JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles. J Struct Biol. 2009;165:1-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W. Validated near-atomic resolution structure of bacteriophage epsilon 15 derived from cryo-EM and modeling. Proc. Natl Acad Sci. 2013;110:12301-12306. [RCA] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 15. | Tang L, Marion WR, Cingolani G, Prevelige PE, Johnson JE. Three-dimensional structure of the bacteriophage P22 tail machine. EMBO J. 2005;24:2087-2095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Lander GC, Khayat R, Li R, Prevelige PE, Potter CS, Carragher B, Johnson JE. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure. 2009;17:789-799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 17. | Hall DH, Tessman I. T4 mutants unable to induce deoxycytidylate deaminase activity. Virology. 1966;29:339-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 69] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | McConnell M, Wright A. An anaerobic technique for increasing bacteriophage plaque size. Virology. 1975;65:588-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 19. | Israel JV, Anderson TF, Levine M. In vitro morphogenesis of phage P22 from heads and baseplate parts. Proc Natl Acad Sci. 1967;57:284-291. |
| 20. | Lundberg KS, Shoemaker DD, Adams MW, Short JM, Sorge JA, Mathur EJ. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991;108:1-6. [PubMed] |
| 21. | Chang JT, Schmid MF, Haase-Pettingell C, Weigele PR, King JA, Chiu W. Visualizing the structural changes of bacteriophage Epsilon15 and its Salmonella host during infection. J Mol Biol. 2010;402:731-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 22. | Israel V. E proteins of bacteriophage P22. I. Identification and ejection from wild-type and defective particles. J Virol. 1977;23:91-97. |
| 23. | Perez GL, Huynh B, Slater M, Maloy S. Transport of phage P22 DNA across the cytoplasmic membrane. J Bacteriol. 2009;191:135-140. [RCA] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791-1795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 251] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 25. | Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865-880. [PubMed] |
| 26. | Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 27. | Villafane R, Zayas M, Gilcrease EB, Kropinski AM, Casjens SR. Genomic analysis of bacteriophage epsilon 34 of Salmonella enterica serovar Anatum (15+). BMC Microbiol. 2008;8:227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Bhardwaj A, Olia AS, Walker-Kopp N, Cingolani G. Domain organization and polarity of tail needle gp26 in the portal vertex structure of bacteriophage P22. J Mol Biol. 2007;371:374-387. [PubMed] |