Published online Dec 24, 2017. doi: 10.5500/wjt.v7.i6.339
Peer-review started: July 14, 2017
First decision: August 7, 2017
Revised: August 22, 2017
Accepted: October 17, 2017
Article in press: October 17, 2017
Published online: December 24, 2017
Processing time: 163 Days and 5 Hours
Renal transplantation remains the best option for patients suffering from end stage renal disease (ESRD). Given the worldwide shortage of organs and growing population of patients with ESRD, those waitlisted for a transplant is ever expanding. Contemporary crossmatch methods and human leukocyte antigen (HLA) typing play a pivotal role in improving organ allocation and afford better matches to recipients. Understanding crossmatch as well as HLA typing for renal transplantation and applying it in clinical practice is the key step to achieve a successful outcome. Interpretation of crossmatch results can be quite challenging where clinicians have not had formal training in applied transplant immunology. This review aims to provide a worked example using a clinical vignette. Furthermore, each technique is discussed in detail with its pros and cons. The index case is that of a young male with ESRD secondary to Lupus nephritis. He is offered a deceased donor kidney with a 1-0-0 mismatch. His complement dependent cytotoxicity (CDC) crossmatch reported positive for B lymphocyte, but flow cytometry crossmatch (FCXM) was reported negative for both B and T lymphocytes. Luminex-SAB (single antigen bead) did not identify any donor specific antibodies (DSA). He never had a blood transfusion. The positive CDC-crossmatch result is not concordant with DSA status. These implausible results are due to underlying lupus erythematosus, leading to false-positive B-lymphocyte crossmatch as a result of binding immune complexes to Fc-receptors. False positive report of CDC crossmatch can be caused by the underlying autoimmune diseases such as lupus erythematosus, that may lead to inadvertent refusal of adequate kidney grafts. Detailed study of DSA by molecular technique would prevent wrong exclusion of such donors. Based on these investigations this patient is deemed to have “standard immunological risk” for renal transplantation.
Core tip: Understanding crossmatch for renal transplantation and applying it in clinical practice is the fundamental step to achieve a successful outcome. At times, interpreting an ambivalent report of crossmatch can be very challenging for clinicians since they have not been trained formally in applied transplant immunology. While there are several published reviews, this is presented as a worked example and is aimed to discuss immunological risk stratification by using an example of an index case.
- Citation: Althaf MM, El Kossi M, Jin JK, Sharma A, Halawa AM. Human leukocyte antigen typing and crossmatch: A comprehensive review. World J Transplant 2017; 7(6): 339-348
- URL: https://www.wjgnet.com/2220-3230/full/v7/i6/339.htm
- DOI: https://dx.doi.org/10.5500/wjt.v7.i6.339
Renal transplantation is the best option in suitable and fit patients who have end stage renal disease (ESRD) as a result of lupus nephritis. Several studies have shown that five- and ten-year allograft survival are similar to that of recipients with other causes of ESRD[1-3]. It is also worth noting that lupus nephritis can recur in the allograft. The risk of recurrence of clinically apparent disease in renal transplantation is between 2%-30% of cases[1,4,5]. Systemic Lupus Erythematosus and its complications predominantly occur in women. However, it is worth noting that clinical manifestations are slightly different in men who have poorer outcomes[6,7]. Understanding crossmatch for renal transplantation and applying it in clinical practice is the key step to achieve a successful outcome. At times, interpreting an ambivalent report of crossmatch can be very challenging for clinicians since they have not been trained formally in applied transplant immunology. The following review is aimed to discuss an immunological risk stratification by using an example of an index case.
We are presented with a 30-year-old male patient who had been on maintenance haemodialysis for five years. His primary disease was Systemic Lupus Erythematosus which was complicated with lupus nephritis which eventually progressed to end-stage renal disease. He was offered a kidney from a deceased donor with a 1-0-0 mismatch. His complement dependent cytotoxicity (CDC) crossmatch reported positive for B lymphocyte, and flow cytometry crossmatch (FCXM) was reported negative for both B and T lymphocytes. His Luminex-SAB did not identify any donor specific antibodies (DSA). He never had a blood transfusion.
HLA typing is a crucial step in renal transplantation, as recognition of foreign HLA by recipient T lymphocytes would trigger an immune response. T lymphocyte activation initiates a cascade of mediators that direct the immune system against the allograft[8]. HLA laboratories currently perform serologic as well as molecular typing methods.
In this approach, a tray containing sera with antibodies to a multitude of known HLA alleles is used. These are commercially available. For typing, recipient lymphocytes are introduced into the tray wells contacting sera, complement and dye. In tray wells where antibodies can bind to the antigens on the surface of lymphocytes; complement is activated. This results in complement pathways triggered resulting in cell death, ultimately allowing the dye to enter the cell. Tray wells with significant cell death are then identified under phase contrast microscopy. Through a process of comparison and elimination of positive wells the HLA type is assigned. The key benefit of serologic typing is that results are available in a short period. This is particularly important in deceased donor renal transplantation. Quick results mean less cold ischemia times. This method also offers the ability to differentiate HLA alleles that have identifiable DNA sequences with molecular typing but with no cell surface antigen expression. These alleles termed “null” HLA alleles are of less immunological significance[9]. The downside of this method is the lack of sera with antibody specificities that are capable of identifying the ever-growing number of HLA alleles[10]. The HLA-Cw, DQ, and DP antigen may have clinically significant effects on the outcomes of allografts. However, serologic assays are scarce for these loci. Furthermore, serologic methods do not readily detect differences in HLA protein small amino acids. These may be antigenic enough to trigger potent immunological responses[11,12]. With more advanced methods of typing currently available serological typing has fallen into disuse.
Sequence-specific primer polymerase chain reaction: In this approach extracted DNA from the subject is amplified in several wells. Each well has primers that are complementary to specific HLA alleles. In wells where DNA probes are complementary to the specific sequence of the HLA molecule, an amplification product is formed. This is then instilled into an agarose gel and undergoes electrophoresis where they appear as a band. HLA typing is then allocated by matching the primers of the amplification product to DNA sequences of several candidate alleles.
Sequence specific oligonucleotide probes: Amplified DNA is mixed with oligonucleotide probes that are complementary to specific segments of the DNA of different alleles. Unique HLA alleles are then identified using fluorescent tags. For a particular gene of interest, the precise order of nucleotides is determined through sequencing. HLA type is then assigned using available HLA allele sequences[10].
Direct DNA sequencing: This method determines the precise order of nucleotides in the gene of interest. Using published HLA allele sequences, HLA type is subsequently assigned by comparison.
Molecular typing regardless of the method can clearly identify differences in HLA antigen between donor and recipient. Often with detail to the amino acid level that can provide insight to the risk accompanying mismatched donor-recipient antigens, epitopes and amino acid[12,13]. HLA typing based on polymerase chain reaction (PCR) is highly specific where specific alleles are identified with no cross-reactivity. However, a gene may occur in two or more forms called alleles. Cross-reactivity is the identification of an allele which is essentially similar to the allele of interest. While this feature is a key advantage of this method its acts as a double-edged sword. The disadvantage it poses is that new alleles not currently on the HLA sequence databank will fail to be identified. Primers used in HLA-typing are constructed on an HLA sequence databank that contains alleles available when the databank was designed[14].
HLA typing of the donor kidney and our patient revealed a 1-0-0-mismatch that corresponds to the pair of alleles mismatched, respectively, at HLA-A, HLA-B and HLA-DR. These three antigens are the considered as the most important ones in kidney transplantation. Logically the fewer the mismatches; the better the match between donor and recipient resulting in a successful transplant outcome. The dissimilarity in the HLA antigen reflects the alloimmune burden that a donor kidney presents to the recipient. In this case, there is 1 HLA mismatch which is that of HLA-A. Mismatch for different HLA antigens does not have equal weight. We know from the initial Collaborative Transplant Study (CTS) analysis that HLA-DR and HLA-B antigens offer the most alloimmune burden with less so from HLA-A[15]. Eurotransplant and old United Kingdom transplant data suggest that HLA-DR matching has a far greater effect than HLA-A or HLA-B[16,17]. Interestingly one study demonstrated that the influence of HLA-DR mismatching had the most effect during the first six months post-transplant while the maximal effect of HLA-B mismatching occurred two years post-transplant[18]. Data from the United Network for Organ Sharing (UNOS) registry further highlighted the significance of paying attention to having the least number of mismatches. They looked at quantifying the risk of transplant failure with HLA mismatch in patients who had their first adult kidney allografts from deceased donors. This study revealed that having six HLA mismatches translated to a 64% higher risk while the risk was down to 13% with just one HLA mismatch. Furthermore, these results were independent of locus[19]. Another study identified seven specific HLA mismatch combinations that were associated with decreased renal allograft survival. These were termed “taboo mismatches”. A taboo mismatch translated to 81% one-year survival and 50% five-year survival[20].
In recent times, the HLA mismatching in deceased donor kidney transplants is of lesser significance due to the use of more potent immunosuppression and better identification of non-immunological determinants of transplantation[21]. Nonetheless, HLA matching continues to have a significant impact on allograft survival.
Almost a third of patients who are waitlisted for transplantation may have a degree of anti-HLA antibodies detected. The usual route for sensitisation towards HLA antigens occurs in three instances; pregnancy, post blood transfusion and prior transplantation. Preformed antibodies increase the chances of immunological failure of the allograft by causing positive crossmatches and, thereby, result in the exclusion of donors[9]. The index patient did not have a history of prior transplantation or blood transfusions. Both sensitive and specific detection of anti-HLA antibodies is crucial. Where crossmatch is negative, even low titres of DSA can lead to early as well as late antibody mediated rejection[22,23]. For sensitised patients, successful transplantation is possible by employing strategies such as desensitisation, paired exchange and acceptable mismatching[13,24,25]. There are different methods used for HLA antibody screening as shown below.
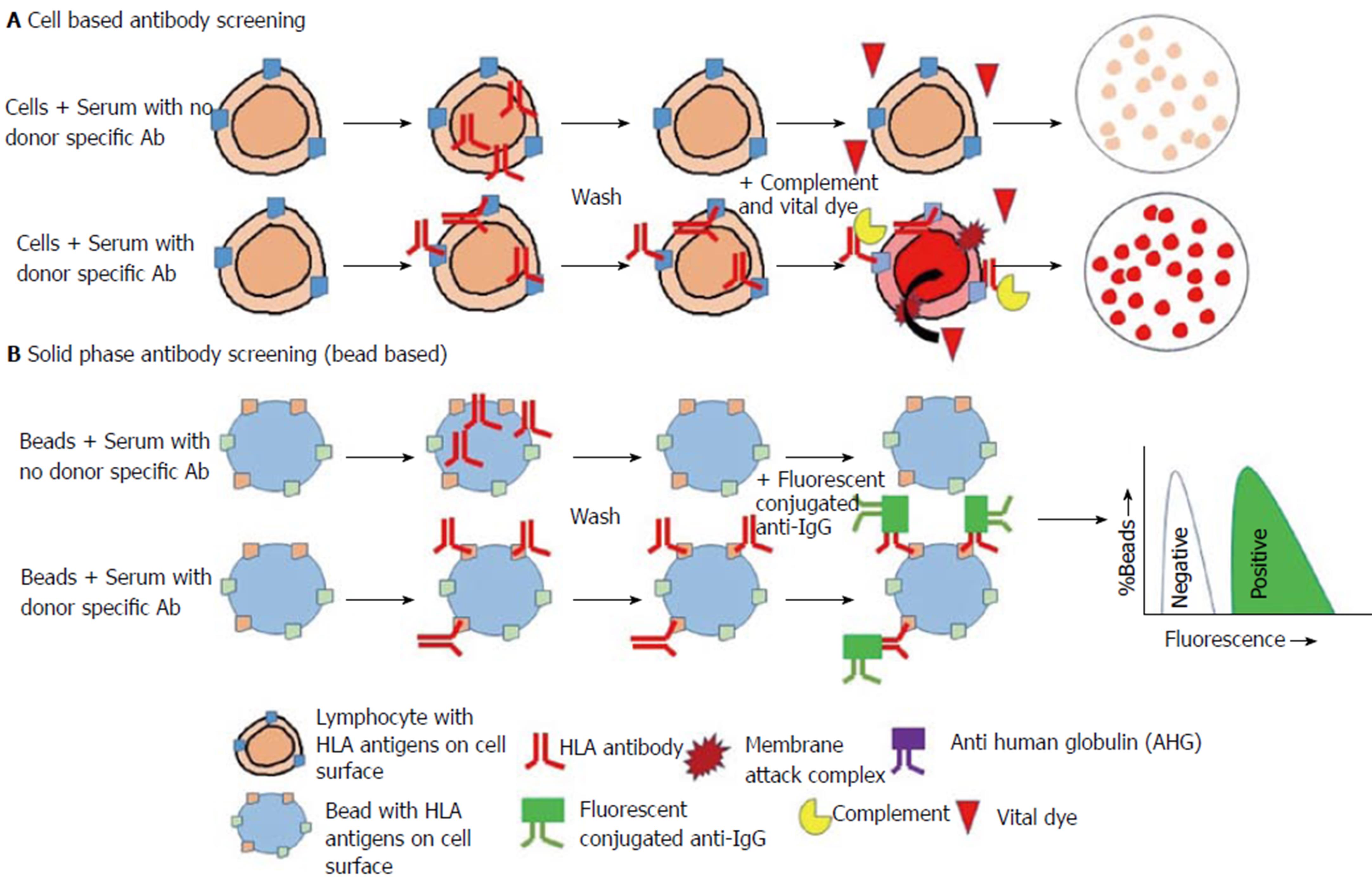
A set of cell donors are randomly selected to be representative of a population. This should be representative of the population of potential deceased donors. Each panel consists of around 30 to 40 different donor lymphocytes. The method is similar to that of serologic typing however here recipient serum is mixed with “cell donor” lymphocytes in individual wells along with complement and dye. Where the serum contains antibodies that bind to the cell surface with adequate density complement pathways are activated which results in cell death and uptake of the dye (Figure 1A). The degree of cytotoxicity is expressed as percentage PRA (panel reactive antibody). It is a tool that can be employed to approximate the risk of a given recipient of having a positive crossmatch. This is to a likely organ donor taken from a similar population.
The limitations of this method are that PRA percent can be different numerically without a corresponding change in the type or amount of antibody. This largely depends on the cell panel used which are commercially produced and may not truly represent the population. HLA frequencies and racial differences need to be factored in but cannot be done. Moreover, significant false positive results can be produced due to non-HLA antibodies, autoantibodies and nonspecific IgM antibodies. Similarly, false negative results are possible as this is purely complement dependent that requires higher antibody titres to be activated[26-28]. The lack of a complement activation simply due to low titres allows a true antibody to be hidden[29]. Precise, complete lists of antibody specificities and unacceptable antigens cannot be identified using this method as there are several antigens in each well[9].
This method employs soluble or recombinant HLA molecules instead of lymphocytes targets - as lymphocytes present both HLA as well as non-HLA molecules (Figure 1B). The variants of these methods are:
Enzyme-linked immunosorbent assay platform: In this method, purified HLA molecules are applied to enzyme-linked immunosorbent assay (ELISA) platforms and will bind individually to HLA antibody after the addition of recipient serum[30,31]. Enzyme conjugated antibodies to IgG (human) is then added to detect the presence of HLA antibody in the serum which is bound to the antigen. Detection is performed by optical density reading.
Microbead platform/single-antigen beads: Pooled panel beads with several different class I or II HLA antigens on a bead yield a positive or negative result and are utilised for screening[32]. The phenotype or also known as ID beads are individually coated with class I or II HLA antigens of an individual patient-derived cell line. Microbead that is fluorescent dye conjugated is then added to detect the presence of HLA antibody in the serum which is bound to the antigen. Fluorescence detection can be done traditionally using a flow cytometer (Flow PRA®) or via the single-antigen beads (SAB) Luminex® platform. These estimate PRA by the proportion of positive beads. SAB are individually coated with a single HLA antigen and yield a list of distinct antibody specificities[33]. Specificities are subsequently compared with HLA frequencies in the donor population to determine the calculated panel-reactive antibody (cPRA)[34]. This yields the best estimate of the likelihood of a positive crossmatch/donor specific antibody to a randomly selected donor[35,36].
It is important to understand the difference between PRA and cPRA. A high traditional PRA value translated to a high probability of a positive crossmatch. cPRA is based on unacceptable HLA antigens - those that the patient has been sensitized to. Furthermore, if these were present in a donor, would represent an unacceptable risk to the potential recipient or organ transplantation program. cPRA is calculated from HLA antigen frequencies among approximately twelve thousand kidney donors in the United States during the period between 2003 and 2005. This, therefore, represents the proportion of actual organ donors who express one or more of the unacceptable HLA antigens[36]. cPRA is useful in the allocation of kidney and pancreas transplants. cPRA estimates the proportion of donors with whom a particular recipient would be incompatible. An offer for a recipient with a high cPRA is a high probability of a positive crossmatch. Formerly, the same highly sensitised potential recipient would be higher on the list of each match run for donors with their blood group. Renal transplant programs were hesitant to set up final crossmatches for more highly sensitised patients for fear of not allocating the kidneys[37-39].
We are able to better discriminate immunologically relevant positive crossmatches from false-positive results when traditional cell based methods are complemented with solid phase assays[40]. Microbead assays (both Flow PRA® and Luminex®) are ten percent more sensitive for lower titre antibody than ELISA. ELISA is ten percent more sensitive compared to anti-human globulin (AHG) enhanced cytotoxicity based assays. Being in control of the antigens places on the beads, these assays are specific for anti-HLA antibodies. SAB assays are rapid with results available in 3-4 h. The assay is also quite efficient in a single reaction chamber up to one hundred unique antigen beads can be tested. Its additional multiplexing ability permits testing many patients simultaneously[41]. Results from SAB enable virtual crossmatching (VXM) to identify DSA pre-transplant, thereby enabling organ allocation and risk stratification[37]. SAB assays permit identification of anti-HLA antibodies for all common and numerous rare antigens and alleles. Its range of identification is up to eleven HLA loci[33].
Despite the fact that solid phase antibody screening addresses most of the short comings with cellular assays they have limitations as well. They detect both complement and non-complement binding simultaneously. Being too sensitive they can detect antibody that is below the threshold associated with a positive crossmatch. The detected antibody may not always have clinical implications but can preclude a potential donor. Non-HLA antibodies are also increasingly being recognised as clinically relevant predictors, and these cannot be accounted for utilising this method solely[42,43]. With the ever-growing list of HLA alleles, the complete spectrum of unique HLA antigens cannot be fully presented on solid phase assays.
The SAB - Luminex® assay has been shown to be susceptible to an artefact known as the prozone phenomenon[44]. This phenomenon is recognised when sera with high titer anti-HLA antibodies give negative results when tested neat, however, react strongly positive after 1:10 dilution[45,46]. The complement-mediated prozone effect is most likely caused by complement component 1 (C1) by competitively displacing the detection antibodies in the confined spaces between antibodies bound to HLA molecules. This in turn prevents HLA antibody binding to the HLA antigen on the bead. A similar scenario can arise with the binding of IgM antibodies or other serum factors to the beads. This can be resolved by treatment with dithiothreitol (DTT) and serum dilution. It is worth noting that nonspecific binding by serum proteins as well as drugs such as intravenous immunoglobulin (IVIG) could also interfere with the specific binding of anti-HLA antibodies to the HLA antigens on beads. Another cause for a false negative result is epitope sharing. Different HLA antigens on different beads share mutual antibody binding epitopes leading to the binding of an anti-HLA antibody to more than one bead. This leads to a reduction in the mean fluorescence intensity (MFI) on a single bead[41].
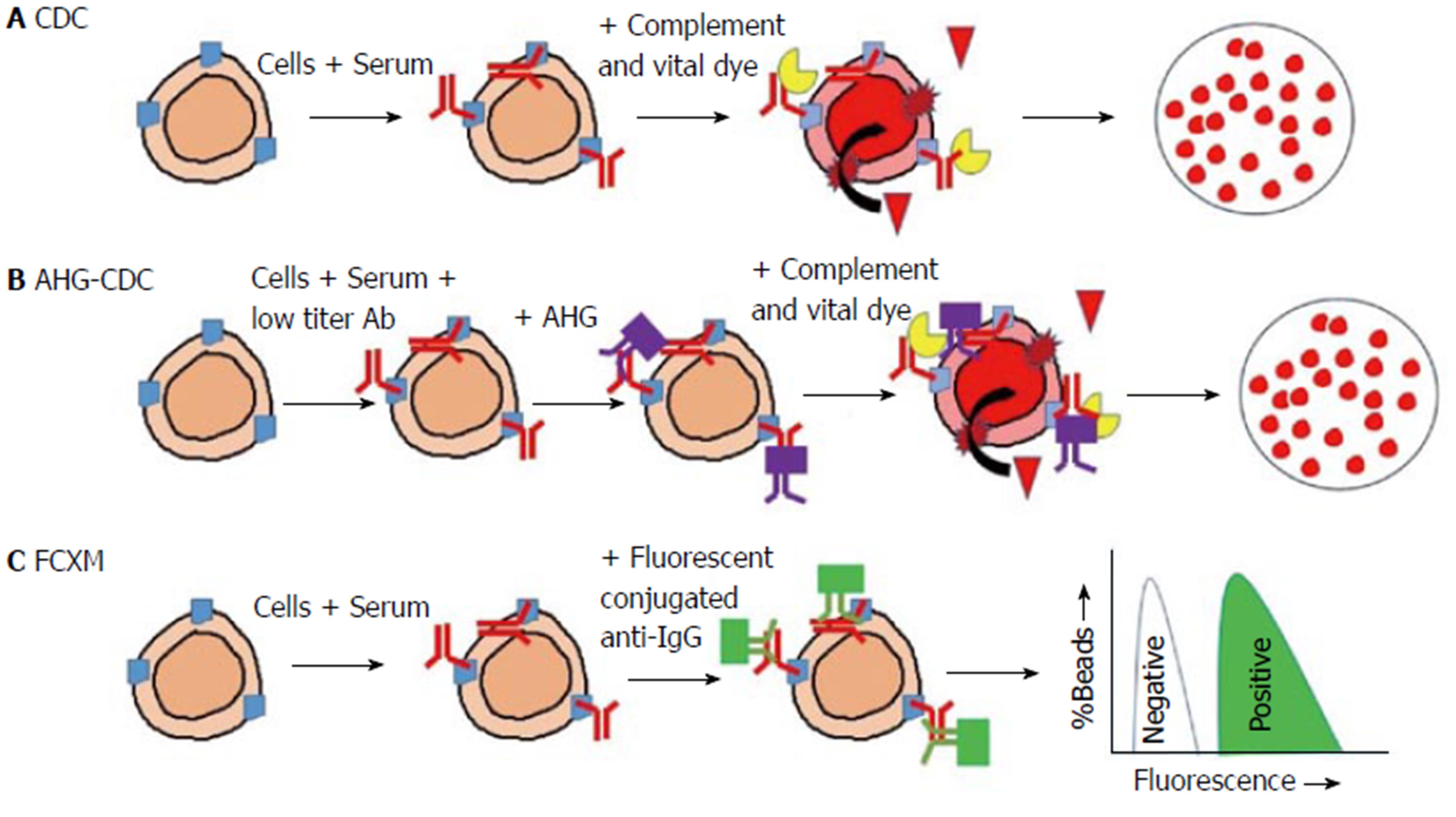
The cytotoxic assay was implemented as the requisite test prior to transplantation when it was shown that recipients with DSA had significantly higher rates of allograft failure due to hyperacute rejection as well as primary failure[47,48]. The presence of donor-specific cytotoxic antibodies depicted as a positive crossmatch was a contraindication to transplantation. With PRA that identifies several antibodies to a potential cluster of donors, the crossmatch will identify if a recipient had antibodies to a specific donor of interest. Despite the obvious benefits of testing the T cell cytotoxic crossmatch had a twenty percent false positive rate and a four percent false negative rate. Therefore, it is insufficient to identify all relevant antibodies, and in addition to that, it may needlessly exclude patients from transplant. The solid-phase antibody test should be used together with crossmatch results to identify those that are immunologically relevant[49].
Similar to cytotoxic assay the complement-dependent cytotoxicity crossmatch is interpreted as positive if a considerable number of lymphocytes are destroyed after the incorporation of complement (Figure 2A). This suggests that a significant DSA has been bound to the cell surface. Complement-dependent cytotoxicity crossmatch (CDC-XM) can be done for B and T lymphocytes. Sensitivity is limited if the relevant antibody is in low titres, but this can be overcome by increasing the incubation time, the use of the AHG-enhanced method as well as additional wash steps[26,27]. The complement fixing antibody to anti-human immunoglobulin (AHG) will bind to any DSA present on lymphocytes (Figure 2B). This increases the chances of activating complement and thus raises the sensitivity of the test.
The antibodies that are present in lower titres are clinically significant as a negative test has an 18% graft loss in 1 year compared to a positive test that is associated with 36%[28]. Similar to cytotoxic PRA this method could miss low titre antibody resulting in false negatives. CDC-XM can also give false positives by detecting autoantibody, IgM/IgG HLA or non-HLA.
Flow cytometry crossmatch (FCXM) detects DSA independent of complement fixation. It precisely detects the presence or lack of IgG DSA on donor lymphocytes. In this method, recipient serum is mixed with donor lymphocytes and then tagged with a fluorochrome-conjugated anti-IgG antibody. Several antibodies with separate fluorochromes particular to B and T lymphocyte surface proteins can be added (Figure 2C). With the use of flow-cytometry, B and T lymphocytes can be readily identified and have their DSA individually interrogated. Compared to complement-dependent cytotoxicity crossmatch this offers greater sensitivity[9]. Different laboratories use different methods, and this can result in a difference in the results between them[50]. However, this approach is not widely available, and its role in assessing immunological risk is still unclear.
In virtual crossmatch (VXM), both donor HLA typing and solid phase antibody screening are utilised together. It is not precisely a crossmatch in the sense of mixing serum and lymphocytes. The data is used to forecast the actual in vitro crossmatch results by “mixing” identified antibody specificities of recipient serum with donor HLA antigens[9]. The use of VXM can lead to shorter wait times and improved outcomes for sensitised transplant recipients. The speed of results generated allows a VXM to be performed at the time of donor identification owing to the fact that there is progressively sensitive and specific flow cytometry technology. VXM permits transplant physicians to consider donor organs that would not otherwise be available by means of a prospective crossmatch strategy, and thereby, allows to consider a potentially positive crossmatch a risk factor for donor selection[51,52].
Titres, specificities, and presence or absence of antibodies could significantly vary over time. Thus, the use of antibody specificity from historical serum sample (earlier than six months) could not predict a crossmatch with certainty. Other factors that can influence antibody specificities should be considered, and these include pregnancies, transplants and blood transfusions. The VXM should, therefore, be done considering all available serum results including at least one recent within less than 3-6 mo for a given patient. False positive results of VXM may arise where there are significantly low titre and/or non-complement binding antibodies, thereby, resulting in the wrong exclusion of potential donors[9]. The VXM can also give false negative results due to the fact that the list of all potential HLA donor antigens have been classed differently and, therefore, can not be correctly represented[53]. The results from VXM are not a hundred percent accurate and current practice mandates an actual crossmatch be performed as well[37]. Furthermore, VXM does not identify the HLA “Null” alleles. Null HLA alleles are ones have identifiable DNA sequences with molecular typing but do not express HLA products on the cell surface. In excess of 190 null alleles have been identified across HLA class I and II. There is a significant risk where a null allele is misidentified for its fully expressed counterpart in stem cell transplantation. However, the risk is slightly lower in solid organ transplantation. A recipient will have the risk of developing DSA for the mismatch where the null allele is misidentified as a fully expressed product and, therefore, transplanted with a donor bearing the expressed antigen. This mismatch is not life threatening but can affect future transplantation. In contrast, where a donor null allele is misidentified as a fully expressed product and subsequently transplanted into a recipient bearing the expressed antigen results in no humoral rejection and is well tolerated[54].
Gebel et al[49] stratified the prospective renal transplant patients into various categories according to immunological risk in renal transplantations. On the basis of this with further additions the principles of risk assessment are as follows:
At the time of transplantation, there are high titres of circulating antibodies specific for mismatched donor HLA (DSA). This can lead to hyperacute rejection. The presence of DSA precludes transplantation. However, there are reports of innovative pre-transplant desensitisation regimens to reduce this risk.
At the time of transplantation, there is a low titer of DSA, and historic DSA is not detectable. It may be acceptable to consider intensified immunosuppression as well as immunological monitoring in the post-transplant period.
Where there is no evidence of donor directed sensitisation to HLA. Refer to Table 1 that gives a summary of the immunological risk assessment pre-transplant based on donor crossmatch and antibody screening outcomes[55].
| Donor crossmatch result | Crossmatchmethod | Current or historical | Antibody screening results | Interpretation ofimmunological risk |
| Positive T and B lymphocyte | CDC (DTT) | C | IgG HLA class I DSA | High risk1 |
| Hyperacute rejection | ||||
| (veto to transplantation) | ||||
| Positive B lymphocyte | CDC (DTT) | C | IgG HLA class II DSA | High risk1 |
| Positive B lymphocyte | CDC (DTT) | C | Weak IgG HLA class I DSA | Intermediate risk2 |
| Positive T and B lymphocyte | FCXM (CDC neg) | C | IgG HLA class I DSA | Intermediate risk2 |
| Positive B lymphocyte | FCXM (CDC neg) | C | IgG HLA class II DSA | Intermediate risk2 |
| Positive T and B lymphocyte | CDC (DTT) | H | IgG HLA class I DSA | High risk3 |
| Positive B cell | CDC (DTT) | H | IgG HLA class II DSA | High risk3 |
| Positive B lymphocyte | CDC (DTT) | H | Weak IgG HLA class I DSA | Intermediate risk2 |
| Positive T and B lymphocyte | FCXM (CDC neg) | H | IgG HLA class I DSA | Intermediate risk2 |
| Positive B lymphocyte | FCXM (CDC neg) | H | IgG HLA class II DSA | Intermediate risk2 |
| Positive T and B lymphocyte | CDC (neg DTT) | C or H | IgM HLA class I DSA | Standard risk |
| Positive B lymphocyte | CDC (neg DTT) | C or H | IgM HLA class II DSA | Standard risk |
| Positive T and B lymphocyte | CDC (neg DTT) | C or H | IgM non-HLA (often autoreactive) | Standard risk |
| Positive B lymphocyte | CDC (neg DTT) | C or H | IgM non-HLA (often autoreactive) | Standard risk |
| Negative T and B lymphocyte | FCXM | C or H | IgG HLA class I or II DSA (detected by Luminex SAB alone) | Standard risk |
| Positive T and/or B lymphocyte | CDC and/or FCXM | C or H | Negative (Luminex Ab detection and/or SAB) | Standard risk (IgM/IgG non-HLA, often showing in vitro autoreactivity) |
| Positive T; Nnegative B lymphocyte | CDC and/or FCXM | C or H | Positive (Luminex SAB-not donor-specific) or negative | Standard risk (results suggest antibody is not HLA-specific) |
| Negative T and B lymphocyte | FCXM | C or H | Positive (Luminex SAB) not donor HLA-specific | Standard risk |
| Negative T and B lymphocyte | CDC and/or FCXM | C or H | Negative (Luminex Ab detection and/or SAB) | Standard risk |
| Donor crossmatch result | Crossmatch method | Current or historical | Antibody screening results | Interpretation of immunological risk |
| Positive T and B lymphocyte | CDC (DTT) | C | IgG HLA class I DSA | High risk1 |
| Hyperacute rejection | ||||
| (veto to transplantation) | ||||
| Positive B lymphocyte | CDC (DTT) | C | IgG HLA class II DSA | High risk1 |
| Positive B lymphocyte | CDC (DTT) | C | Weak IgG HLA class I DSA | Intermediate risk2 |
| Positive T and B lymphocyte | FCXM (CDC neg) | C | IgG HLA class I DSA | Intermediate risk2 |
| Positive B lymphocyte | FCXM (CDC neg) | C | IgG HLA class II DSA | Intermediate risk2 |
| Positive T and B lymphocyte | CDC (DTT) | H | IgG HLA class I DSA | High risk3 |
| Positive B lymphocyte | CDC (DTT) | H | IgG HLA class II DSA | High risk3 |
| Positive B lymphocyte | CDC (DTT) | H | Weak IgG HLA class I DSA | Intermediate risk2 |
| Positive T and B lymphocyte | FCXM (CDC neg) | H | IgG HLA class I DSA | Intermediate risk2 |
| Positive B lymphocyte | FCXM (CDC neg) | H | IgG HLA class II DSA | Intermediate risk2 |
| Positive T and B lymphocyte | CDC (neg DTT) | C or H | IgM HLA class I DSA | Standard risk |
| Positive B lymphocyte | CDC (neg DTT) | C or H | IgM HLA class II DSA | Standard risk |
| Positive T and B lymphocyte | CDC (neg DTT) | C or H | IgM non-HLA (often autoreactive) | Standard risk |
| Positive B lymphocyte | CDC (neg DTT) | C or H | IgM non-HLA (often autoreactive) | Standard risk |
| Negative T and B lymphocyte | FCXM | C or H | IgG HLA class I or II DSA (detected by Luminex SAB alone) | Standard risk |
| Positive T and/or B lymphocyte | CDC and/or FCXM | C or H | Negative (Luminex Ab detection and/or SAB) | Standard risk (IgM/IgG non-HLA, often showing in vitro autoreactivity) |
| Positive T; negative B lymphocyte | CDC and/or FCXM | C or H | Positive (Luminex SAB-not donor-specific) or negative | Standard risk (results suggest antibody is not HLA-specific) |
| Negative T and B lymphocyte | FCXM | C or H | Positive (Luminex SAB) not donor HLA-specific | Standard risk |
| Negative T and B lymphocyte | CDC and/or FCXM | C or H | Negative (Luminex Ab detection and/or SAB) | Standard risk |
Our patient had CDC-XM reported positive for B and T lymphocytes but FCXM was reported negative for both B and T lymphocytes. His Luminex-SAB did not identify any DSA. These results can be risk stratified as “standard immunological risk”, and we can proceed with transplantation. Positive CDC-XM result is not in accordance with DSA status. These implausible results are due to underlying lupus erythematosus, leading to false-positive B- lymphocyte crossmatches as a result of binding immune complexes to Fc-receptors.
Interpretation and clinical application of transplant immunology are crucial steps to a successful outcome. Understanding of crossmatch results and the caveats of individual tests can be quite challenging where clinicians have not had formal training in applied transplant immunology. This case illustrated a common scenario and detailed the approach to testing and its interpretation. If we were to rely simply on the CDC-XM, we would have made an erroneous conclusion. It is crucial to realise that false positive report of CDC-XM can be due to autoimmune diseases where type III hypersensitivity occurs such as in Systemic Lupus Erythematosus. The false-positive B-lymphocyte crossmatch result from immune complexes binding to Fc-receptors[56,57]. Such a result may lead to inadvertent refusal of adequate kidney grafts. It has been previously reported that false positive CDC-XM could also be a result of medications such as Isoniazid and Hydralazine[58,59]. Detailed study of DSA by molecular technique would prevent erroneous exclusion of such donors. This can eventually lead to improved organ allocation and shorter waiting times in transplant lists.
Manuscript source: Unsolicited manuscript
Specialty type: Transplantation
Country of origin: United Kingdom
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Nagata T, Sanchez-Zapardiel E S- Editor: Ji FF L- Editor: A E- Editor: Yan JL
| 1. | Mojcik CF, Klippel JH. End-stage renal disease and systemic lupus erythematosus. Am J Med. 1996;101:100-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 96] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 2. | Nossent HC, Swaak TJ, Berden JH. Systemic lupus erythematosus after renal transplantation: patient and graft survival and disease activity. The Dutch Working Party on Systemic Lupus Erythematosus. Ann Intern Med. 1991;114:183-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Ponticelli C, Moroni G. Renal transplantation in lupus nephritis. Lupus. 2005;14:95-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Contreras G, Mattiazzi A, Guerra G, Ortega LM, Tozman EC, Li H, Tamariz L, Carvalho C, Kupin W, Ladino M. Recurrence of lupus nephritis after kidney transplantation. J Am Soc Nephrol. 2010;21:1200-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 5. | Stone JH, Amend WJ, Criswell LA. Outcome of renal transplantation in systemic lupus erythematosus. Semin Arthritis Rheum. 1997;27:17-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 46] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 6. | Costenbader KH, Feskanich D, Stampfer MJ, Karlson EW. Reproductive and menopausal factors and risk of systemic lupus erythematosus in women. Arthritis Rheum. 2007;56:1251-1262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 224] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 7. | Gilbert EL, Ryan MJ. Estrogen in cardiovascular disease during systemic lupus erythematosus. Clin Ther. 2014;36:1901-1912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | von Andrian UH, Mackay CR. T-cell function and migration. Two sides of the same coin. N Engl J Med. 2000;343:1020-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1101] [Cited by in RCA: 1080] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 9. | Tinckam KJ. Basic histocompatibility testing methods. Core concepts in renal transplantation. New York: Springer Science + Business Media, LLC 2012; 21-42. |
| 10. | Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 681] [Cited by in RCA: 671] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 11. | Claas FH, Witvliet MD, Duquesnoy RJ, Persijn GG, Doxiadis II. The acceptable mismatch program as a fast tool for highly sensitized patients awaiting a cadaveric kidney transplantation: short waiting time and excellent graft outcome. Transplantation. 2004;78:190-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 156] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 12. | Duquesnoy RJ, Claas FH. Is the application of HLAMatchmaker relevant in kidney transplantation? Transplantation. 2005;79:250-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Claas FH, Roelen DL, Oudshoorn M, Doxiadis II. Future HLA matching strategies in clinical transplantation. Dev Ophthalmol. 2003;36:62-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 14. | Gyllensten U, Allen M. PCR-based HLA class II typing. PCR Methods Appl. 1991;1:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 15. | Opelz G. Correlation of HLA matching with kidney graft survival in patients with or without cyclosporine treatment. Transplantation. 1985;40:240-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 100] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Doxiadis II, de Fijter JW, Mallat MJ, Haasnoot GW, Ringers J, Persijn GG, Claas FH. Simpler and equitable allocation of kidneys from postmortem donors primarily based on full HLA-DR compatibility. Transplantation. 2007;83:1207-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Gilks WR, Bradley BA, Gore SM, Klouda PT. Substantial benefits of tissue matching in renal transplantation. Transplantation. 1987;43:669-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Danovitch GM, Nast C. Dialysis and Transplantation. Owen WF, Pereira BJ, Sayegh MH, ed. Philadelphia: W.B.Saunders 2000; 504. |
| 19. | Williams RC, Opelz G, McGarvey CJ, Weil EJ, Chakkera HA. The Risk of Transplant Failure With HLA Mismatch in First Adult Kidney Allografts From Deceased Donors. Transplantation. 2016;100:1094-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 135] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 20. | Doxiadis II, Smits JM, Schreuder GM, Persijn GG, van Houwelingen HC, van Rood JJ, Claas FH. Association between specific HLA combinations and probability of kidney allograft loss: the taboo concept. Lancet. 1996;348:850-853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 86] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Su X, Zenios SA, Chakkera H, Milford EL, Chertow GM. Diminishing significance of HLA matching in kidney transplantation. Am J Transplant. 2004;4:1501-1508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 109] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 22. | van den Berg-Loonen EM, Billen EV, Voorter CE, van Heurn LW, Claas FH, van Hooff JP, Christiaans MH. Clinical relevance of pretransplant donor-directed antibodies detected by single antigen beads in highly sensitized renal transplant patients. Transplantation. 2008;85:1086-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 115] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 23. | Gupta A, Iveson V, Varagunam M, Bodger S, Sinnott P, Thuraisingham RC. Pretransplant donor-specific antibodies in cytotoxic negative crossmatch kidney transplants: are they relevant? Transplantation. 2008;85:1200-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 24. | Stegall MD, Gloor J, Winters JL, Moore SB, Degoey S. A comparison of plasmapheresis versus high-dose IVIG desensitization in renal allograft recipients with high levels of donor specific alloantibody. Am J Transplant. 2006;6:346-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 322] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 25. | Gentry SE, Segev DL, Simmerling M, Montgomery RA. Expanding kidney paired donation through participation by compatible pairs. Am J Transplant. 2007;7:2361-2370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 26. | Amos DB, Cohen I, Klein WJ Jr. Mechanisms of immunologic enhancement. Transplant Proc. 1970;2:68-75. [PubMed] |
| 27. | Fuller TC, Fuller AA, Golden M, Rodey GE. HLA alloantibodies and the mechanism of the antiglobulin-augmented lymphocytotoxicity procedure. Hum Immunol. 1997;56:94-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 40] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Kerman RH, Kimball PM, Van Buren CT, Lewis RM, DeVera V, Baghdahsarian V, Heydari A, Kahan BD. AHG and DTE/AHG procedure identification of crossmatch-appropriate donor-recipient pairings that result in improved graft survival. Transplantation. 1991;51:316-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 48] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Gebel HM, Bray RA. Sensitization and sensitivity: defining the unsensitized patient. Transplantation. 2000;69:1370-1374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 157] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 30. | Zachary AA, Ratner LE, Graziani JA, Lucas DP, Delaney NL, Leffell MS. Characterization of HLA class I specific antibodies by ELISA using solubilized antigen targets: II. Clinical relevance. Hum Immunol. 2001;62:236-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Zachary AA, Delaney NL, Lucas DP, Leffell MS. Characterization of HLA class I specific antibodies by ELISA using solubilized antigen targets: I. Evaluation of the GTI QuikID assay and analysis of antibody patterns. Hum Immunol. 2001;62:228-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Pei R, Wang G, Tarsitani C, Rojo S, Chen T, Takemura S, Liu A, Lee J. Simultaneous HLA Class I and Class II antibodies screening with flow cytometry. Hum Immunol. 1998;59:313-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 175] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 33. | Pei R, Lee JH, Shih NJ, Chen M, Terasaki PI. Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation. 2003;75:43-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 34. | Zachary AA, Braun WE. Calculation of a predictive value for transplantation. Transplantation. 1985;39:316-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 35. | Cecka JM, Kucheryavaya AY, Reinsmoen NL, Leffell MS. Calculated PRA: initial results show benefits for sensitized patients and a reduction in positive crossmatches. Am J Transplant. 2011;11:719-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 130] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 36. | Cecka JM. Calculated PRA (CPRA): the new measure of sensitization for transplant candidates. Am J Transplant. 2010;10:26-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 165] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 37. | Bray RA, Nolen JD, Larsen C, Pearson T, Newell KA, Kokko K, Guasch A, Tso P, Mendel JB, Gebel HM. Transplanting the highly sensitized patient: The emory algorithm. Am J Transplant. 2006;6:2307-2315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 166] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 38. | Tambur AR, Leventhal J, Kaufman DB, Friedewald J, Miller J, Abecassis MM. Tailoring antibody testing and how to use it in the calculated panel reactive antibody era: the Northwestern University experience. Transplantation. 2008;86:1052-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 39. | Bingaman AW, Murphey CL, Palma-Vargas J, Wright F. A virtual crossmatch protocol significantly increases access of highly sensitized patients to deceased donor kidney transplantation. Transplantation. 2008;86:1864-1868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 40. | Bray RA, Nickerson PW, Kerman RH, Gebel HM. Evolution of HLA antibody detection: technology emulating biology. Immunol Res. 2004;29:41-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 87] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 41. | Konvalinka A, Tinckam K. Utility of HLA Antibody Testing in Kidney Transplantation. J Am Soc Nephrol. 2015;26:1489-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 147] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 42. | Sumitran-Holgersson S. Relevance of MICA and other non-HLA antibodies in clinical transplantation. Curr Opin Immunol. 2008;20:607-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 43. | Tait BD, Süsal C, Gebel HM, Nickerson PW, Zachary AA, Claas FH, Reed EF, Bray RA, Campbell P, Chapman JR. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation. 2013;95:19-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 620] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 44. | Weinstock C, Schnaidt M. The complement-mediated prozone effect in the Luminex single-antigen bead assay and its impact on HLA antibody determination in patient sera. Int J Immunogenet. 2013;40:171-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 45. | Lowe D. The high-dose hook effect in the detection and monitoring of HLA specific antibody by Luminex assay. Int J Immunogenetics. 2007;34:288. |
| 46. | Stenzel A, Bringmann G, Koch K, Strathmann K, Zeiler T. Prozone phenomenon of anti-HLA-A2 in Lifecodes Luminex single antigen class I, Tepnel (LSA Class I). 42nd Annual Meeting of German Society for Transfusion Medicine and Immunohematology (DGTI). Transfus Med Hemotherapy. 2009;36:24. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 47. | Patel R, Terasaki PI. Significance of the positive crossmatch test in kidney transplantation. N Engl J Med. 1969;280:735-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1129] [Cited by in RCA: 1088] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 48. | Stiller CR, Sinclair NR, Abrahams S, Ulan RA, Fung M, Wallace AC. Lymphocyte-dependent antibody and renal graft rejection. Lancet. 1975;1:953-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 49. | Gebel HM, Bray RA, Nickerson P. Pre-transplant assessment of donor-reactive, HLA-specific antibodies in renal transplantation: contraindication vs. risk. Am J Transplant. 2003;3:1488-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 270] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 50. | Scornik JC, Bray RA, Pollack MS, Cook DJ, Marrari M, Duquesnoy R, Langley JW. Multicenter evaluation of the flow cytometry T-cell crossmatch: results from the American Society of Histocompatibility and Immunogenetics-College of American Pathologists proficiency testing program. Transplantation. 1997;63:1440-1445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 51. | Pescovitz MD. B cells: a rational target in alloantibody-mediated solid organ transplantation rejection. Clin Transplant. 2006;20:48-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (1)] |
| 52. | Zangwill S, Ellis T, Stendahl G, Zahn A, Berger S, Tweddell J. Practical application of the virtual crossmatch. Pediatr Transplant. 2007;11:650-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 53. | Schreuder GM, Hurley CK, Marsh SG, Lau M, Fernandez-Vina M, Noreen HJ, Setterholm M, Maiers M; World Marrow Donor Associations Quality Assurance and IT Working Groups; WHO Nomenclature Committee for Factors of the HLA system; 13th International Histocompatibility Workshop Serology Component; International Cell Exchange, UCLA; US National Marrow Donor Program. The HLA Dictionary 2004: a summary of HLA-A, -B, -C, -DRB1/3/4/5 and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR and -DQ antigens. Tissue Antigens. 2005;65:1-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 54. | Kallon D. Histocompatibility Immunogenetics. A collection of brief revision notes. 2001: 38. Available from: http://www.histocompatibilityandimmunogenetics.com/. |
| 55. | Guidelines BTS. Detection and characterisation of clinically relevant antibodies in allotransplantation (joint with BSHI). British Society for Histocompatibility Immunogenetics and British Transplantation Society, 2016. Available from: https://bts.org.uk/guidelines-standards/. |
| 56. | Schlaf G, Mauz-Körholz C, Ott U, Leike S, Altermann W. General insufficiency of the classical CDC-based crossmatch to detect donor-specific anti-HLA antibodies leading to invalid results under recipients’ medical treatment or underlying diseases. Histol Histopathol. 2012;27:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 57. | Eggleton P. Hypersensitivity: Immune Complex Mediated (Type III). : John Wiley Sons, Ltd 2001; 1-14. |
| 58. | Poli F, Innocente A, Cagni N, Brambilla C, Crespiatico L, Colombo MB, Scalamogna M. Isoniazid in patient plasma may cause a false-positive result on the complement-dependent cytotoxicity test. Hum Immunol. 2009;70:758-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 59. | Kucharz EJ, Goodwin JS. Hydralazine causes nonspecific binding of antibodies to human lymphocytes in vitro. Immunopharmacology. 1990;19:1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |