Published online Nov 25, 2022. doi: 10.5501/wjv.v11.i6.496
Peer-review started: May 19, 2022
First decision: August 22, 2022
Revised: August 23, 2022
Accepted: October 4, 2022
Article in press: Octobe 4, 2022
Published online: November 25, 2022
Processing time: 182 Days and 21.3 Hours
BA.2 is a novel omicron offshoot of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that has gone viral. There is limited knowledge regarding this variant of concern. Current evidence suggests that this variant is more contagious but less severe than previous SARS-CoV-2 variants. However, there is concern regarding the virus mutations that could influence pathogenicity, transmissibility, and immune evasion.
Core Tip: BA.2 is novel omicron offshoot that goes viral. There is limit knowledge regarding this variant of concern. Current evidence suggested this variant is more contagious but less severe than previous severe acute respiratory syndrome coronavirus 2 previous variants. However, there is concern regarding the virus mutations that could be influenced pathogenicity, transmissibility as well as immune evasion.
- Citation: Roohani J, Keikha M. Global challenge with the SARS-CoV-2 omicron BA.2 (B.1.1.529.2) subvariant: Should we be concerned? World J Virol 2022; 11(6): 496-501
- URL: https://www.wjgnet.com/2220-3249/full/v11/i6/496.htm
- DOI: https://dx.doi.org/10.5501/wjv.v11.i6.496
On November 25, 2021, 22 patients in Gauteng province, South Africa, were diagnosed with atypical pneumonia caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Afterward, the Technical Advisory Group of the World Health Organization (WHO) designated the novel SARS-CoV-2 variant as B.1.1.529[1]. Omicron has been identified as the fifth variant of concern (VOCs); the B.1.1.529 genome contains over 50 mutations that increase transmissibility, infectivity, immune system evasion, and vaccine inefficacy[2].
In late November 2021, the South African National Institute of Communicable Diseases reported a 22.4% increase in infections with this variant in a single day[1]. Based on a retrospective study conducted in South Africa, the VOC omicron significantly increased the risk of reinfection (× 2.39) compared to the beta and delta variants, indicating that this variant has a higher potential to evade the immune system[3]. Belgium, Hong Kong, Israel, Germany, the Netherlands, and the United Kingdom reported the detection of the omicron variant shortly after.
On November 27, 2021 due to the omicron variant’s spread to more than 50 countries, the United States and Australia banned all transportation to those countries. However, on November 29, 2021 the first omicron case in Australia was identified in a traveler who traveled to Johannesburg, South Africa (https://www.abc.net.au/news/2021-11-29/nt-covid-outbreakkatherine-traveller-positive-for-omicron/100657690). Subsequently, the VOC omicron was identified in populations that were unvaccinated, partially or fully vaccinated, and immune to previous natural infection.
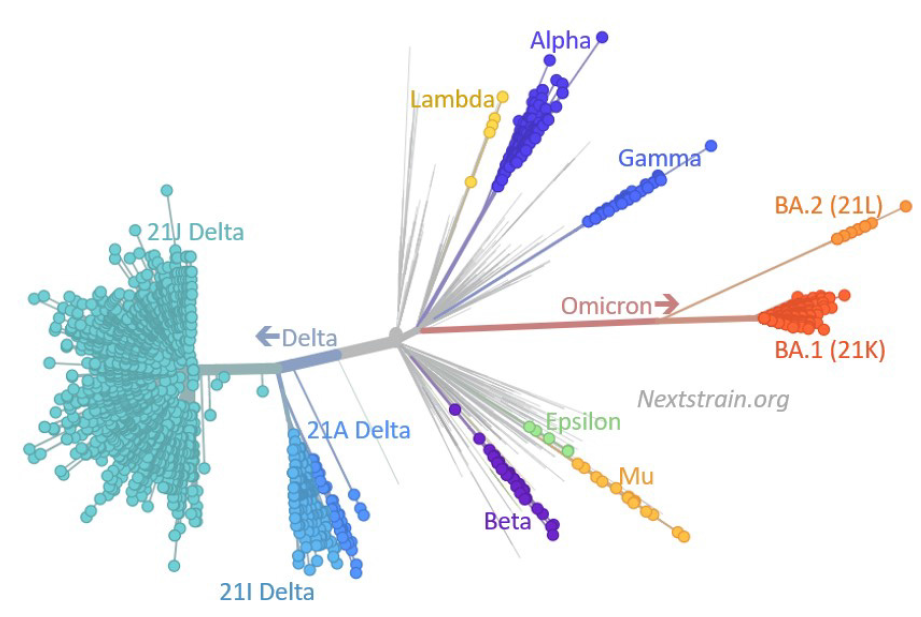
According to the WHO classification, the omicron variant has three to four subvariants, namely BA.1, BA.1.1, BA.2, and BA.3; nucleotide sequencing analysis revealed that BA.1, BA.1.1, BA.2, and BA.3 subvariants possess 39, 40, 31, and 34 mutations, respectively. All three subvariants evolved simultaneously in South Africa[4] (Figure 1). According to the hypothesis of Gao et al[5], omicron subvariants have emerged in the unvaccinated African population with compromised immune systems. Immunocompromised individuals are unable to fight SARS-CoV-2 infection effectively, and they have the best opportunity for multiplication, mutagenesis, and the emergence of new surge variants. Furthermore, animals can act as reservoirs for the evolution of novel variants. Nonetheless, there has been an increase in cases of BA.2 in recent days, to the point where the BA.2 variant is now the most common SARS-CoV-2 variant in most European countries.
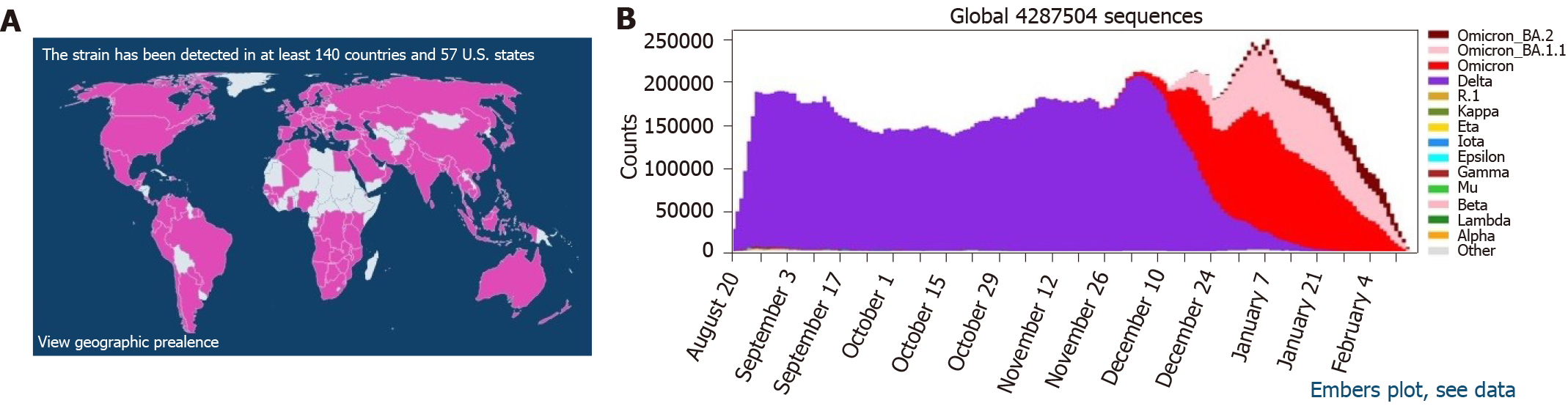
BA.2 is also known as the “stealth variant” because it lacks a deletion signature at positions 69-70 that was not detected by the spike (S) gene target failure assay; therefore, BA.2 was underestimated with the current reverse transcription polymerase chain reaction setup, and its detection is only possible via whole genome sequencing[6,7]. According to viral genome sequences uploaded to the global GISAID database, the United Kingdom, the United States, Denmark, Germany, and Canada are the five countries with the highest prevalence of BA.2 (https://www.gisaid.org/). By February 18, 2022, BA.2 was reported in 153 countries (https://www.gisaid.org).
Beginning in 2022, BA.2 has been on the rise in European countries. On January 1, 2022, the prevalence of BA.2 in the United Kingdom was approximately 5%, and it has been steadily rising[8]. According to preliminary studies conducted in Denmark, the first reports of BA.1 and BA.2 occurred on November 25, 2021 and December 5, 2021, respectively. During the 52nd wk of 2021, BA.2 prevalence in Denmark was 20%, while more than 45% of circulating SARS-CoV-2 strains in Denmark during the 2nd wk of 2022 were the BA.2 subvariant[9]. To this end, BA.2 is spreading at an alarming rate across the globe (Figure 2).
Lyngse et al[9] demonstrated that the BA.2 subvariant was able to infect unvaccinated individuals (odds ratio = 2.19; 95% confidence interval: 1.58-3.14) and individuals vaccinated with a third booster (odds ratio = 2.99; 95% confidence interval: 2.11-4.20). According to the Danish Staten’s Serum Institute, BA.2 is approximately 1.5 and 4.2 times more contagious than BA.1 and the delta variant[9,10]. Yu et al[11] observed that the neutralizing antibody titer against BA.1 and BA.2 is 23-fold and 27-fold lower than that of WA1/2020. According to their research, the mean neutralizing antibody titers after the third booster of the BNT162b2 mRNA vaccine were approximately 1.4-fold lower than BA.1, indicating the capacity of BA.2 to confer neutralizing antibodies and evade humoral immunity[11].
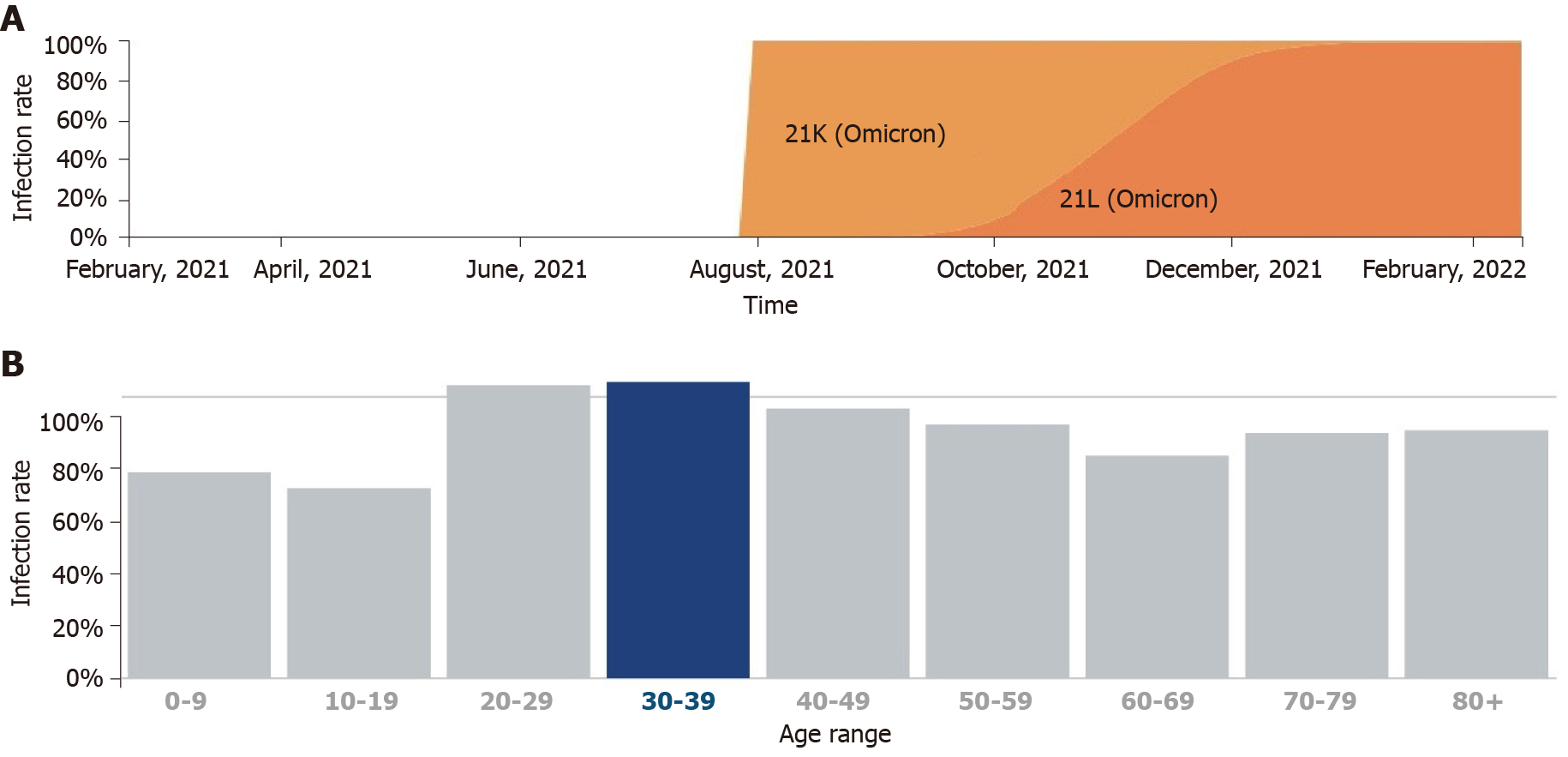
Chen and Wei[10] hypothesized that BA.2 mutations caused the ability of the immune evasion to be approximately 30% and 17-fold greater than that of BA.1 and the delta variants, respectively, and resistant to most monoclonal antibodies except for sotrovimab. Evidently, BA.2 will quickly become the next dominant global variant. In addition, the United Kingdom Health Security Agency (UKHSA) cautioned that contact tracing data from the United Kingdom estimates that BA.2 is more likely to infect household contacts than BA.1 (10.3%). UKHSA estimated that the increase in the number of BA.2 patients after a third booster dose vaccination was more significant than that of BA.1-infected population (63% for BA.1 vs 70% for BA.2)[12]. According to Covglobe data, the incidence of BA.2 infections has been on the rise since October 2021 and has recently supplanted the VOC B.1.1.529; according to existing databases, BA.2 typically affects younger age groups (Figure 3).
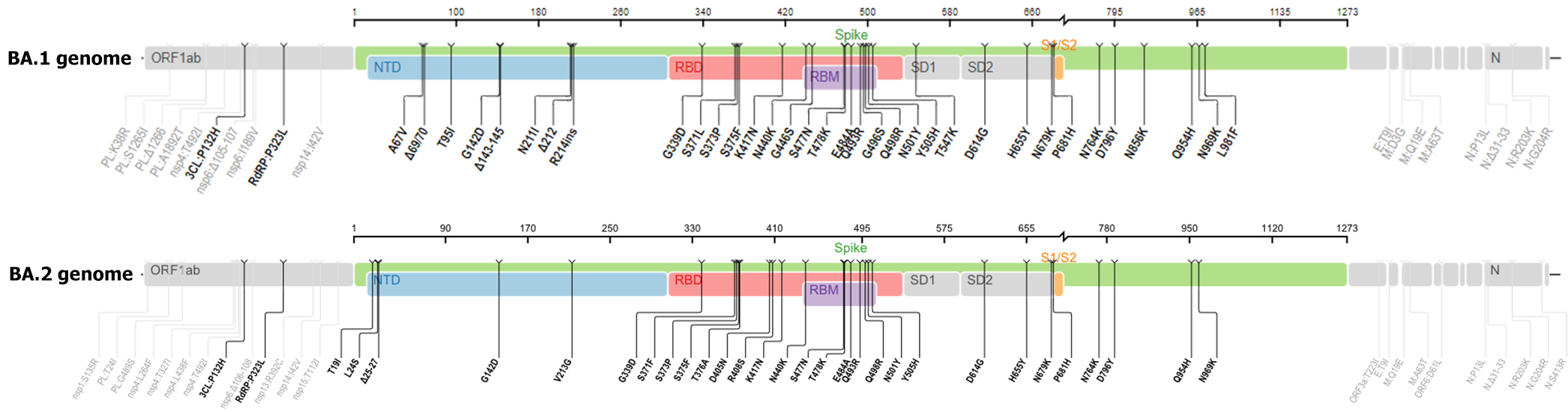
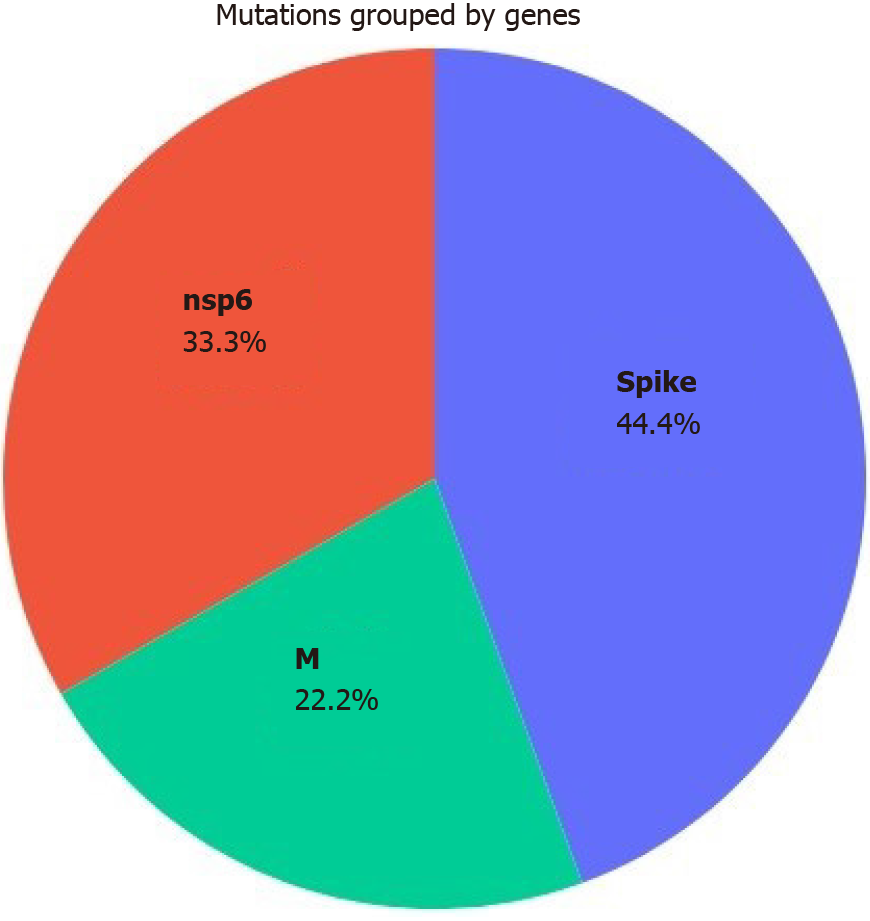
According to Nextstrain online server data, the omicron variant comprises three distinct branches: 21K (or Pangolin lineage12 BA.1), 21L (or BA.2), and BA.3 of the 21M omicron clade[4] (Figure 4). The BA.2 genome contains 20 shared and 6 unique mutations in the S protein compared to B.1.1.529 (archetypical) variant. BA.2 has significantly more mutations than BA1; most of these mutations are non-synonymous. By analyzing and comparing the genomes of BA.1 and BA.2, Wiegand et al[13] observed that BA.2 possesses the most genetic variation in the S protein (24-35 mutations, mean = 30.7) and that the effect of these mutations on virulence, viral transmission, and immune evasion has been identified in previous research. After the S protein, the nucleoprotein has the most genetic changes, but the majority of these changes are non-synonymous, affecting the sensitivity and specificity of diagnostic methods[13]. According to the CoronaTrend online server, the S, nsp6, and M proteins exhibited the most BA.2 mutations (Figure 5).
Kumar et al[4] demonstrated in a recent study that multiple alignments of four omicron subvariants revealed that BA.1 comprised 39 mutations, BA.1.1 comprised 40 mutations, BA.2 comprised 31 mutations, and BA.3 comprised 34 mutations. BA.1.1 has a single unique mutation of R346K, and BA.2 has eight mutations. Only one unique mutation exists in T19I, L24del (deletion), P25del, P26del, A27S, V213G, T376A, R408S, and BA.3 (R216del). Meanwhile, all subvariants comprise eleven shared mutations in their second receptor binding domain, including G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, and N501Y. By increasing the positive electrostatic surface, these mutations improve the interaction between the receptor binding domain motif and human angiotensin-converting enzyme 2[4]. They also observed that R400, R490, and R495 mutations in BA.2 formed new salt bridges and hydrogen, resulting in higher viral transmission than the BA.1 and BA.1.1 subvariants[4].
Desingu and Nagarajan[14] deduced that the BA.2 subvariant consisted of five distinct phylogenetically based original geographic regions, namely Sweden/Denmark, Philippines, Hong Kong, India, and China. They demonstrated that each of these clades exhibited unique mutations, such as the H78Y mutation in Denmark, the substitutions of ORF1a: A2909V and ORF3a: L140F in isolates from the Philippines and Hong Kong, and ORF1a: -3677L, ORF1b: S959P, and ORF7b: H42Y in the Indian subgroup. Each distinct mutation in the subpopulations modified the characteristics of BA.2 in terms of viral transmission, infectivity, disease severity, vaccine efficacy, and clinical outcomes in different geographic regions[14,15]. Recent studies suggest that mutations of P681H, H655Y, and N679K at the furin cleavage site increase the replication of omicron variants and as a result the transmissibility of omicron subvariants[4].
The 21L/BA.2 omicron variant has become predominant even after the third dose of the Pfizer-BioNTech vaccine, according to reports from Denmark (www.news-medical.net/news/20220214/First-survey-on-Omicron-BA2-in-France.aspx). Moreover, on January 26, 2022, Tyra Grove Krause stated, “There is some evidence that it is more contagious, particularly among the unvaccinated, but it can also infect vaccinated individuals to a greater extent” (https://www.gavi.org/vaccineswork/stealth-omicron-everything-you-need-know-about-new-ba2-subvariant-coronavirus).
Lyngse et al[9] showed that the viral load of unvaccinated individuals is significantly greater than that of fully immunized populations. Thus, non-immunized individuals can more effectively release BA.2. Initial UKHSA surveys indicated that the effectiveness of the BA.2 vaccine against symptomatic BA.2 infection was greater than that of the BA.1 vaccine (13% for BA.2 vs 9% for BA.1); additionally, a third booster dose may increase the effectiveness of the BA.2 vaccine (70% for BA.2 vs 63% for BA.1)[12,16]. However, Peiris et al[17] evaluated the effect of the third dose of BNT162b2 or CoronaVac vaccines against BA.2; they concluded that three doses with BNT162b2 or vaccination with two doses of CoronaVac and a third booster dose with BNT162b2 increased plaque reduction neutralization antibody titer above the threshold for protection against symptomatic BA.2 infection.
Although BA.2 mutations confer resistance to neutralizing antibodies, recent research shows that a third booster causes antibodies to cross-react with the omicron variant[18]. In addition, Lippi et al[19] revealed that the third booster vaccination increases neutralizing antibodies against the omicron variant and is a safe strategy until a new, more effective vaccine is introduced.
Despite the recent increase in BA.2 cases, the WHO has not yet given BA.2 a special designation. However, on January 21, 2022 the UKHSA designated BA.2 as a “variant under investigation” (https://www.gavi.org/vaccineswork/stealth-omicron-everything-you-need-know-about-new-ba2-subvariant-coronavirus). The WHO official Dr. Maria Van Kerkhove stated that BA.2 is significantly more contagious than BA.1 (https://www.cnbc.com/2022/02/08/who-says-omicron-bapoint2-subvariant-will-rise-globally.html); nonetheless, preliminary studies indicate that there is no difference between BA.2 and BA.1 in terms of hospitalization risk.
The remarkable increase in BA.2 infection cases and the rapidity with which it has spread in a short period of time is perplexing. In addition, intensive care unit admissions and mortality rates are rising, causing worldwide concern in healthcare facilities. According to the Infectious Diseases Society of America, the most effective treatments for SARS-CoV-2 B1.1.529 are monoclonal antibodies such as sotrovimab, evusheld, convalescent sera, and Oxford-AstraZeneca and Pfizer-BioNTech vaccines (https://www.idsociety.org/covid-19-real-time-learning-network/emerging-variants/emerging-covid-19-variants/#). Hand hygiene, physical distance, mask use, and mass vaccination, particularly a third booster dose, are recommended countermeasures to control the global spread of the BA.2 variant, as is the consideration of nationwide lockdowns.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Virology
Country/Territory of origin: Iran
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cazzato G, Italy S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Wang JJ
| 1. | National Institute for Communicable Diseases. Latest Confirmed Cases of COVID-19 in South Africa (2 December 2021). [cited 20 April 2022]. https://www.nicd.ac.za/Latest-confirmed-cases-of-covid-19-in-southafrica-2-december-2021/. |
| 2. | Rahimi F, Talebi Bezmin Abadi A. The Omicron subvariant BA.2: Birth of a new challenge during the COVID-19 pandemic. Int J Surg. 2022;99:106261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 3. | Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, Dushoff J, Mlisana K, Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376:eabn4947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 461] [Cited by in RCA: 521] [Article Influence: 173.7] [Reference Citation Analysis (0)] |
| 4. | Kumar S, Karuppanan K, Subramaniam G. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment. J Med Virol. 2022;94:4780-4791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 124] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 5. | Gao SJ, Guo H, Luo G. Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert! J Med Virol. 2022;94:1255-1256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 144] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 6. | Li K, Zheng Z, Zhao X, Zeng Q, Zhou T, Guo Q, Hu Y, Xu W, Zhang Z, Li B. An Imported Case and an Infected Close Contact of the Omicron Variant of SARS-CoV-2 - Guangdong Province, China, December 13, 2021. China CDC Wkly. 2022;4:96-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | Carrazco-Montalvo A, Armendáriz-Castillo I, Tello CL, Morales D, Armas-Gonzalez R, Guizado-Herrera D, León-Sosa A, Ramos-Sarmiento D, Fuertes B; USFQ-Consortium, Patino L. First detection of SARS-CoV-2 variant B.1.1.529 (Omicron) in Ecuador. New Microbes New Infect. 2022;45:100951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Leeman D, Campos-Matos I, Dabrera G. Inequalities associated with emergence of Delta SARS-CoV-2 variant of concern (B.1.617.2) in England: awareness for future variants. Public Health. 2022;205:e14-e15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 9. | Lyngse FP, Kirkeby CT, Denwood M, Christiansen LE, Mølbak K, Møller CH, Skov RL, Krause TG, Rasmussen M, Sieber RN, Johannesen TB, Lillebaek T, Fonager J, Fomsgaard A, Møller FT, Stegger M, Overvad M, Spiess K, Mortensen LH. Transmission of SARS-CoV-2 Omicron VOC subvariants BA. 1 and BA. 2: Evidence from Danish Households. 2022 Preprint. Available from: medRxiv:2022.01.28.22270044. [RCA] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 92] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 10. | Chen J, Wei GW. Omicron BA.2 (B.1.1.529.2): high potential to becoming the next dominating variant. ArXiv. 2022;. [PubMed] |
| 11. | Yu J, Collier AY, Rowe M, Mardas F, Ventura JD, Wan H, Miller J, Powers O, Chung B, Siamatu M, Hachmann NP, Surve N, Nampanya F, Chandrashekar A, Barouch DH. Comparable Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 Variants. N Engl J Med. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 12. | Mahase E. Omicron sub-lineage BA.2 may have "substantial growth advantage," UKHSA reports. BMJ. 2022;376:o263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 13. | Wiegand TR, McVey A, Nemudraia A, Nemudryi A, Wiedenheft B. The rise and fall of SARS-CoV-2 variants and the emergence of competing Omicron lineages. 2022 Preprint. Available from: bioRxiv:2022.02.09.479842. [DOI] [Full Text] |
| 14. | Desingu PA, Nagarajan K. Omicron BA.2 Lineage spreads in clusters and is concentrated in Denmark. J Med Virol. 2022;94:2360-2364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 15. | Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol. 2022;94:1808-1810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 107] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 16. | Mahase E. Covid-19: What do we know about omicron sublineages? BMJ. 2022;376:o358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 17. | Peiris M, Cheng S, Chan K, Luk L, Li J, Tsang L, Poon L, Mok CKP, Ng SS, Ko F, Chen C, Yiu K, Hui D, Lam B. Virus neutralization of SARS-CoV-2 Omicron variant BA. 2 in those vaccinated with three doses of BNT162b2 or CoronaVac vaccines. 2022 Preprint. Available from: Research Square:2022.10.21203/rs.3.rs-1331606/v1. [DOI] [Full Text] |
| 18. | Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, Berrios C, Ofoman O, Chang CC, Hauser BM, Feldman J, Roederer AL, Gregory DJ, Poznansky MC, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457-466.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 832] [Cited by in RCA: 765] [Article Influence: 255.0] [Reference Citation Analysis (0)] |
| 19. | Lippi G, Mattiuzzi C, Henry BM. Neutralizing potency of COVID-19 vaccines against the SARS-CoV-2 Omicron (B.1.1.529) variant. J Med Virol. 2022;94:1799-1802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |