Published online Dec 9, 2014. doi: 10.5497/wjp.v3.i4.120
Revised: September 1, 2014
Accepted: September 23, 2014
Published online: December 9, 2014
Processing time: 166 Days and 6.5 Hours
Prostate cancer is a major public health concern worldwide, being one of the most prevalent cancers in men. Great improvements have been made both in terms of early diagnosis and therapeutics. However, there is still an urgent need for reliable biomarkers that could overcome the lack of cancer-specificity of prostate-specific antigen, as well as alternative therapeutic targets for advanced metastatic cases. Reversible phosphorylation of proteins is a post-translational modification critical to the regulation of numerous cellular processes. Phosphoprotein phosphatase 1 (PPP1) is a major serine/threonine phosphatase, whose specificity is determined by its interacting proteins. These interactors can be PPP1 substrates, regulators, or even both. Deregulation of this protein-protein interaction network alters cell dynamics and underlies the development of several cancer hallmarks. Therefore, the identification of PPP1 interactome in specific cellular context is of crucial importance. The knowledge on PPP1 complexes in prostate cancer remains scarce, with only 4 holoenzymes characterized in human prostate cancer models. However, an increasing number of PPP1 interactors have been identified as expressed in human prostate tissue, including the tumor suppressors TP53 and RB1. Efforts should be made in order to identify the role of such proteins in prostate carcinogenesis, since only 26 have yet well-recognized roles. Here, we revise literature and human protein databases to provide an in-depth knowledge on the biological significance of PPP1 complexes in human prostate carcinogenesis and their potential use as therapeutic targets for the development of new therapies for prostate cancer.
Core tip: Protein kinases and phosphatases are challenging and valuable therapeutic targets for cancer. Here, we revise the relevance of phosphoprotein phosphatase 1 and its interactors for prostate carcinogenesis. Although only 4 complexes are characterized in human prostate cancer models, 81 additional interactors are expressed in human prostate tissue and, at least, 29 of which are involved in prostate carcinogenesis. This complex network has promising roles in the development of new therapies for prostate cancer. Therefore, efforts should be made in order to characterize their biological significance in prostate carcinogenesis.
- Citation: Felgueiras J, Fardilha M. Phosphoprotein phosphatase 1-interacting proteins as therapeutic targets in prostate cancer. World J Pharmacol 2014; 3(4): 120-139
- URL: https://www.wjgnet.com/2220-3192/full/v3/i4/120.htm
- DOI: https://dx.doi.org/10.5497/wjp.v3.i4.120
Prostate cancer (PCa) is the second most commonly diagnosed non-cutaneous cancer and a leading cause of cancer-related death in men worldwide[1]. In spite of the recent advances in early diagnosis and therapeutic management of the disease, prognoses are still poor once the disease progresses to castration-resistant and metastasizes, mainly to bone[2,3]. The urgent need for a panel of reliable biomarkers that could overcome the lack of cancer-specificity of prostate-specific antigen (PSA), as well as alternative therapeutic targets is challenging the scientific community[4].
Reversible phosphorylation of proteins regulates more than 70% of all eukaryotic cellular processes[5]. Phosphorylation at serine (Ser) and threonine (Thr) residues is accomplished by protein Ser/Thr kinases (PSTKs) and reversed by protein Ser/Thr phosphatases (PSTPs)[6]. Deregulation of the counterbalanced action between PSTKs and PSTPs is frequently associated with system-wide disruption of signal transduction and malignant transformation of cells[7,8]. For this reason, emergent studies have been focused on large-scale examination of PCa phosphoproteome[9-12]. Androgen receptor (AR), for instance, is a phosphoprotein vital to the development and progression of PCa that presents, at least, 15 Ser and Thr phosphorylated residues. Phosphorylation of such residues modulates the transcriptional activity, subcellular localization, and stability of the AR[13].
The mammalian genome encodes considerably more PSTKs than PSTPs-nearly 10 to 1[14]. Hence, the success of the protein reversible phosphorylation system depends on the ability of PSTPs to form stable complexes with other proteins, giving rise to a huge number of distinct holoenzymes. This is particularly true for phosphoprotein phosphatase 1 (PPP1), a major PSTP of eukaryotic cells that controls a myriad of processes: glycogen metabolism, muscle contraction, RNA splicing, apoptosis, protein synthesis, cell cycle, among others[15-18]. PPP1 exhibits an effective catalytic machinery, but lacks substrate specificity. Therefore, a number of regulatory subunits, also known as PPP1-interacting proteins (PIPs), have been associated with the spatiotemporal regulation of PPP1 activity[19]. Given the key roles of PIPs, efforts have been made to characterize PPP1 interactomes in human tissues and to identify disease relevant PIPs[20-24].
In contrast to PSTKs, whose therapeutic benefits have been largely explored, PSTPs had been considered not “drug-targetable” for years and, thus, remain understudied[25,26]. In the case of PPP1, this vision is changing due to the increasing number of PPP1 holoenzymes that have been described, and which seem to be attractive targets for the development of new therapies[27]. In fact, pharmaceutical companies are being encouraged to pursue approaches that aim the inhibition or activation of PPP1 holoenzymes.
Here, we revise literature and human protein databases to provide an in-depth knowledge on the relevance of PIPs, expressed in human prostate tissue, for prostate carcinogenesis. Moreover, we address the biological significance of their interaction with PPP1 and consider their potential use as therapeutic targets for the management of human PCa.
A comprehensive literature search of studies involving human samples or human cell lines was performed to identify articles on PPP1 and its interactors in human PCa. The Pubmed database was searched until May 2014 using the Medical Subject Heading (MeSH) whenever possible-for terms not included in MeSH (e.g., “PP1-interacting protein” or “PP1 interactor”) a basic Pubmed search was employed instead. MeSH terms included: (“protein phosphatase 1” or “(PIP abbreviation) protein, human”) and (“prostate” or “prostatic neoplasms”). Reference lists of included studies and review articles were manually searched. The search was restricted to English-language literature.
For the sake of completeness, databases were reviewed: TissueNet and HIPPIE were used to identify additional PIPs expressed in human prostate tissue; BioGPS and The Human Protein Atlas were used to assess mRNA and protein expression levels, respectively; Gene Ontology Consortium was used to identify the biological processes in which proteins are involved; and, ScanProsite was used to identify PPP1 binding motifs for each PIP.
PPP1 is one of the most conserved proteins in eukaryotic species[28,29]. In mammals, three genes encode the catalytic isoforms of the enzyme-PPP1CA, PPP1CB, and PPP1CC-which are ubiquitously expressed. Additionally, PPP1CC gene can generate two splice variants-PPP1CC1 and PPP1CC2-with the latter one being testis-enriched. This catalytic core is analogous, both in terms of structure and mechanism of action, to all members of the phosphoprotein phosphatase superfamily[30]. The major divergences among PPP1 isoforms are found at NH2- and COOH-terminal sequences[15]. Interestingly, the N-terminal was shown to influence the properties of the active site and, consequently, the function of the enzyme and its sensitivity to inhibitors[31,32].
PPP1 catalytic isoforms are not found freely in cells. PPP1 catalytic subunit (PPP1C) interacts with diverse regulatory subunits, known as PIPs, thus enabling the formation of distinct PPP1 multimeric holoenzymes. The nature of the relationship between PPP1 and PIPs greatly varies: (1) PIPs can be substrates for PPP1, with their functions being directly controlled through dephosphorylation by PPP1; (2) PIPs can determine the substrate specificity of PPP1 by either targeting PPP1C to specific subcellular compartments or enhancing/suppressing PPP1C activity towards different substrates; and (3) some PIPs are simultaneously substrates and regulators of PPP1[15,19]. More than 200 holoenzymes have already been identified and characterized, but thousand more remain unknown[15,33].
PPP1 is not able to recognize a consensus sequence near the phosphorylated residue of its phosphotarget. Instead, PPP1C binding is mostly mediated by a short sequence (usually four to eight residues long), remote from the active site, commonly referred to as docking motif[19]. A number of novel PPP1 binding sites have been mapped in PIPs, such as SILK and MyPhoNE, but the RVxF motif (x = any amino acid except proline) is still the most frequent, described for more than 70% of PIPs[34,35]. It was also reported that some PIPs display isoform-specificity, suggesting that they possess isoform-specific docking motifs with putative location at the N- or C-terminal[19].
Imbalances in the protein phosphorylation system strongly contribute to carcinogenesis. In addition to the constitutive activation of oncogenic protein kinases, there is also evidence that the gain and loss of phosphorylation sites in relevant signaling proteins occur in human cancers[36,37].
PPP1 has been shown to take part of various complexes that control cancer hallmarks[24,38]. However, whether the role of PPP1 is pro- or anti-cancer largely depends on its interacting partners and cellular context. For instance, the interaction between PPP1CA and tensin1 impairs the migration and invasion of cancer cells, and the interaction between PPP1CC and hScrib downregulates extracellular signal-regulated kinase (ERK) signaling, thus suppressing oncogene-induced transformation of primary rodent cells[39,40]. On the other hand, the interplay between PPP1 and transforming growth factor-β (TGFβ) drives malignant transformation of premalignant oral lesion cells[41].
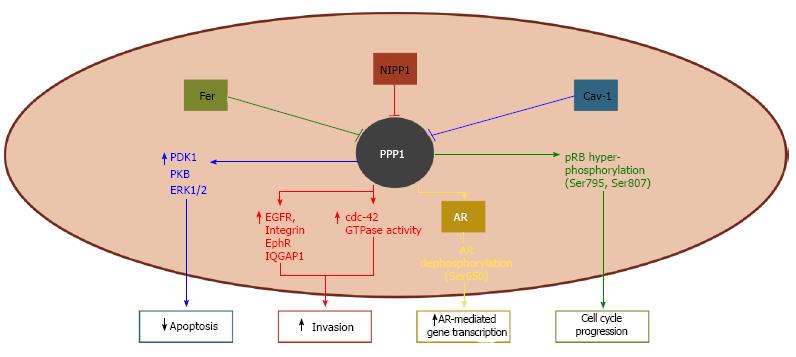
The knowledge on the involvement of PPP1 complexes in human PCa remains scarce. Few complexes have actually been characterized and even those are not fully understood; nonetheless, such complexes seem to have central roles in prostate carcinogenesis (Figure 1).
AR plays a key role in the development of PCa and, accordingly, androgen deprivation therapy is the standard hormonal treatment for the disease[42,43]. AR is regulated by phosphorylation at multiple Ser residues and PPP1CA was shown to specifically reverse phosphorylation at Ser650[44]. Since phosphoSer650 mediates AR nuclear export, PPP1CA dephosphorylation increases the stability and the transcriptional activity of the AR (Figure 1)[44,45]. Besides being a substrate for PPP1, AR may also regulate the phosphatase activity by targeting it to chromatin, where PPP1 can modulate transcription and splicing events[44].
Nuclear inhibitor of protein phosphatase 1 (NIPP1) is a ubiquitously expressed scaffold protein that was firstly identified as a PPP1 inhibitor[46]. The interaction between NIPP1 and PPP1 was shown to orientate cell migration by regulating the expression of integrin and growth factor receptors, and the activity of Cdc42 GTPase (Figure 1). Genetic disruption of this complex decreases the directional migration of PC-3 cells and impairs their migratory potential[47].
Fer tyrosine kinase is highly expressed in human malignant prostate tissues compared to normal or benign tissues, which suggests its involvement in PCa progression[48]. Fer interacts with signal transducer and activator of transcription 3 (STAT3) and phosphorylates AR at Tyr223, thus contributing to interleukin-6 (IL-6)-mediated AR activation and cell growth[49,50]. Downregulation of Fer results in the activation of PPP1CA and consequent hypo-phosphorylation and activation of retinoblastoma protein (RB1), which, in turn, leads to cells arrest at the G0/G1 phase (Figure 1)[51]. Accordingly, downregulation of Fer impairs the proliferation of PCa cells and their ability to form colonies in soft agar [48].
Caveolin-1 (Cav-1) is overexpressed in human PCa and correlates positively with Gleason score, thus being suggested as a potential prognostic marker[52-55]. It has also been proposed as biomarker to monitor the response to treatments with dasatinib and sunitinib[56]. Cav-1-mediated cell survival depends on its interaction with and inhibition of PPP1 (and also PPP2), leading to the increased activity of phosphoinositide-dependent kinase-1, v-akt murine thymoma viral oncogene homolog (AKT), and ERK1/2 (Figure 1)[57].
In spite of the limited number of PPP1 complexes experimentally characterized in human PCa models, several additional PIPs have already been identified as expressed in human prostate tissue. The human prostate proteome includes a total of 81 hitherto experimentally detected PIPs (Table 1)[58,59], but many more may remain unknown. None of the interactors is prostate-specific; however, 28 are highly expressed in prostate tissue, namely BAD, BCL2, CCND1, CUEDC2, GABARAPL2, HCFC1, HDAC1, HDAC10, HEYL, IKBKB, LMTK2, MAP1LC3B, MYC, NOM1, PPP1R3D, PPP1R7, PPP1R11, PPP1R13B, PPP1R37, RB1, RRP1B, RYR2, SH2D4A, STAM, STAU1, SYTL2, TRIM28, and ZFYVE9 (Table 1)[60].
| PIP | Uniprot ID | Biological processes | PPP1 specificity | PPP1 binding motif |
| AKAP11 | Q9UKA4 | Intracellular signal transduction | PPP1CB, PPP1CC | RVxF, other motifs |
| APAF1 | O14727 | Apoptosis | PPP1CA | RVxF, other motifs |
| ATM | Q13315 | Cell cycle; response to DNA damage; protein phosphorylation | PPP1CA | RVxF, SILK |
| AXIN1 | O15169 | Intracellular signal transduction; apoptosis; regulation of protein phosphorylation; transcription | PPP1CA | - |
| BAD1 | Q92934 | Apoptosis | PPP1CA | other motifs |
| BCL21 | P10415 | Apoptosis; response to DNA damage; transmembrane transport | PPP1CA, PPP1CB | RVxF, other motifs |
| BCL2L2 | Q92843 | Apoptosis | PPP1CA | RVxF, other motifs |
| BRCA1 | P38398 | Cell cycle; DNA repair; lipid metabolism | PPP1CA, PPP1CB, PPP1CC | RVxF, other motifs |
| CCND11 | P24385 | Cell cycle; response to DNA damage; transcription | PPP1CB | - |
| CCND3 | P30281 | Cell cycle; intracellular signal transduction; protein phosphorylation | PPP1CB | - |
| CDC5L | Q99459 | Cell cycle; transcription; mRNA splicing | PPP1CA | RVxF |
| CDC34 | P49427 | Cell cycle; protein ubiquitination; intracellular signal transduction | PPP1CB | other motifs |
| CDK2 | P24941 | Cell cycle; DNA repair; meiosis; mitosis; intracellular signal transduction; cell proliferation | PPP1CA | - |
| CDK4 | P11802 | Cell cycle; protein phosphorylation; cell proliferation | PPP1CA | - |
| CSRNP2 | Q9H175 | Apoptosis; transcription; protein phosphorylation | PPP1CA | RVxF, SILK |
| CUEDC21 | Q9H467 | Intracellular signal transduction | PPP1CA | - |
| CUL1 | Q13616 | Host-virus interaction; intracellular signal transduction | PPP1CA | - |
| EED | O75530 | Transcription | PPP1CA | RVxF |
| EIF2AK2 | P19525 | Transcription; immunity; host-virus interaction | PPP1CA | other motifs |
| GABARAP | O95166 | Apoptosis; autophagy; transport | PPP1CC | other motifs |
| GABARAPL21 | P60520 | Autophagy; transport | PPP1CC | - |
| GRB2 | P62993 | Host-virus interaction | PPP1CB | RVxF |
| GSK3B | P49841 | Carbohydrate metabolism; differentiation; intracellular signal transduction | PPP1CA | - |
| HCFC11 | P51610 | Cell cycle; host-virus interaction | PPP1CA | RVxF, RARA |
| HDAC11 | Q13547 | Transcription; host-virus interaction; biological rhythms | PPP1CC | RVxF |
| HDAC6 | Q9UBN7 | Transcription; autophagy | PPP1CC | RVxF |
| HDAC8 | Q9BY41 | Transcription | PPP1CC | - |
| HDAC101 | Q969S8 | Transcription | PPP1CC | - |
| HEYL1 | Q9NQ87 | Transcription; intracellular signal transduction | PPP1CA | RVxF |
| HSPA8 | P11142 | Host-virus interaction; mRNA processing; transcription | PPP1CA | other motifs |
| IKBKB1 | O14920 | Intracellular signal transduction | PPP1CA | other motifs |
| IKBKG | Q9Y6K9 | Transcription; host-virus interaction | PPP1CB, PPP1CC | RARA |
| LMTK21 | Q8IWU2 | Protein phosphorylation; intracellular transport; receptor recycling | PPP1CA | RVxF, other motifs |
| MAP1LC3A | Q9H492 | Autophagy; intracellular signal transduction | PPP1CC | other motifs |
| MAP1LC3B1 | Q9GZQ8 | Autophagy; intracellular signal transduction | PPP1CC | - |
| MAP3K3 | Q99759 | Intracellular signal transduction; protein phosphorylation | PPP1CA, PPP1CC | RVxF |
| MAX | P61244 | Transcription | PPP1CA, PPP1CB | - |
| MDM4 | O15151 | Cell cycle; cell proliferation; apoptosis; response to DNA damage and hypoxia; protein stabilization; protein complex assembly | PPP1CA, PPP1CB, PPP1CC | - |
| MPHOSPH10 | O00566 | Ribosome biogenesis; RNA processing | PPP1CA | RVxF |
| MYC1 | P01106 | Transcription | PPP1CA | RVxF |
| NCL | P19338 | Transcription; angiogenesis | PPP1CB | other motifs |
| NCOR1 | O75376 | Transcription | PPP1CA, PPP1CB, PPP1CC | RVxF, other motifs |
| NOC2L | Q9Y3T9 | Apoptosis; transcription | PPP1CA | RVxF |
| NOM11 | Q5C9Z4 | Targets PPP1CA to the nucleolus | PPP1CA | RVxF, SILK |
| PAK6 | Q9NQU5 | Transcription; protein phosphorylation; cytoskeleton organization; apoptosis | - | Other motifs |
| PLCL2 | Q9UPR0 | Intracellular signal transduction; lipid metabolic process | PPP1CA | RVxF |
| PPP1R2 | P41236 | Regulation of phosphoprotein phosphatase activity; regulation of signal transduction; carbohydrate and glycogen metabolism | PPP1CB, PPP1CC | SILK, other motifs |
| PPP1R3B | Q86XI6 | Carbohydrate and glycogen metabolism | PPP1CA | RVxF |
| PPP1R3D1 | O95685 | Regulation of protein dephosphorylation; carbohydrate and glycogen metabolism | PPP1CC | RVxF |
| PPP1R71 | Q15435 | Regulation of protein dephosphorylation; regulation of catalytic activity | PPP1CB | other motifs |
| PPP1R10 | Q96QC0 | Regulation of catalytic activity; transcription; protein import into nucleus | PPP1CA | RVxF |
| PPP1R111 | O60927 | Regulation of catalytic activity | PPP1CB | RVxF |
| PPP1R12A | O14974 | Regulation of catalytic activity; intracellular transport; cell cycle; regulation of cell adhesion; protein dephosphorylation; intracellular signal transduction | PPP1CB | RVxF, MyPhoNE |
| PPP1R13B1 | Q96KQ4 | Cell cycle; apoptosis | PPP1CA | RVxF |
| PPP1R14B | Q96C90 | Regulation of phosphorylation; regulation of catalytic activity | PPP1CC | RVxF |
| PPP1R15A | O75807 | Apoptosis; regulation of translation; stress response | PPP1CA, PPP1CB, PPP1CC | RVxF, RARA |
| PPP1R15B | Q5SWA1 | Regulation of translation; stress response; dephosphorylation | PPP1CA | RVxF, other motifs |
| PPP1R26 | Q5T8A7 | Regulation of phosphatase activity | PPP1CA | RVxF |
| PPP1R371 | O75864 | Regulation of phosphatase activity | PPP1CA | RVxF |
| PTEN | P60484 | Lipid metabolism; apoptosis; neurogenesis | PPP1CA | RVxF |
| PTK2 | Q05397 | Angiogenesis | PPP1CB | SILK |
| RB11 | P06400 | Transcription; cell cycle; host-virus interaction | PPP1CA | RVxF, SILK, other motifs |
| RIPK3 | Q9Y572 | Necrosis | PPP1CB, PPP1CC | RVxF |
| RPAP2 | Q8IXW5 | Transcription | PPP1CA | RVxF |
| RPAP3 | Q9H6T3 | Alternative splicing; polymorphism | PPP1CA | other motifs |
| RRP1B1 | Q14684 | Regulation of phosphatase activity; RNA processing | PPP1CA | RVxF, other motifs |
| RUVBL2 | Q9Y230 | DNA repair; growth regulation; transcription | PPP1CA | - |
| RYR21 | Q92736 | Intracellular transport | PPP1CA, PPP1CB, PPP1CC | RVxF, other motifs |
| SF3A2 | Q15428 | mRNA processing and splicing | PPP1CA | - |
| SH2D4A1 | Q9H788 | Regulation of phosphatase activity | PPP1CB | RVxF, MyPhoNE |
| SKP1 | P63208 | Intracellular signal transduction | PPP1CA | RVxF |
| SMARCB1 | Q12824 | Transcription; cell cycle; host-virus interaction; neurogenesis | PPP1CA, PPP1CB, PPP1CC | RVxF |
| SPRED1 | Q7Z699 | Regulation of protein phosphorylation; regulation of protein deacetylation; response to DNA damage; development | PPP1CA | RVxF |
| STAM1 | Q92783 | Protein transport | PPP1CA | RVxF |
| STAU11 | O95793 | Intracellular mRNA localization | PPP1CA | RVxF, other motifs |
| SYTL21 | Q9HCH5 | Intracellular transport; exocytosis; regulation of phosphatase activity | PPP1CA | RVxF, SILK, other motifs |
| TMEM33 | P57088 | - | PPP1CB | - |
| TP53 | P04637 | Apoptosis; cell cycle; host-virus interaction; necrosis; transcription | PPP1CA | - |
| TP53BP2 | Q13625 | Intracellular signal transduction; apoptosis; cell cycle; embryo development; heart development; response to ionizing radiation | PPP1CA, PPP1CC | RVxF, other motifs |
| TRIM281 | Q13263 | Transcription; DNA repair; protein ubiquitination; protein sumoylation; protein oligomerization; gene expression; epithelial to mesenchymal transition; innate immune response; regulation of viral release from host cell | PPP1CA, PPP1CB, PPP1CC | RVxF |
| TUSC3 | Q13454 | Intracellular transport | PPP1CA | RVxF, other motifs |
| USF1 | P22415 | Transcription | PPP1CC | RVxF, other motifs |
| ZFYVE91 | O95405 | Intracellular signal transduction | PPP1CA, PPP1CB, PPP1CC | RVxF, other motifs |
| ZFYVE16 | Q7Z3T8 | Intracellular signal transduction; regulation of endocytosis; protein targeting | PPP1CA | RVxF, other motifs |
Of the interactions identified, 67 were described for a specific PPP1 isoform, while 6 seem to be common to all isoforms (Table 1)[58,59]. In the vast majority, binding to PPP1 is assured via RVxF motif, although less described SILK, MyPhoNE, RARA, and other motifs (e.g., apoptotic signature motifs and inhibitor-2 degenerate motif) are also present in some PIPs (Table 1).
PIPs expressed in human prostate tissue are key mediators of several signaling pathways and cellular processes, such as apoptosis, transcription, cell cycle, development/differentiation, and immunology/inflammation.
In the context of human prostate carcinogenesis, only 31 of these proteins have well-recognized functions. In this section, we revise the contribution of these PIPs to prostate carcinogenesis, focusing on studies involving human tissue samples or cell lines.
Apoptotic protease-activating factor 1 (APAF1) is responsible for the cleavage of procaspase-9 and mitochondria-mediated activation of caspases-9, being a major effector of apoptosis[61].
An alternative splicing product of APAF1, known as APAF1-ALT, was found in LNCaP cells. APAF1-ALT exhibits a defective pro-apoptotic function and its expression was shown to be increased under infective conditions. Therefore, this spliced form may be particularly involved in inflammation and carcinogenesis, since it compromises the apoptotic pathway[62].
Resveratrol, sulforaphane, and vitamin D3 exert their tumor suppressive functions through changes in the gene that encodes for APAF1, at least in part[63-65]. APAF1 apoptosome is also involved in malignant cells-selective induction of apoptosis by apoptin[66].
Ataxia telangiectasia mutated (ATM) is a ubiquitously expressed Ser/Thr kinase with a wide spectrum of downstream targets involved in cell-cycle control, DNA repair after radiation-induced damage, and apoptosis[67].
The expression levels of ATM are similar or higher in PCa samples compared to normal prostate tissue; however, its activation is higher in precursor stages of prostate tumorigenesis, like PIN[68,69].
Variants of the ATM gene have been associated with the risk of PCa development, and might be useful predictive markers of adverse responses to radiotherapy[70-72]. ATM maintains telomeres’ length and mediates tumor surveillance[68,69,73].
Downregulation of ATM increases LNCaP, DU-145, and PC-3 cells’ sensitivity to radiation-induced apoptosis[74-77]. The molecular events that arose from ATM inhibition include increased mitotic index, augmented expression of E2F transcription factor and proliferating cell nuclear antigen, and inhibition of G2 arrest in response to DNA damage[78]. Therefore, ATM gene therapy and the use of ATM inhibitors have been explored as adjuvants to radiation therapy in PCa.
Axis inhibition protein 1 (AXIN1) is a tumor suppressor that integrates the β-catenin destruction complex, along with adenomatous polyposis coli protein and glycogen synthase kinase-3 β (GSK3B).
Wnt/β-catenin signaling pathway has been extensively explored due to its impact on development, proliferation, and tumorigenesis[79]. Mutations in the signaling mediators of such system are reported in several types of cancer. For instance, 7 variations in the DNA sequence of axin-1 were found in specimens with abnormal β-catenin immunohistochemistry and 4 different polymorphisms were observed in LNCaP, DU145, PC-3, 22Rv1, and P69SV40T cell lines, as well as in the sublines M12, M2182, M2205[80].
Members of the Bcl-2 family of proteins are pivotal regulators of apoptosis. Bcl-2 (BCL2) and Bcl-2-like protein 2 (BCL2L2) are anti-apoptotic proteins, while Bcl-2 antagonist of cell death (BAD) has proapoptotic functions[81].
The expression of BCL2 is not observed in normal prostate epithelial cells, but is found in PIN and increases in advanced PCa (further details in Catz et al[82]). Higher BCL2 expression is also found in patients that underwent radiotherapy before surgery than those who received surgical treatment as first choice[83]. BCL2 upregulation is required for the acquisition of castration-resistance, in part by suppressing TGFβ and dihydrotestosterone-mediated induction of caspase-1 expression and activation[84,85].
In conformity with BCL2, the expression of BAD is found elevated in highly proliferative states, in spite of not being helpful in the discrimination between benign and malignant prostate tissues[86]. The overexpression of proapoptotic proteins in highly proliferative states seems paradoxical since cancer cells normally take advantage of the molecular machinery to evade apoptosis. However, BAD overexpression was shown to stimulate PCa cells proliferation and enhance tumor growth[87]. On the other hand, overexpression of BAD in LNCaP cells, which are resistant to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis, renders the cells sensitive to TRAIL effects[88].
Several studies have reported the great value of targeting apoptotic molecules in order to increase the sensitivity to apoptosis-inducing agents. Polygene therapy and other combinatorial approaches are receiving increased attention due to their effectiveness[89]. Since BCL2 is associated with increased resistance to androgen deprivation in LNCaP cells, a number of approaches aim to decrease its expression and phosphorylation state[90,91]. The targeting of BCL2 has been explored not only in already settled castration-resistant cases, but also to delay the progression to this advanced state[92,93]. In the case of BCL2L2, it was shown to be a target of miR-205-modulated chemosensitivity[94]. Pharmacological interventions targeting BAD intent to increase its expression and decrease its phosphorylation state[95-97].
Breast cancer type 1 susceptibility protein (BRCA1) has long been described as a tumor suppressor that regulates gene transcription and DNA damage repair[98]. In spite of the relevance of BRCA1 mutations in other types of cancers, evidences of their association with PCa development have been inconsistent and at times contradictory. While some studies point to their irrelevance in PCa development, others state that carriers of BRCA1 mutations have more aggressive phenotype and are more prone to develop distant metastasis[99-102].
The expression profile of BRCA1 during PCa progression is also very heterogeneous, although it tends to be higher in PCa compared to normal prostate epithelium[103,104]. Some works actually suggest that its expression correlates with increased tumor proliferative index and development of lethal cancer, being, therefore, considered a potential prognostic marker[105].
The mode of action of BRCA1 in PCa is complex and seeks clarification[102]. BRCA1 mediates apoptosis, cell-cycle arrest, and the response to doxorubicin treatment in PC-3 cells by targeting a wide variety of genes (e.g., CCND1, BLM, BRCA2, DDB2, FEN1, H3F3B, CCNB2, MAD2L1, and GADD153)[106]. It was also shown to negatively regulate the transcription of insulin-like growth factor I receptor in an AR-dependent manner[104].
The use of anticancer drugs that inhibit poly ADP-ribose polymerase (PARP), such as niraparib and olaparib, has demonstrated efficacy in PCa patients with BRCA1 mutations[107,108].
Cyclins are key mediators of the cell cycle. Cyclin D1 (CCND1) is scarcely found in non-neoplastic tissues, but its levels are increased in the majority of localized tumors, where distinct subcellular localizations are observed according to tumor grading[109]. Likewise, CCND3 displays higher expression in PCa than in BPH and its expression correlates positively with PSA serum levels[110].
In addition to the roles of cyclins in the regulation cell cycle, CCND1/3 interact with AR. CCND1 suppresses the activity of the AR either directly, with the main mediator being the repressor domain of CCND1, or indirectly via histone deacetylases[111]. In this fashion, CCND1 differentially regulates the expression of several androgen-sensitive genes-it represses some genes, such as KLK3/PSA, while induces the transcription of others, as CDC6 and MCM2. Further effects of CCND1 include alteration of transcription factor-chromatin interactions, restraining of TGFβ, Snail, Twist, and Goosecoid signaling pathways, enhancement of Wnt and ES gene expression, and enlargement of a prostate stem cell population[112,113]. The association between CCND3 and the AR represses ligand-dependent activation through cyclin-dependent kinase 4 (CDK4)-independent mechanisms and appeases androgen-dependent proliferation[114].
CCND1 has been proposed as a prognostic marker for poor clinical outcome in PCa biochemical-free recurrence. A number of strategies targets CCND1, including miR-153 and perhaps miR-449a, piperine, and L-mimosine, with the effect of the latter being only observed in PC-3 cells[115-118].
CDK family of proteins regulates cell cycle progression and is involved in AR-mediated cell proliferation. CDK2 mRNA levels decrease after castration, increase after testosterone propionate treatment, and are expressed at high levels in recurrent human xenograft CWR22 tumors[119]. The expression of CDK2 and CDK4 is up-regulated within hours of androgen treatment; nevertheless, castration-resistant PC-3 cells, which do not respond to androgen stimulation, show constitutively high basal expression of both kinases[120].
The activity of CDK2 kinase is stimulated by androgen[119,120]. Increased CDK2 activity correlates with PCa cells insensitivity to TGF-β1, while even modest depletion of CDK2 in LNCaP cells results in strong growth repression[121,122].
CDK4 protein expression was not found elevated in localized prostate tumors, but its overexpression overcome 3,9-dihydroxy-2-prenylcoumestan-induced G0/G1 arrest in castration-resistant cells[123].
Decreased expression and inhibition of the activity of CDK2 and CDK4 are observed upon treatment with anti-proliferative agents, such as resveratrol, BZL 101, and inositol hexaphosphate[124-126]. As phosphorylation of CDK2 on Thr160 is essential for the kinase activity, the manipulation of this phospho-residue has also been analyzed[127].
The multifunctional GSK3B exhibits potent tumor suppressor qualities and is upregulated in many types of tumor, including PCa. The expression pattern of GSK3B differs between normal prostate and PCa cells - nuclear GSK3B is higher in normal prostate, whereas cytoplasmic GSK3B is higher in PCa. Increased GSK3B cytoplasmic levels might determine PCa development and progression due to their correlation with high Ki-67 labeling index, low apoptotic index by TUNEL, high levels of AR and phosphorylated AKT, extracapsular extension, high Gleason score, lymph node metastasis, and biochemical recurrence-free survival[128].
GSK3B mediates both estrogen and AR signaling (for details see Mulholland et al[129]). GSK3B phosphorylates AR and represses AR-mediated transcription and growth[130,131]. GSK3B/AR complex, which locates within the cytoplasm and nucleus, contributes to AR stability, nuclear translocation, and consequent modulation of PCa cells’ response to androgen[132]. The signaling pathway AKT/GSK3B is also involved in nuclear factor α (NFα)-induced epithelial–mesenchymal transition (EMT) in PC-3 cells by contributing to Snail stability[133]. On the other hand, suppression of GSK3B sensitizes PCa cells to TRAIL-induced apoptosis, which might suggest its involvement during resistance acquisition[134]. The suppression of GSK3B expression or phosphorylation state has also demonstrated positive results in inhibiting proliferation of PCa cells[135,136].
AKT/GSK3B signaling pathway has been targeted by a number of antiproliferative and apoptosis-induced agents, namely thiazolidenediones and isoflavone, as well as agents that impair cell migration and invasion, such as fenretinide[137-139].
Histone deacetylases (HDACs) are a large family of enzymes that regulates the nucleosomal histone acetylation. Members of HDACs’ family are divided into four classes (classes I-IV), according to their homology with yeast proteins, with class II being further subdivided into class IIa and IIb. HDAC1 and -8 belong to the class I histone deacetylases, whereas HDAC6 belongs to the class IIb. All members of class I were found to be deregulated in many types of cancers (for review see[140]).
HDAC1 is expressed in normal prostate tissues, where it locates exclusively in the nucleus, cancer precursor lesions, and PCa, and its expression was shown to be lower in stromal cells[141,142]. Conversely, HDAC8 is primarily found in the cytoplasm of stromal cells[142].
HDAC1 expression levels correlate with tumor dedifferentiation, high Gleason score, high pT stage, and high biochemical recurrence rates[141,143]. HDAC1 is a major repressor of AR and E-cadherin, thereby regulating AR-transcriptional activity, cell proliferation and motility, and invasion[144-146].
HDAC6 deacetylates and activates HSP90 chaperone protein, which, in turn, binds to AR[147]. Indeed, HDAC6 regulates AR hypersensitivity to androgens, nuclear localization, and attenuation of its degradation[147,148]. HDAC6 might establish important interactions with other proteins since its decrease is also observed in PC-3 cells, which are castration-resistant[149,150].
The use of HDAC inhibitors in the prevention and treatment of cancer has become an area of intense research. The repression of HDAC1 expression by miR-449a induces growth arrest in PCa and its inhibition by maspin prevents pathologic gene silencing, increasing tumor cell’s sensitivity to drug-induced apoptosis[151,152]. The deacetylase activity of HDAC6 decreases after sulforaphane treatment and it might be responsible for the selective effects of this agent in both hormone-sensitive and castration-resistant cells, while normal cells remain intact[149,150].
Heyl is a member of the hairy/enhancer-of-split-related with YRPW-like motif family of transcriptional repressors. Of the three members of the referred protein family, Heyl is the more potent AR corepressor and reduces the growth of LNCaP cells. The repression of AR activity by Heyl occurs through HDAC1/2-independent mechanisms. Heyl was shown to be excluded from the nucleus in malignant cells but not in benign tissue, thus nuclear exclusion of the protein might be involved in tumor progression[153].
Inhibitor of nuclear factor-κB kinase subunit beta and gamma
Inhibitor of nuclear factor-κB (NF-κB) kinase subunit β and γ (IKBKB and IKBKG, respectively) are involved in the activation of NF-κB[154], a transcription factor that regulates cell growth, apoptosis, inflammation, angiogenesis, and metastasis (for review see[155]).
Studies on human prostate cell lines have not revealed significant differences between primary prostate cells, hormone-sensitive, and castration-resistant PCa cells[156]. However, IKBKB expression is higher in PCa tissue than in benign non-atrophic and atrophic glands[157].
The effects of sulforaphane and phenethyl isothiocyanate are mediated, at least in part, through the inhibition of IKBKB phosphorylation[158]. The loss of IKBKB and IKBKG are also involved in proteasome inhibitors-induced apoptosis[159].
Lemur tyrosine kinase 2 (LMTK2) is a Ser/Thr transmembrane protein kinase mainly involved in endosomal membrane trafficking[160]. In LNCaP cells, this function is achieved, at least in part, by the interaction with myosin VI and consequent recruitment of this protein to the surface of endosomes[161]. Interestingly, the gene that encodes for LMTK2 is one of the novel common alleles associated with PCa[162]. LMTK2 is underexpressed in PCa tissue compared to non-malignant BPH tissue due to alterations in intron 9; however, the mechanism by which this alteration leads to the increased risk of PCa is not properly understood[163]. LMTK2 functions depend on its interaction with other proteins, which includes CDK/P35 complex and PPP1, besides the already mentioned myosin VI[164,165].
MYC deregulation is a well-established mechanism in carcinogenesis[166]. The role of MYC in PCa, nevertheless, is not fully understood. MYC overexpression is frequently observed in PCa, which can be partially explained by locus amplification, mainly in advanced cancers[167]. MYC stabilizes the length of telomeres and is required for EMT[168,169]. MYC and MAX interact with and regulate the AR[170].
MYC amplification status and its overexpression have been suggested as a valuable prognostic tool[171,172]. The existence of a panel of markers that encompass MYC, PTEN, and Ki67 shown benefits in predicting progression-free survival in men receiving adjuvant docetaxel after prostatectomy[173]. Cells that exhibit resistance to treatment with docetaxel have constitutive activation of MYC signaling[174].
Nucleolin (NCL) is an abundant nucleolar phosphoprotein involved in various stages of ribosome synthesis. The expression and phosphorylation of NCL are extremely sensitive to androgens-with both decreasing following androgen deprivation. Thus, the control of NCL expression and phosphorylation by androgens may be an important nucleolar control mechanisms involved in the growth of prostate cells[175]. NCL can also be found in the cell surface, where it may function as a hepatocyte growth factor receptor. Cell surface NCL was shown to be upregulated during PCa progression[176].
Nuclear corepressor 1 (NCOR1), an AR co-repressor, is overexpressed in PCa cell lines compared to normal prostate cells. NCOR1 expression is confined to the S phase of cell cycle; therefore, during this time NCOR1 represses the expression of AR target genes[177]. In PCa cells, the activity of NCOR1 is positively regulated by protein kinase A (PKA)[178].
The increased expression and activity of NCOR1 impair peroxisome proliferator activated receptor α/γ-mediated expression of key target genes, such as CDKN1A and TGFBRAP1, thus contributing to the loss of ligands anti-proliferative responsiveness in PCa cells[179]. NCOR1 might also be important during the process of castration-resistant acquisition[180].
The expression of PAK6 is increased in primary and metastatic PCa, and correlates with cells’ sensitivity to androgens[181,182]. PAK6 co-localizes with AR in the cytoplasm of normal prostate epithelium and translocates into the nucleus in malignant phenotypes, where it represses both AR- and ER-mediated gene transcription[181,183,184]. It was also shown that PAK6 phosphorylates the AR at Ser578, promoting the association of AR-E3 ligase murine double minute-2 (Mdm2) and guiding AR degradation[185].
The knockdown of PAK6 impairs PCa growth and improves chemosensitivity of docetaxel and sensitivity to radiation[186,187].
Phosphatase and tensin homolog (PTEN) is a dual specificity phosphatase and a recognized tumor suppressor. Inactivation of PTEN has been associated with many different types of cancer, including PCa, and assumes preponderant roles (further details on[188,189]). A number of molecules have been recently shown to contribute to PTEN downregulation, including lamin A/C and a subset of microRNAs (e.g., miR-19b, miR-23b, miR-26a, miR-92a, and miR-153)[117,190,191]. Loss of PTEN determines PCa progression through several downstream effectors and signaling pathways, including PI3K/AKT, BIM1, CXCL12/CXCR4, and PDGF D/β-PDGFR[192-195]. The loss of PTEN is also associated with increased risk of capsular penetration[196]. Interestingly, it was recently shown that PTEN is incorporated in the cargo of exosomes prevenient from cancer cells, but not in those derived from non-malignant cells. Exosomes are able to transfer PTEN to other cells, which in turn recover the tumor-suppressor activity[197].
Recent evidences support the usefulness of PTEN in PCa management. Blood exosomes of PCa patients contain PTEN, contrarily to the exosomes isolated from normal subjects, which may indicate exosomal PTEN as a putative diagnostic tool[197]. The loss of cytoplasmic PTEN was shown to accurately distinct intraductal carcinoma from prostatic intraepithelial neoplasia, since the latter does not manifest PTEN loss at all[198]. PTEN status might also be useful in the prognostic evaluation of men with localized PCa[199,200].
Moreover, a phase II clinical trial reported that the activity of PTEN determines the improvement of progression-free survival and is potentially required for the efficacy of cetuximab in metastatic castration-resistant PCa[201]. PTEN expression is enhanced by resveratrol-mediated AR inhibition[202].
Protein tyrosine kinase 2 (PTK2) regulates adhesion and motility of cells. Its upregulation and activation was observed in localized and castration-resistant PCa, in spite of being more evident in the latter case[203,204]. The complexes that PTK2 forms with paxillin and p50csk are mainly observed in metastatic PCa and contribute to the metastatic behavior[204]. PTK2 is also involved in the migration and invasion mediated by IL-8 and CXCL13-CXCR5[205,206].
Treatments with FTY720 and the combinatorial therapy with curcumin and methylseleninic acid compromises PTK2 activity, and PTK2 inhibition was shown to delay the progression of PCa[203,207]. PTK2 is also a target of genistein-mediated morphologic changes[208].
Tripartite motif-containing protein 28 (TRIM28) is a substrate of ATM kinase involved in the maintenance of chromatin in condensed states[209]. The expression of TRIM28 is observed in prostate cancer lines, despite being lower in castration-resistant cell lines[210]. TRIM28 was recently identified as an activator of the AR and is also involved in the response of prostate cells to DNA damage[210,211].
Retinoblastoma-associated protein (RB1) and cellular tumor antigen p53 (TP53) are major tumor suppressors whose functions in PCa have been broadly explored. Loss of RB1 and TP53 is strictly associated with AR misregulation and progression to castration-resistant disease. Both their mechanism and their potential roles in managing PCa are extensively revised in Aparicio et al[212], Dean et al[213] and Lee et al[214].
Ryanodine receptor 2 (RYR2) is expressed in PWR-1E non-tumor cells, as well as in LNCaP and DU145 PCa cells, with the latter registering the lowest expression[215,216]. RYR2 mobilizes Ca2+ from intracellular stores, which is essential to the regulation of apoptosis[215].
SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 (SMARCB1) is a core subunit of the SWI/SNF family of nucleosome-remodeling complexes[217]. In aggressive PCa, the expression of SWI/SNF target genes is impaired by the binding to SChLAP1, which was shown to be aberrantly upregulated[218].
The relationship between PPP1 and the majority of the PIPs here referred, as well as possible alterations in the dynamics of such complexes during prostate carcinogenesis, require further elucidation.
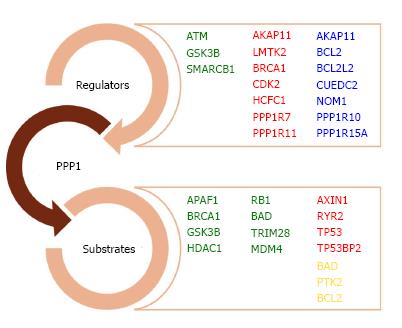
Some of the PIPs are already characterized as PPP1 regulators, substrates, or both (Figure 2). For a significant number of PIPs identified in protein-protein interaction screenings, nevertheless, the functional significance of their interaction with PPP1 remains poorly understood. Therefore, efforts should be made in order to understand PPP1 interaction with CCND1, CCND3, CDC5L, CDC34, CDK4, EED, EIF2AK2, GABARAP, GABARAPL2, GRB2, HDAC8, HDAC10, HSPA8, HEYL, MAX, NCL, IKBKB, IKBKG, MAP1LC3A, MAP1LC3B, MAP3K3, MPHOSPH10, MYC, NCOR1, NOC2L, PAK6, PLCL2, PPP1R3B, PPP1R12A, PPP1R13B, PTEN, RIPK3, RPAP2, RPAP3, RRP1B, RUVBL2, SF3A2, SH2D4A, SKP1, SPRED1, STAM, STAU1, SYTL2, and USF1.
In other cases, the biological significance of the complex is partially known, although it is not established whether the PIP is the regulator or the substrate (or even both). For instance, HDAC6 directly binds to PPP1 and the complex controls microtubule dynamics by maintaining α-tubulin in a deacetylated state, but the exact mechanism remains poorly understood[219].
AKAP11 acts as a targeting subunit of PPP1 and it can also inhibit the phosphatase activity[220,221]. BCL2L2 and CUEDC2 target PPP1 to protein complexes: BCL2L2 recruits PPP1 to BAD, forming a complex that is involved in the control of apoptosis[222]; and, CUEDC2 targets PPP1 to IKK, thereby promoting the dephosphorylation and inactivation of the kinase[223]. NOM1 acts as a PPP1 nucleolar targeting subunit, PPP1R10 targets PPP1 to the nucleus, and PPP1R15A targets PPP1 to the endoplasmic reticulum[224-227]. PPP1R10/PPP1 holoenzyme is known to regulate chromosome decondensation and apoptosis in response to cellular stresses[226,228].
PPP1C positive regulators include ATM, GSK3B, and SMARCB1. In response to ionizing radiation, PPP1 is dephosphorylated and activated by ATM[229]. ATM-mediated activation of PPP1 could occur, at least, via two mechanisms: (1) phosphorylation of I-2 and consequent dissociation of the complex I-2/PPP1; or, (2) dephosphorylation of PPP1C at Thr320 to amplify its activity[230]. As a result, PPP1 dephosphorylates HDAC1, leading to the dissociation of the HDAC1-PPP1-Rb complex[231,232]. In similar way to ATM, GSK3B activates PPP1 via phosphorylation of I-2 and consequent disruption of the I-2/PPP1 complex[233]. SMARCB1 forms a tricomplex with PPP1R15A and PPP1, and weakly stimulates PPP1 activity[234].
AKAP11, BRCA1, CDK2, LMTK2, HCFC1, PPP1R7, PPP1R11, and TP53BP2 inhibit the activity of PPP1[229,235-239].
PPP1C is a key regulator of the two major tumor suppressors: it inhibits TP53 and activates RB1 (further details on[24]). The apoptotic process is strictly controlled by reversible phosphorylation[240]. The phosphorylation of APAF1 by the 90-kDa ribosomal S6 kinase (RSK) compromises the formation of the apoptosome, impairs cells’ sensitivity to cytochrome c, and inhibits apoptosis. PPP1CA was shown to reverse the RSK-mediated phosphorylation of APAF1, thus enhancing its pro-apoptotic activities[241]. BAD is also dephosphorylated by PPP1CA in a dependent way of the anti-apoptotic members BCL2, BCL2L2, and BCL-XL[222,242,243]. While BAD overexpression provides proliferative advantage to tumor cells, BAD dephosphorylation increases their sensitivity to apoptosis[87].
PPP1 exerts a positive control on Wnt signaling through dephosphorylation of AXIN1. As a result, β-catenin destruction complex dissociates, the free phospho-β-catenin accumulates in the cytoplasm, and the transcriptional activity of β-catenin is promoted[244].
In addition of being regulators, BRCA1 and GSK3B are also substrates for PPP1C. PPP1C dephosphorylates BRCA1 and enhances its DNA repair function[235,245,246]. GSK3B is also dephosphorylated and disinhibited by PPP1-mediated dephosphorylation[247].
HDAC1 activity is promoted through dephosphorylation of Ser133 by PPP1, which leads to dissociation of HDAC1-PP1-Rb complex, and consequent increase of HDAC1 activity[232,248]. Similarly, HDAC6 and -8, which can be phosphorylated, might also be PPP1 substrates, although this fact has not been confirmed yet.
RYRs are regulated through reversible phosphorylation, and PPP1 reverts PKA-mediated phosphorylation and activation of RYR2[249-251]. PPP1 dephosphorylates TRIM28 (Ser824), enhancing its sumoylation state, and MDM4 (Ser367), enhancing its stability and leading to the consequent inhibition of TP53 activity[252]. PPP1 also reverts the phosphorylation of PTK2 and BCL2[253-255].
Since PPP1 is a Ser/Thr phosphatase with major roles in several pathological processes, the manipulation of its activity is a valuable therapeutic tool that had been misjudged for years. PPP1 activity could be manipulated through direct or indirect inhibition of the catalytic site (for review see[27]).
The dissociation of PPP1 complexes through the targeting of PIPs is challenging and might overcome the problems that arise from direct inhibition of PPP1C. However, this area remains understudied and only two complexes are currently being targeted: PPP1C/HDAC and PPP1C/PPP1R15A. Trichostatin A disrupts PPP1C/HDAC and is used in the treatment of glioblastoma and PCa cells. LBH589, an inhibitor of HDAC, was also shown to be able to dissociate this complex[256]. As a consequence, AKT is dephosphorylated and its activity decreases. PPP1C/PPP1R15A complex is disrupted upon salubrinal treatment, thereby dephosphorylating eIF2α[257,258].
The increasing number of PPP1 docking motifs identified offers excellent opportunities for targeting specific complexes. In fact, the docking motif found in Bad has inspired the designing of a peptide that interferes with PPP1/BAD complex and is able to induce cell death[259]. Also, the PPP1 docking motif R/Kx(0,1)V/IxFxxR/KxR/K, a new PPP1C-dependent apoptotic signature, might be a useful tool for drug design[260].
The disruption or enhancement of several other complexes might contribute to the enhancement of PCa management, enabling more efficient therapies for advanced castration-resistant PCa. Therefore, the identification of PPP1 complexes in human prostate and their characterization in prostate carcinogenesis is imperative for the search of new therapeutic targets.
P- Reviewer: Choi CY, Moens U S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25543] [Article Influence: 1824.5] [Reference Citation Analysis (7)] |
| 2. | Bubendorf L, Schöpfer A, Wagner U, Sauter G, Moch H, Willi N, Gasser TC, Mihatsch MJ. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol. 2000;31:578-583. [PubMed] |
| 3. | Kapoor A. What’s new in prostate cancer research? Highlights of GU-ASCO 2014. Can Urol Assoc J. 2014;8:S8-S12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Felgueiras J, Silva JV, Fardilha M. Prostate cancer: the need for biomarkers and new therapeutic targets. J Zhejiang Univ Sci B. 2014;15:16-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1174] [Cited by in RCA: 1219] [Article Influence: 81.3] [Reference Citation Analysis (0)] |
| 6. | Brautigan DL. Protein Ser/Thr phosphatases--the ugly ducklings of cell signalling. FEBS J. 2013;280:324-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 7. | Stebbing J, Lit LC, Zhang H, Darrington RS, Melaiu O, Rudraraju B, Giamas G. The regulatory roles of phosphatases in cancer. Oncogene. 2014;33:939-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 8. | Lim YP. Mining the tumor phosphoproteome for cancer markers. Clin Cancer Res. 2005;11:3163-3169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Chen L, Giorgianni F, Beranova-Giorgianni S. Characterization of the phosphoproteome in LNCaP prostate cancer cells by in-gel isoelectric focusing and tandem mass spectrometry. J Proteome Res. 2010;9:174-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Lescarbeau R, Kaplan DL. Correlating phosphoproteomic signaling with castration resistant prostate cancer survival through regression analysis. Mol Biosyst. 2014;10:605-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 11. | Lescarbeau RM, Kaplan DL. Quantitative analysis of castration resistant prostate cancer progression through phosphoproteome signaling. BMC Cancer. 2014;14:325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Giorgianni F, Zhao Y, Desiderio DM, Beranova-Giorgianni S. Toward a global characterization of the phosphoproteome in prostate cancer cells: identification of phosphoproteins in the LNCaP cell line. Electrophoresis. 2007;28:2027-2034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 13. | van der Steen T, Tindall DJ, Huang H. Posttranslational modification of the androgen receptor in prostate cancer. Int J Mol Sci. 2013;14:14833-14859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 14. | Moorhead GB, Trinkle-Mulcahy L, Ulke-Lemée A. Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol. 2007;8:234-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 267] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 15. | Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 511] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 16. | Bollen M. Combinatorial control of protein phosphatase-1. Trends Biochem Sci. 2001;26:426-431. [PubMed] |
| 17. | Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241-256. [PubMed] |
| 18. | Flores-Delgado G, Liu CW, Sposto R, Berndt N. A limited screen for protein interactions reveals new roles for protein phosphatase 1 in cell cycle control and apoptosis. J Proteome Res. 2007;6:1165-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 19. | Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci. 2010;35:450-458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 442] [Cited by in RCA: 407] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 20. | Fardilha M, Esteves SL, Korrodi-Gregório L, Vintém AP, Domingues SC, Rebelo S, Morrice N, Cohen PT, da Cruz e Silva OA, da Cruz e Silva EF. Identification of the human testis protein phosphatase 1 interactome. Biochem Pharmacol. 2011;82:1403-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Esteves SL, Domingues SC, da Cruz e Silva OA, Fardilha M, da Cruz e Silva EF. Protein phosphatase 1α interacting proteins in the human brain. OMICS. 2012;16:3-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Fardilha M, Esteves SL, Korrodi-Gregório L, Pelech S, da Cruz E Silva OA, da Cruz E Silva E. Protein phosphatase 1 complexes modulate sperm motility and present novel targets for male infertility. Mol Hum Reprod. 2011;17:466-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 23. | Esteves SL, Korrodi-Gregório L, Cotrim CZ, van Kleeff PJ, Domingues SC, da Cruz e Silva OA, Fardilha M, da Cruz e Silva EF. Protein phosphatase 1γ isoforms linked interactions in the brain. J Mol Neurosci. 2013;50:179-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 24. | Figueiredo J, da Cruz E Silva OA, Fardilha M. Protein phosphatase 1 and its complexes in carcinogenesis. Curr Cancer Drug Targets. 2014;14:2-29. [PubMed] |
| 25. | Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1941] [Cited by in RCA: 1959] [Article Influence: 122.4] [Reference Citation Analysis (0)] |
| 26. | McConnell JL, Wadzinski BE. Targeting protein serine/threonine phosphatases for drug development. Mol Pharmacol. 2009;75:1249-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 131] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 27. | Chatterjee J, Köhn M. Targeting the untargetable: recent advances in the selective chemical modulation of protein phosphatase-1 activity. Curr Opin Chem Biol. 2013;17:361-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 28. | Sangrador A, Andrés I, Eguiraun A, Lorenzo ML, Ortiz JM. Growth arrest of Schizosaccharomyces pombe following overexpression of mouse type 1 protein phosphatases. Mol Gen Genet. 1998;259:449-456. [PubMed] |
| 29. | Ceulemans H, Stalmans W, Bollen M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. Bioessays. 2002;24:371-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 121] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 30. | Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1044] [Cited by in RCA: 1161] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 31. | Xie X, Huang W, Xue C, Wei Q. The nonconserved N-terminus of protein phosphatases 1 influences its active site. BMB Rep. 2008;41:881-885. [PubMed] |
| 32. | Xie XJ, Huang W, Xue CZ, Wei Q. The N-terminal domain influences the structure and property of protein phosphatase 1. Mol Cell Biochem. 2009;327:241-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 33. | Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876-1887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 527] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 34. | Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol. 2009;16:365-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 256] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 35. | Meiselbach H, Sticht H, Enz R. Structural analysis of the protein phosphatase 1 docking motif: molecular description of binding specificities identifies interacting proteins. Chem Biol. 2006;13:49-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 97] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 36. | Radivojac P, Baenziger PH, Kann MG, Mort ME, Hahn MW, Mooney SD. Gain and loss of phosphorylation sites in human cancer. Bioinformatics. 2008;24:i241-i247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 37. | Bellacosa A, Kumar CC, Di Cristofano A, Testa JR. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv Cancer Res. 2005;94:29-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 631] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 38. | Korrodi-Gregório L, Silva JV, Santos-Sousa L, Freitas MJ, Felgueiras J, Fardilha M. TGF-β cascade regulation by PPP1 and its interactors -impact on prostate cancer development and therapy. J Cell Mol Med. 2014;18:555-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 39. | Hall EH, Daugherty AE, Choi CK, Horwitz AF, Brautigan DL. Tensin1 requires protein phosphatase-1alpha in addition to RhoGAP DLC-1 to control cell polarization, migration, and invasion. J Biol Chem. 2009;284:34713-34722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 40. | Nagasaka K, Seiki T, Yamashita A, Massimi P, Subbaiah VK, Thomas M, Kranjec C, Kawana K, Nakagawa S, Yano T. A novel interaction between hScrib and PP1γ downregulates ERK signaling and suppresses oncogene-induced cell transformation. PLoS One. 2013;8:e53752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Walsh JE, Young MR. TGF-beta regulation of focal adhesion proteins and motility of premalignant oral lesions via protein phosphatase 1. Anticancer Res. 2011;31:3159-3164. [PubMed] |
| 42. | Grossmann M, Cheung AS, Zajac JD. Androgens and prostate cancer; pathogenesis and deprivation therapy. Best Pract Res Clin Endocrinol Metab. 2013;27:603-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 43. | Wen S, Niu Y, Lee SO, Chang C. Androgen receptor (AR) positive vs negative roles in prostate cancer cell deaths including apoptosis, anoikis, entosis, necrosis and autophagic cell death. Cancer Treat Rev. 2014;40:31-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 44. | Chen S, Kesler CT, Paschal BM, Balk SP. Androgen receptor phosphorylation and activity are regulated by an association with protein phosphatase 1. J Biol Chem. 2009;284:25576-25584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 45. | Gioeli D, Black BE, Gordon V, Spencer A, Kesler CT, Eblen ST, Paschal BM, Weber MJ. Stress kinase signaling regulates androgen receptor phosphorylation, transcription, and localization. Mol Endocrinol. 2006;20:503-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 137] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 46. | Beullens M, Van Eynde A, Stalmans W, Bollen M. The isolation of novel inhibitory polypeptides of protein phosphatase 1 from bovine thymus nuclei. J Biol Chem. 1992;267:16538-16544. [PubMed] |
| 47. | Martin-Granados C, Prescott AR, Van Dessel N, Van Eynde A, Arocena M, Klaska IP, Görnemann J, Beullens M, Bollen M, Forrester JV. A role for PP1/NIPP1 in steering migration of human cancer cells. PLoS One. 2012;7:e40769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Allard P, Zoubeidi A, Nguyen LT, Tessier S, Tanguay S, Chevrette M, Aprikian A, Chevalier S. Links between Fer tyrosine kinase expression levels and prostate cell proliferation. Mol Cell Endocrinol. 2000;159:63-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 49. | Zoubeidi A, Rocha J, Zouanat FZ, Hamel L, Scarlata E, Aprikian AG, Chevalier S. The Fer tyrosine kinase cooperates with interleukin-6 to activate signal transducer and activator of transcription 3 and promote human prostate cancer cell growth. Mol Cancer Res. 2009;7:142-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 50. | Rocha J, Zouanat FZ, Zoubeidi A, Hamel L, Benidir T, Scarlata E, Brimo F, Aprikian A, Chevalier S. The Fer tyrosine kinase acts as a downstream interleukin-6 effector of androgen receptor activation in prostate cancer. Mol Cell Endocrinol. 2013;381:140-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (1)] |
| 51. | Pasder O, Shpungin S, Salem Y, Makovsky A, Vilchick S, Michaeli S, Malovani H, Nir U. Downregulation of Fer induces PP1 activation and cell-cycle arrest in malignant cells. Oncogene. 2006;25:4194-4206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | Yang G, Truong LD, Timme TL, Ren C, Wheeler TM, Park SH, Nasu Y, Bangma CH, Kattan MW, Scardino PT. Elevated expression of caveolin is associated with prostate and breast cancer. Clin Cancer Res. 1998;4:1873-1880. [PubMed] |
| 53. | Yang G, Truong LD, Wheeler TM, Thompson TC. Caveolin-1 expression in clinically confined human prostate cancer: a novel prognostic marker. Cancer Res. 1999;59:5719-5723. [PubMed] |
| 54. | Karam JA, Lotan Y, Roehrborn CG, Ashfaq R, Karakiewicz PI, Shariat SF. Caveolin-1 overexpression is associated with aggressive prostate cancer recurrence. Prostate. 2007;67:614-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 107] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 55. | Thompson TC, Tahir SA, Li L, Watanabe M, Naruishi K, Yang G, Kadmon D, Logothetis CJ, Troncoso P, Ren C. The role of caveolin-1 in prostate cancer: clinical implications. Prostate Cancer Prostatic Dis. 2010;13:6-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 56. | Tahir SA, Kurosaka S, Tanimoto R, Goltsov AA, Park S, Thompson TC. Serum caveolin-1, a biomarker of drug response and therapeutic target in prostate cancer models. Cancer Biol Ther. 2013;14:117-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 57. | Li L, Ren CH, Tahir SA, Ren C, Thompson TC. Caveolin-1 maintains activated Akt in prostate cancer cells through scaffolding domain binding site interactions with and inhibition of serine/threonine protein phosphatases PP1 and PP2A. Mol Cell Biol. 2003;23:9389-9404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 234] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 58. | Barshir R, Basha O, Eluk A, Smoly IY, Lan A, Yeger-Lotem E. The TissueNet database of human tissue protein-protein interactions. Nucleic Acids Res. 2013;41:D841-D844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 59. | Schaefer MH, Fontaine JF, Vinayagam A, Porras P, Wanker EE, Andrade-Navarro MA. HIPPIE: Integrating protein interaction networks with experiment based quality scores. PLoS One. 2012;7:e31826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 241] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 60. | Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1039] [Cited by in RCA: 1137] [Article Influence: 71.1] [Reference Citation Analysis (0)] |
| 61. | Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549-11556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1507] [Cited by in RCA: 1505] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 62. | Ogawa T, Shiga K, Hashimoto S, Kobayashi T, Horii A, Furukawa T. APAF-1-ALT, a novel alternative splicing form of APAF-1, potentially causes impeded ability of undergoing DNA damage-induced apoptosis in the LNCaP human prostate cancer cell line. Biochem Biophys Res Commun. 2003;306:537-543. [PubMed] |
| 63. | Choi S, Lew KL, Xiao H, Herman-Antosiewicz A, Xiao D, Brown CK, Singh SV. D,L-Sulforaphane-induced cell death in human prostate cancer cells is regulated by inhibitor of apoptosis family proteins and Apaf-1. Carcinogenesis. 2007;28:151-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 94] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 64. | Narayanan BA, Narayanan NK, Re GG, Nixon DW. Differential expression of genes induced by resveratrol in LNCaP cells: P53-mediated molecular targets. Int J Cancer. 2003;104:204-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 106] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 65. | Guzey M, Luo J, Getzenberg RH. Vitamin D3 modulated gene expression patterns in human primary normal and cancer prostate cells. J Cell Biochem. 2004;93:271-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 66. | Burek M, Maddika S, Burek CJ, Daniel PT, Schulze-Osthoff K, Los M. Apoptin-induced cell death is modulated by Bcl-2 family members and is Apaf-1 dependent. Oncogene. 2006;25:2213-2222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 67. | Rotman G, Shiloh Y. ATM: a mediator of multiple responses to genotoxic stress. Oncogene. 1999;18:6135-6144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 195] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 68. | Fan C, Quan R, Feng X, Gillis A, He L, Matsumoto ED, Salama S, Cutz JC, Kapoor A, Tang D. ATM activation is accompanied with earlier stages of prostate tumorigenesis. Biochim Biophys Acta. 2006;1763:1090-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 69. | Angèle S, Falconer A, Foster CS, Taniere P, Eeles RA, Hall J. ATM protein overexpression in prostate tumors: possible role in telomere maintenance. Am J Clin Pathol. 2004;121:231-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 70. | Angèle S, Falconer A, Edwards SM, Dörk T, Bremer M, Moullan N, Chapot B, Muir K, Houlston R, Norman AR. ATM polymorphisms as risk factors for prostate cancer development. Br J Cancer. 2004;91:783-787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 71. | Meyer A, Wilhelm B, Dörk T, Bremer M, Baumann R, Karstens JH, Machtens S. ATM missense variant P1054R predisposes to prostate cancer. Radiother Oncol. 2007;83:283-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 72. | Cesaretti JA, Stock RG, Lehrer S, Atencio DA, Bernstein JL, Stone NN, Wallenstein S, Green S, Loeb K, Kollmeier M. ATM sequence variants are predictive of adverse radiotherapy response among patients treated for prostate cancer. Int J Radiat Oncol Biol Phys. 2005;61:196-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 71] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 73. | Farooqi AA, Fayyaz S, Rashid S. Upon the tightrope in prostate cancer: two acrobats on the same tightrope to cross the finishline. Mol Cell Biochem. 2012;364:53-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 74. | Fan Z, Chakravarty P, Alfieri A, Pandita TK, Vikram B, Guha C. Adenovirus-mediated antisense ATM gene transfer sensitizes prostate cancer cells to radiation. Cancer Gene Ther. 2000;7:1307-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 75. | Collis SJ, Swartz MJ, Nelson WG, DeWeese TL. Enhanced radiation and chemotherapy-mediated cell killing of human cancer cells by small inhibitory RNA silencing of DNA repair factors. Cancer Res. 2003;63:1550-1554. [PubMed] |
| 76. | Truman JP, Gueven N, Lavin M, Leibel S, Kolesnick R, Fuks Z, Haimovitz-Friedman A. Down-regulation of ATM protein sensitizes human prostate cancer cells to radiation-induced apoptosis. J Biol Chem. 2005;280:23262-23272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 77. | Shaheen FS, Znojek P, Fisher A, Webster M, Plummer R, Gaughan L, Smith GC, Leung HY, Curtin NJ, Robson CN. Targeting the DNA double strand break repair machinery in prostate cancer. PLoS One. 2011;6:e20311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 78. | Mukhopadhyay UK, Senderowicz AM, Ferbeyre G. RNA silencing of checkpoint regulators sensitizes p53-defective prostate cancer cells to chemotherapy while sparing normal cells. Cancer Res. 2005;65:2872-2881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 79. | Luo W, Lin SC. Axin: a master scaffold for multiple signaling pathways. Neurosignals. 2004;13:99-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 119] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 80. | Yardy GW, Bicknell DC, Wilding JL, Bartlett S, Liu Y, Winney B, Turner GD, Brewster SF, Bodmer WF. Mutations in the AXIN1 gene in advanced prostate cancer. Eur Urol. 2009;56:486-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 81. | Ola MS, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol Cell Biochem. 2011;351:41-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 714] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 83. | Rosser CJ, Reyes AO, Vakar-Lopez F, Levy LB, Kuban DA, Hoover DC, Lee AK, Pisters LL. Bcl-2 is significantly overexpressed in localized radio-recurrent prostate carcinoma, compared with localized radio-naive prostate carcinoma. Int J Radiat Oncol Biol Phys. 2003;56:1-6. [PubMed] |
| 84. | Lin Y, Fukuchi J, Hiipakka RA, Kokontis JM, Xiang J. Up-regulation of Bcl-2 is required for the progression of prostate cancer cells from an androgen-dependent to an androgen-independent growth stage. Cell Res. 2007;17:531-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 85. | Bruckheimer EM, Kyprianou N. Bcl-2 antagonizes the combined apoptotic effect of transforming growth factor-beta and dihydrotestosterone in prostate cancer cells. Prostate. 2002;53:133-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 86. | Royuela M, Arenas MI, Bethencourt FR, Sánchez-Chapado M, Fraile B, Paniagua R. Immunoexpressions of p21, Rb, mcl-1 and bad gene products in normal, hyperplastic and carcinomatous human prostates. Eur Cytokine Netw. 2001;12:654-663. [PubMed] |
| 87. | Smith AJ, Karpova Y, D’Agostino R, Willingham M, Kulik G. Expression of the Bcl-2 protein BAD promotes prostate cancer growth. PLoS One. 2009;4:e6224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 88. | Taghiyev AF, Guseva NV, Harada H, Knudson CM, Rokhlin OW, Cohen MB. Overexpression of BAD potentiates sensitivity to tumor necrosis factor-related apoptosis-inducing ligand treatment in the prostatic carcinoma cell line LNCaP. Mol Cancer Res. 2003;1:500-507. [PubMed] |
| 89. | Zhang Y, Yu J, Unni E, Shao TC, Nan B, Snabboon T, Kasper S, Andriani F, Denner L, Marcelli M. Monogene and polygene therapy for the treatment of experimental prostate cancers by use of apoptotic genes bax and bad driven by the prostate-specific promoter ARR(2)PB. Hum Gene Ther. 2002;13:2051-2064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 90. | Kajiwara T, Takeuchi T, Ueki T, Moriyama N, Ueki K, Kakizoe T, Kawabe K. Effect of Bcl-2 overexpression in human prostate cancer cells in vitro and in vivo. Int J Urol. 1999;6:520-525. [PubMed] |
| 91. | Tolcher AW. Preliminary phase I results of G3139 (bcl-2 antisense oligonucleotide) therapy in combination with docetaxel in hormone-refractory prostate cancer. Semin Oncol. 2001;28:67-70. [PubMed] |
| 92. | Gleave ME, Miayake H, Goldie J, Nelson C, Tolcher A. Targeting bcl-2 gene to delay androgen-independent progression and enhance chemosensitivity in prostate cancer using antisense bcl-2 oligodeoxynucleotides. Urology. 1999;54:36-46. [PubMed] |
| 93. | Chi KN. Targeting Bcl-2 with oblimersen for patients with hormone refractory prostate cancer. World J Urol. 2005;23:33-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 94. | Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, Guo B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010;1:e105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 168] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 95. | Gapter L, Wang Z, Glinski J, Ng KY. Induction of apoptosis in prostate cancer cells by pachymic acid from Poria cocos. Biochem Biophys Res Commun. 2005;332:1153-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 99] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 96. | Sambantham S, Radha M, Paramasivam A, Anandan B, Malathi R, Chandra SR, Jayaraman G. Molecular mechanism underlying hesperetin-induced apoptosis by in silico analysis and in prostate cancer PC-3 cells. Asian Pac J Cancer Prev. 2013;14:4347-4352. [PubMed] |
| 97. | Cho YS, Cho-Chung YS. Antisense protein kinase A RIalpha acts synergistically with hydroxycamptothecin to inhibit growth and induce apoptosis in human cancer cells: molecular basis for combinatorial therapy. Clin Cancer Res. 2003;9:1171-1178. [PubMed] |
| 98. | Wang Q, Zhang H, Fishel R, Greene MI. BRCA1 and cell signaling. Oncogene. 2000;19:6152-6158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 99. | Fachal L, Gómez-Caamaño A, Celeiro-Muñoz C, Peleteiro P, Blanco A, Carballo A, Forteza J, Carracedo A, Vega A. BRCA1 mutations do not increase prostate cancer risk: results from a meta-analysis including new data. Prostate. 2011;71:1768-1779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 100. | Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, Mahmud N, Dadaev T, Govindasami K, Guy M. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748-1757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 618] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 101. | Douglas JA, Levin AM, Zuhlke KA, Ray AM, Johnson GR, Lange EM, Wood DP, Cooney KA. Common variation in the BRCA1 gene and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1510-1516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 102. | Sundararajan S, Ahmed A, Goodman OB. The relevance of BRCA genetics to prostate cancer pathogenesis and treatment. Clin Adv Hematol Oncol. 2011;9:748-755. [PubMed] |
| 103. | Rabiau N, Déchelotte P, Adjakly M, Kemeny JL, Guy L, Boiteux JP, Kwiatkowski F, Bignon YJ, Bernard-Gallon D. BRCA1, BRCA2, AR and IGF-I expression in prostate cancer: correlation between RT-qPCR and immunohistochemical detection. Oncol Rep. 2011;26:695-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 104. | Schayek H, Haugk K, Sun S, True LD, Plymate SR, Werner H. Tumor suppressor BRCA1 is expressed in prostate cancer and controls insulin-like growth factor I receptor (IGF-IR) gene transcription in an androgen receptor-dependent manner. Clin Cancer Res. 2009;15:1558-1565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 105. | Fiorentino M, Judson G, Penney K, Flavin R, Stark J, Fiore C, Fall K, Martin N, Ma J, Sinnott J. Immunohistochemical expression of BRCA1 and lethal prostate cancer. Cancer Res. 2010;70:3136-3139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 106. | De Luca P, Vazquez ES, Moiola CP, Zalazar F, Cotignola J, Gueron G, Gardner K, De Siervi A. BRCA1 loss induces GADD153-mediated doxorubicin resistance in prostate cancer. Mol Cancer Res. 2011;9:1078-1090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 107. | Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, Hylands L, Riisnaes R, Forster M, Omlin A. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2013;14:882-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 438] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 108. | Olaparib shows promise in multiple tumor types. Cancer Discov. 2013;3:OF5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 109. | Comstock CE, Revelo MP, Buncher CR, Knudsen KE. Impact of differential cyclin D1 expression and localisation in prostate cancer. Br J Cancer. 2007;96:970-979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 110. | Nikoleishvili D, Pertia A, Trsintsadze O, Gogokhia N, Managadze L, Chkhotua A. Expression of p27((Kip1)), cyclin D3 and Ki67 in BPH, prostate cancer and hormone-treated prostate cancer cells. Int Urol Nephrol. 2008;40:953-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 111. | Schiewer MJ, Morey LM, Burd CJ, Liu Y, Merry DE, Ho SM, Knudsen KE. Cyclin D1 repressor domain mediates proliferation and survival in prostate cancer. Oncogene. 2009;28:1016-1027. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 112. | Ju X, Casimiro MC, Gormley M, Meng H, Jiao X, Katiyar S, Crosariol M, Chen K, Wang M, Quong AA. Identification of a cyclin D1 network in prostate cancer that antagonizes epithelial-mesenchymal restraint. Cancer Res. 2014;74:508-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 113. | Comstock CE, Augello MA, Schiewer MJ, Karch J, Burd CJ, Ertel A, Knudsen ES, Jessen WJ, Aronow BJ, Knudsen KE. Cyclin D1 is a selective modifier of androgen-dependent signaling and androgen receptor function. J Biol Chem. 2011;286:8117-8127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 114. | Olshavsky NA, Groh EM, Comstock CE, Morey LM, Wang Y, Revelo MP, Burd C, Meller J, Knudsen KE. Cyclin D3 action in androgen receptor regulation and prostate cancer. Oncogene. 2008;27:3111-3121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 115. | Chung LC, Tsui KH, Feng TH, Lee SL, Chang PL, Juang HH. L-Mimosine blocks cell proliferation via upregulation of B-cell translocation gene 2 and N-myc downstream regulated gene 1 in prostate carcinoma cells. Am J Physiol Cell Physiol. 2012;302:C676-C685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 116. | Ouyang DY, Zeng LH, Pan H, Xu LH, Wang Y, Liu KP, He XH. Piperine inhibits the proliferation of human prostate cancer cells via induction of cell cycle arrest and autophagy. Food Chem Toxicol. 2013;60:424-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 117. | Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73:596-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 118. | Noonan EJ, Place RF, Basak S, Pookot D, Li LC. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget. 2010;1:349-358. [PubMed] |
| 119. | Gregory CW, Johnson RT, Presnell SC, Mohler JL, French FS. Androgen receptor regulation of G1 cyclin and cyclin-dependent kinase function in the CWR22 human prostate cancer xenograft. J Androl. 2001;22:537-548. [PubMed] |
| 120. | Lu S, Tsai SY, Tsai MJ. Regulation of androgen-dependent prostatic cancer cell growth: androgen regulation of CDK2, CDK4, and CKI p16 genes. Cancer Res. 1997;57:4511-4516. [PubMed] |
| 121. | Flores O, Wang Z, Knudsen KE, Burnstein KL. Nuclear targeting of cyclin-dependent kinase 2 reveals essential roles of cyclin-dependent kinase 2 localization and cyclin E in vitamin D-mediated growth inhibition. Endocrinology. 2010;151:896-908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 122. | Cipriano SC, Chen YQ. Insensitivity to growth inhibition by TGF-beta1 correlates with a lack of inhibition of the CDK2 activity in prostate carcinoma cells. Oncogene. 1998;17:1549-1556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 123. | Gulappa T, Reddy RS, Suman S, Nyakeriga AM, Damodaran C. Molecular interplay between cdk4 and p21 dictates G0/G1 cell cycle arrest in prostate cancer cells. Cancer Lett. 2013;337:177-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 124. | Fang Y, DeMarco VG, Nicholl MB. Resveratrol enhances radiation sensitivity in prostate cancer by inhibiting cell proliferation and promoting cell senescence and apoptosis. Cancer Sci. 2012;103:1090-1098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 125. | Marconett CN, Morgenstern TJ, San Roman AK, Sundar SN, Singhal AK, Firestone GL. BZL101, a phytochemical extract from the Scutellaria barbata plant, disrupts proliferation of human breast and prostate cancer cells through distinct mechanisms dependent on the cancer cell phenotype. Cancer Biol Ther. 2010;10:397-405. [PubMed] |
| 126. | Agarwal C, Dhanalakshmi S, Singh RP, Agarwal R. Inositol hexaphosphate inhibits growth and induces G1 arrest and apoptotic death of androgen-dependent human prostate carcinoma LNCaP cells. Neoplasia. 2004;6:646-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 127. | Ukomadu C, Dutta A. Inhibition of cdk2 activating phosphorylation by mevastatin. J Biol Chem. 2003;278:4840-4846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 128. | Li R, Erdamar S, Dai H, Sayeeduddin M, Frolov A, Wheeler TM, Ayala GE. Cytoplasmic accumulation of glycogen synthase kinase-3beta is associated with aggressive clinicopathological features in human prostate cancer. Anticancer Res. 2009;29:2077-2081. [PubMed] |
| 129. | Mulholland DJ, Dedhar S, Wu H, Nelson CC. PTEN and GSK3beta: key regulators of progression to androgen-independent prostate cancer. Oncogene. 2006;25:329-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 168] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 130. | Wang L, Lin HK, Hu YC, Xie S, Yang L, Chang C. Suppression of androgen receptor-mediated transactivation and cell growth by the glycogen synthase kinase 3 beta in prostate cells. J Biol Chem. 2004;279:32444-32452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 71] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 131. | Salas TR, Kim J, Vakar-Lopez F, Sabichi AL, Troncoso P, Jenster G, Kikuchi A, Chen SY, Shemshedini L, Suraokar M. Glycogen synthase kinase-3 beta is involved in the phosphorylation and suppression of androgen receptor activity. J Biol Chem. 2004;279:19191-19200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 132. | Schütz SV, Cronauer MV, Rinnab L. Inhibition of glycogen synthase kinase-3beta promotes nuclear export of the androgen receptor through a CRM1-dependent mechanism in prostate cancer cell lines. J Cell Biochem. 2010;109:1192-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 133. | Wang H, Fang R, Wang XF, Zhang F, Chen DY, Zhou B, Wang HS, Cai SH, Du J. Stabilization of Snail through AKT/GSK-3β signaling pathway is required for TNF-α-induced epithelial-mesenchymal transition in prostate cancer PC3 cells. Eur J Pharmacol. 2013;714:48-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 134. | Liao X, Zhang L, Thrasher JB, Du J, Li B. Glycogen synthase kinase-3beta suppression eliminates tumor necrosis factor-related apoptosis-inducing ligand resistance in prostate cancer. Mol Cancer Ther. 2003;2:1215-1222. [PubMed] |
| 135. | Lu Y, Nie D, Witt WT, Chen Q, Shen M, Xie H, Lai L, Dai Y, Zhang J. Expression of the fat-1 gene diminishes prostate cancer growth in vivo through enhancing apoptosis and inhibiting GSK-3 beta phosphorylation. Mol Cancer Ther. 2008;7:3203-3211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 136. | Ban JO, Oh JH, Son SM, Won D, Song HS, Han SB, Moon DC, Kang KW, Song MJ, Hong JT. Troglitazone, a PPAR agonist, inhibits human prostate cancer cell growth through inactivation of NFκB via suppression of GSK-3β expression. Cancer Biol Ther. 2011;12:288-296. [PubMed] |
| 137. | Li Y, Wang Z, Kong D, Li R, Sarkar SH, Sarkar FH. Regulation of Akt/FOXO3a/GSK-3beta/AR signaling network by isoflavone in prostate cancer cells. J Biol Chem. 2008;283:27707-27716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 138. | Zhu H, Han B, Pan X, Qi H, Xu L. Thiazolidenediones induce tumour-cell apoptosis through the Akt-GSK3β pathway. J Clin Pharm Ther. 2012;37:65-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 139. | Benelli R, Monteghirfo S, Venè R, Tosetti F, Ferrari N. The chemopreventive retinoid 4HPR impairs prostate cancer cell migration and invasion by interfering with FAK/AKT/GSK3beta pathway and beta-catenin stability. Mol Cancer. 2010;9:142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 140. | Barneda-Zahonero B, Parra M. Histone deacetylases and cancer. Mol Oncol. 2012;6:579-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 354] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 141. | Weichert W, Röske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604-610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 352] [Cited by in RCA: 379] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 142. | Waltregny D, North B, Van Mellaert F, de Leval J, Verdin E, Castronovo V. Screening of histone deacetylases (HDAC) expression in human prostate cancer reveals distinct class I HDAC profiles between epithelial and stromal cells. Eur J Histochem. 2004;48:273-290. [PubMed] |
| 143. | Song Y, Shiota M, Tamiya S, Kuroiwa K, Naito S, Tsuneyoshi M. The significance of strong histone deacetylase 1 expression in the progression of prostate cancer. Histopathology. 2011;58:773-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 144. | Kim NH, Kim SN, Kim YK. Involvement of HDAC1 in E-cadherin expression in prostate cancer cells; its implication for cell motility and invasion. Biochem Biophys Res Commun. 2011;404:915-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 145. | Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277:25904-25913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 233] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 146. | Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 378] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 147. | Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 889] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 148. | Ai J, Wang Y, Dar JA, Liu J, Liu L, Nelson JB, Wang Z. HDAC6 regulates androgen receptor hypersensitivity and nuclear localization via modulating Hsp90 acetylation in castration-resistant prostate cancer. Mol Endocrinol. 2009;23:1963-1972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 149. | Gibbs A, Schwartzman J, Deng V, Alumkal J. Sulforaphane destabilizes the androgen receptor in prostate cancer cells by inactivating histone deacetylase 6. Proc Natl Acad Sci USA. 2009;106:16663-16668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 123] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 150. | Clarke JD, Hsu A, Yu Z, Dashwood RH, Ho E. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol Nutr Food Res. 2011;55:999-1009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 151. | Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714-1724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 292] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 152. | Li X, Kaplun A, Lonardo F, Heath E, Sarkar FH, Irish J, Sakr W, Sheng S. HDAC1 inhibition by maspin abrogates epigenetic silencing of glutathione S-transferase pi in prostate carcinoma cells. Mol Cancer Res. 2011;9:733-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 153. | Lavery DN, Villaronga MA, Walker MM, Patel A, Belandia B, Bevan CL. Repression of androgen receptor activity by HEYL, a third member of the Hairy/Enhancer-of-split-related family of Notch effectors. J Biol Chem. 2011;286:17796-17808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 154. | Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860-866. [PubMed] |
| 155. | Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 799] [Cited by in RCA: 857] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 156. | Gasparian AV, Yao YJ, Kowalczyk D, Lyakh LA, Karseladze A, Slaga TJ, Budunova IV. The role of IKK in constitutive activation of NF-kappaB transcription factor in prostate carcinoma cells. J Cell Sci. 2002;115:141-151. [PubMed] |
| 157. | Häggarth L, Hägglöf C, Jaraj SJ, Wester K, Pontén F, Ostman A, Egevad L. Diagnostic biomarkers of prostate cancer. Scand J Urol Nephrol. 2011;45:60-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 158. | Xu C, Shen G, Chen C, Gélinas C, Kong AN. Suppression of NF-kappaB and NF-kappaB-regulated gene expression by sulforaphane and PEITC through IkappaBalpha, IKK pathway in human prostate cancer PC-3 cells. Oncogene. 2005;24:4486-4495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 236] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 159. | Shirley RB, Kaddour-Djebbar I, Patel DM, Lakshmikanthan V, Lewis RW, Kumar MV. Combination of proteasomal inhibitors lactacystin and MG132 induced synergistic apoptosis in prostate cancer cells. Neoplasia. 2005;7:1104-1111. [PubMed] |
| 160. | Inoue T, Kon T, Ohkura R, Yamakawa H, Ohara O, Yokota J, Sutoh K. BREK/LMTK2 is a myosin VI-binding protein involved in endosomal membrane trafficking. Genes Cells. 2008;13:483-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 161. | Puri C, Chibalina MV, Arden SD, Kruppa AJ, Kendrick-Jones J, Buss F. Overexpression of myosin VI in prostate cancer cells enhances PSA and VEGF secretion, but has no effect on endocytosis. Oncogene. 2010;29:188-200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 162. | Guy M, Kote-Jarai Z, Giles GG, Al Olama AA, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI. Identification of new genetic risk factors for prostate cancer. Asian J Androl. 2009;11:49-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 163. | Harries LW, Perry JR, McCullagh P, Crundwell M. Alterations in LMTK2, MSMB and HNF1B gene expression are associated with the development of prostate cancer. BMC Cancer. 2010;10:315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 164. | Kesavapany S, Lau KF, Ackerley S, Banner SJ, Shemilt SJ, Cooper JD, Leigh PN, Shaw CE, McLoughlin DM, Miller CC. Identification of a novel, membrane-associated neuronal kinase, cyclin-dependent kinase 5/p35-regulated kinase. J Neurosci. 2003;23:4975-4983. [PubMed] |
| 165. | Eto M, Elliott E, Prickett TD, Brautigan DL. Inhibitor-2 regulates protein phosphatase-1 complexed with NimA-related kinase to induce centrosome separation. J Biol Chem. 2002;277:44013-44020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 80] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 166. | Tansey WP. Mammalian MYC Proteins and Cancer. New J Sci. 2014;2014:27. [RCA] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 167. | Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, Hicks JL, Morgan J, Cornish TC, Sutcliffe S. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156-1167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 352] [Cited by in RCA: 340] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 168. | Gil J, Kerai P, Lleonart M, Bernard D, Cigudosa JC, Peters G, Carnero A, Beach D. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 2005;65:2179-2185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 91] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 169. | Amatangelo MD, Goodyear S, Varma D, Stearns ME. c-Myc expression and MEK1-induced Erk2 nuclear localization are required for TGF-beta induced epithelial-mesenchymal transition and invasion in prostate cancer. Carcinogenesis. 2012;33:1965-1975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 170. | Grad JM, Dai JL, Wu S, Burnstein KL. Multiple androgen response elements and a Myc consensus site in the androgen receptor (AR) coding region are involved in androgen-mediated up-regulation of AR messenger RNA. Mol Endocrinol. 1999;13:1896-1911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 78] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 171. | Hawksworth D, Ravindranath L, Chen Y, Furusato B, Sesterhenn IA, McLeod DG, Srivastava S, Petrovics G. Overexpression of C-MYC oncogene in prostate cancer predicts biochemical recurrence. Prostate Cancer Prostatic Dis. 2010;13:311-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 122] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 172. | Fromont G, Godet J, Peyret A, Irani J, Celhay O, Rozet F, Cathelineau X, Cussenot O. 8q24 amplification is associated with Myc expression and prostate cancer progression and is an independent predictor of recurrence after radical prostatectomy. Hum Pathol. 2013;44:1617-1623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 173. | Antonarakis ES, Keizman D, Zhang Z, Gurel B, Lotan TL, Hicks JL, Fedor HL, Carducci MA, De Marzo AM, Eisenberger MA. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer. 2012;118:6063-6071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 174. | Hatano K, Yamaguchi S, Nimura K, Murakami K, Nagahara A, Fujita K, Uemura M, Nakai Y, Tsuchiya M, Nakayama M. Residual prostate cancer cells after docetaxel therapy increase the tumorigenic potential via constitutive signaling of CXCR4, ERK1/2 and c-Myc. Mol Cancer Res. 2013;11:1088-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 175. | Tawfic S, Goueli SA, Olson MO, Ahmed K. Androgenic regulation of phosphorylation and stability of nucleolar protein nucleolin in rat ventral prostate. Prostate. 1994;24:101-106. [PubMed] |
| 176. | Tate A, Isotani S, Bradley MJ, Sikes RA, Davis R, Chung LW, Edlund M. Met-Independent Hepatocyte Growth Factor-mediated regulation of cell adhesion in human prostate cancer cells. BMC Cancer. 2006;6:197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 177. | Altintas DM, Vlaeminck V, Angelov D, Dimitrov S, Samarut J. Cell cycle regulated expression of NCoR might control cyclic expression of androgen responsive genes in an immortalized prostate cell line. Mol Cell Endocrinol. 2011;332:149-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 178. | Choi HK, Yoo JY, Jeong MH, Park SY, Shin DM, Jang SW, Yoon HG, Choi KC. Protein kinase A phosphorylates NCoR to enhance its nuclear translocation and repressive function in human prostate cancer cells. J Cell Physiol. 2013;228:1159-1165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 179. | Battaglia S, Maguire O, Thorne JL, Hornung LB, Doig CL, Liu S, Sucheston LE, Bianchi A, Khanim FL, Gommersall LM. Elevated NCOR1 disrupts PPARalpha/gamma signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis. 2010;31:1650-1660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 180. | Wang Y, Li JQ, Shao C, Shi CH, Liu F, Yang ZY, Qiu JX, Li YM, Fu Q, Zhang W. Androgen receptor coregulators NOCR1, TIF2, and ARA70 may account for the hydroxyflutamide insensitivity of prostate cancer cells. Ir J Med Sci. 2011;180:865-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 181. | Schrantz N, da Silva Correia J, Fowler B, Ge Q, Sun Z, Bokoch GM. Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. J Biol Chem. 2004;279:1922-1931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 182. | Kaur R, Yuan X, Lu ML, Balk SP. Increased PAK6 expression in prostate cancer and identification of PAK6 associated proteins. Prostate. 2008;68:1510-1516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 183. | Yang F, Li X, Sharma M, Zarnegar M, Lim B, Sun Z. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J Biol Chem. 2001;276:15345-15353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 122] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 184. | Lee SR, Ramos SM, Ko A, Masiello D, Swanson KD, Lu ML, Balk SP. AR and ER interaction with a p21-activated kinase (PAK6). Mol Endocrinol. 2002;16:85-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 185. | Liu T, Li Y, Gu H, Zhu G, Li J, Cao L, Li F. p21-Activated kinase 6 (PAK6) inhibits prostate cancer growth via phosphorylation of androgen receptor and tumorigenic E3 ligase murine double minute-2 (Mdm2). J Biol Chem. 2013;288:3359-3369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 186. | Wen X, Li X, Liao B, Liu Y, Wu J, Yuan X, Ouyang B, Sun Q, Gao X. Knockdown of p21-activated kinase 6 inhibits prostate cancer growth and enhances chemosensitivity to docetaxel. Urology. 2009;73:1407-1411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 187. | Zhang M, Siedow M, Saia G, Chakravarti A. Inhibition of p21-activated kinase 6 (PAK6) increases radiosensitivity of prostate cancer cells. Prostate. 2010;70:807-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 188. | Uzoh CC, Perks CM, Bahl A, Holly JM, Sugiono M, Persad RA. PTEN-mediated pathways and their association with treatment-resistant prostate cancer. BJU Int. 2009;104:556-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 189. | Shen MM, Abate-Shen C. Pten inactivation and the emergence of androgen-independent prostate cancer. Cancer Res. 2007;67:6535-6538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 105] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 190. | Tian L, Fang YX, Xue JL, Chen JZ. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS One. 2013;8:e75885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 191. | Kong L, Schäfer G, Bu H, Zhang Y, Zhang Y, Klocker H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 2012;33:751-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 192. | Conley-LaComb MK, Saliganan A, Kandagatla P, Chen YQ, Cher ML, Chinni SR. PTEN loss mediated Akt activation promotes prostate tumor growth and metastasis via CXCL12/CXCR4 signaling. Mol Cancer. 2013;12:85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 193. | Conley-LaComb MK, Huang W, Wang S, Shi D, Jung YS, Najy A, Fridman R, Bonfil RD, Cher ML, Chen YQ. PTEN regulates PDGF ligand switch for β-PDGFR signaling in prostate cancer. Am J Pathol. 2012;180:1017-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 194. | Fan C, He L, Kapoor A, Rybak AP, De Melo J, Cutz JC, Tang D. PTEN inhibits BMI1 function independently of its phosphatase activity. Mol Cancer. 2009;8:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 195. | Chetram MA, Odero-Marah V, Hinton CV. Loss of PTEN permits CXCR4-mediated tumorigenesis through ERK1/2 in prostate cancer cells. Mol Cancer Res. 2011;9:90-102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 196. | Nagle RB, Algotar AM, Cortez CC, Smith K, Jones C, Sathyanarayana UG, Yun S, Riley J, Nagy D, Dittamore R. ERG overexpression and PTEN status predict capsular penetration in prostate carcinoma. Prostate. 2013;73:1233-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 197. | Gabriel K, Ingram A, Austin R, Kapoor A, Tang D, Majeed F, Qureshi T, Al-Nedawi K. Regulation of the tumor suppressor PTEN through exosomes: a diagnostic potential for prostate cancer. PLoS One. 2013;8:e70047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 198. | Lotan TL, Gumuskaya B, Rahimi H, Hicks JL, Iwata T, Robinson BD, Epstein JI, De Marzo AM. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol. 2013;26:587-603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 199. | Cuzick J, Yang ZH, Fisher G, Tikishvili E, Stone S, Lanchbury JS, Camacho N, Merson S, Brewer D, Cooper CS. Prognostic value of PTEN loss in men with conservatively managed localised prostate cancer. Br J Cancer. 2013;108:2582-2589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 200. | Chaux A, Peskoe SB, Gonzalez-Roibon N, Schultz L, Albadine R, Hicks J, De Marzo AM, Platz EA, Netto GJ. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol. 2012;25:1543-1549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 201. | Cathomas R, Rothermundt C, Klingbiel D, Bubendorf L, Jaggi R, Betticher DC, Brauchli P, Cotting D, Droege C, Winterhalder R. Efficacy of cetuximab in metastatic castration-resistant prostate cancer might depend on EGFR and PTEN expression: results from a phase II trial (SAKK 08/07). Clin Cancer Res. 2012;18:6049-6057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 202. | Wang Y, Romigh T, He X, Orloff MS, Silverman RH, Heston WD, Eng C. Resveratrol regulates the PTEN/AKT pathway through androgen receptor-dependent and -independent mechanisms in prostate cancer cell lines. Hum Mol Genet. 2010;19:4319-4329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 203. | Menon R, Deng M, Rüenauver K, Queisser A, Peifer M, Offermann A, Boehm D, Vogel W, Scheble V, Fend F. Somatic copy number alterations by whole-exome sequencing implicates YWHAZ and PTK2 in castration-resistant prostate cancer. J Pathol. 2013;231:505-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 204. | Tremblay L, Hauck W, Aprikian AG, Begin LR, Chapdelaine A, Chevalier S. Focal adhesion kinase (pp125FAK) expression, activation and association with paxillin and p50CSK in human metastatic prostate carcinoma. Int J Cancer. 1996;68:164-171. [PubMed] |
| 205. | Lee LF, Louie MC, Desai SJ, Yang J, Chen HW, Evans CP, Kung HJ. Interleukin-8 confers androgen-independent growth and migration of LNCaP: differential effects of tyrosine kinases Src and FAK. Oncogene. 2004;23:2197-2205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 139] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 206. | El Haibi CP, Sharma PK, Singh R, Johnson PR, Suttles J, Singh S, Lillard JW. PI3Kp110-, Src-, FAK-dependent and DOCK2-independent migration and invasion of CXCL13-stimulated prostate cancer cells. Mol Cancer. 2010;9:85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 207. | Guo X, Yin S, Dong Y, Fan L, Ye M, Lu J, Hu H. Enhanced apoptotic effects by the combination of curcumin and methylseleninic acid: potential role of Mcl-1 and FAK. Mol Carcinog. 2013;52:879-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 208. | Bergan R, Kyle E, Nguyen P, Trepel J, Ingui C, Neckers L. Genistein-stimulated adherence of prostate cancer cells is associated with the binding of focal adhesion kinase to beta-1-integrin. Clin Exp Metastasis. 1996;14:389-398. [PubMed] |
| 209. | Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 576] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 210. | Van Tilborgh N, Spans L, Helsen C, Clinckemalie L, Dubois V, Lerut E, Boonen S, Vanderschueren D, Claessens F. The transcription intermediary factor 1β coactivates the androgen receptor. J Endocrinol Invest. 2013;36:699-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 211. | Zhang Z, Yang Z, Jäämaa S, Liu H, Pellakuru LG, Iwata T, af Hällström TM, De Marzo AM, Laiho M. Differential epithelium DNA damage response to ATM and DNA-PK pathway inhibition in human prostate tissue culture. Cell Cycle. 2011;10:3545-3553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 212. | Aparicio A, Den RB, Knudsen KE. Time to stratify? The retinoblastoma protein in castrate-resistant prostate cancer. Nat Rev Urol. 2011;8:562-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 213. | Dean JL, Knudsen KE. The role of tumor suppressor dysregulation in prostate cancer progression. Curr Drug Targets. 2013;14:460-471. [PubMed] |
| 214. | Lee JT, Lehmann BD, Terrian DM, Chappell WH, Stivala F, Libra M, Martelli AM, Steelman LS, McCubrey JA. Targeting prostate cancer based on signal transduction and cell cycle pathways. Cell Cycle. 2008;7:1745-1762. [PubMed] |
| 215. | Mariot P, Prevarskaya N, Roudbaraki MM, Le Bourhis X, Van Coppenolle F, Vanoverberghe K, Skryma R. Evidence of functional ryanodine receptor involved in apoptosis of prostate cancer (LNCaP) cells. Prostate. 2000;43:205-214. [PubMed] |
| 216. | Kobylewski SE, Henderson KA, Eckhert CD. Identification of ryanodine receptor isoforms in prostate DU-145, LNCaP, and PWR-1E cells. Biochem Biophys Res Commun. 2012;425:431-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 217. | Tang L, Nogales E, Ciferri C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol. 2010;102:122-128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 165] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 218. | Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392-1398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 483] [Cited by in RCA: 548] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 219. | Brush MH, Guardiola A, Connor JH, Yao TP, Shenolikar S. Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J Biol Chem. 2004;279:7685-7691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 105] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 220. | Schillace RV, Scott JD. Association of the type 1 protein phosphatase PP1 with the A-kinase anchoring protein AKAP220. Curr Biol. 1999;9:321-324. [PubMed] |
| 221. | Schillace RV, Voltz JW, Sim AT, Shenolikar S, Scott JD. Multiple interactions within the AKAP220 signaling complex contribute to protein phosphatase 1 regulation. J Biol Chem. 2001;276:12128-12134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 222. | Ayllón V, Cayla X, García A, Fleischer A, Rebollo A. The anti-apoptotic molecules Bcl-xL and Bcl-w target protein phosphatase 1alpha to Bad. Eur J Immunol. 2002;32:1847-1855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 223. | Li HY, Liu H, Wang CH, Zhang JY, Man JH, Gao YF, Zhang PJ, Li WH, Zhao J, Pan X. Deactivation of the kinase IKK by CUEDC2 through recruitment of the phosphatase PP1. Nat Immunol. 2008;9:533-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 224. | Gunawardena SR, Ruis BL, Meyer JA, Kapoor M, Conklin KF. NOM1 targets protein phosphatase I to the nucleolus. J Biol Chem. 2008;283:398-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 225. | Kim YM, Watanabe T, Allen PB, Kim YM, Lee SJ, Greengard P, Nairn AC, Kwon YG. PNUTS, a protein phosphatase 1 (PP1) nuclear targeting subunit. Characterization of its PP1- and RNA-binding domains and regulation by phosphorylation. J Biol Chem. 2003;278:13819-13828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 226. | Landsverk HB, Kirkhus M, Bollen M, Küntziger T, Collas P. PNUTS enhances in vitro chromosome decondensation in a PP1-dependent manner. Biochem J. 2005;390:709-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 227. | Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292-1303. [PubMed] |
| 228. | Lee SJ, Lim CJ, Min JK, Lee JK, Kim YM, Lee JY, Won MH, Kwon YG. Protein phosphatase 1 nuclear targeting subunit is a hypoxia inducible gene: its role in post-translational modification of p53 and MDM2. Cell Death Differ. 2007;14:1106-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 229. | Guo CY, Brautigan DL, Larner JM. Ionizing radiation activates nuclear protein phosphatase-1 by ATM-dependent dephosphorylation. J Biol Chem. 2002;277:41756-41761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 230. | Tang X, Hui ZG, Cui XL, Garg R, Kastan MB, Xu B. A novel ATM-dependent pathway regulates protein phosphatase 1 in response to DNA damage. Mol Cell Biol. 2008;28:2559-2566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 231. | Shimada M, Haruta M, Niida H, Sawamoto K, Nakanishi M. Protein phosphatase 1γ is responsible for dephosphorylation of histone H3 at Thr 11 after DNA damage. EMBO Rep. 2010;11:883-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 232. | Guo C, Mi J, Brautigan DL, Larner JM. ATM regulates ionizing radiation-induced disruption of HDAC1: PP1: Rb complexes. Cell Signal. 2007;19:504-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 233. | Sakashita G, Shima H, Komatsu M, Urano T, Kikuchi A, Kikuchi K. Regulation of type 1 protein phosphatase/inhibitor-2 complex by glycogen synthase kinase-3beta in intact cells. J Biochem. 2003;133:165-171. [PubMed] |
| 234. | Wu DY, Tkachuck DC, Roberson RS, Schubach WH. The human SNF5/INI1 protein facilitates the function of the growth arrest and DNA damage-inducible protein (GADD34) and modulates GADD34-bound protein phosphatase-1 activity. J Biol Chem. 2002;277:27706-27715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 235. | Liu Y, Virshup DM, White RL, Hsu LC. Regulation of BRCA1 phosphorylation by interaction with protein phosphatase 1alpha. Cancer Res. 2002;62:6357-6361. [PubMed] |
| 236. | Manser C, Guillot F, Vagnoni A, Davies J, Lau KF, McLoughlin DM, De Vos KJ, Miller CC. Lemur tyrosine kinase-2 signalling regulates kinesin-1 light chain-2 phosphorylation and binding of Smad2 cargo. Oncogene. 2012;31:2773-2782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 237. | Ajuh PM, Browne GJ, Hawkes NA, Cohen PT, Roberts SG, Lamond AI. Association of a protein phosphatase 1 activity with the human factor C1 (HCF) complex. Nucleic Acids Res. 2000;28:678-686. [PubMed] |
| 238. | Lesage B, Beullens M, Pedelini L, Garcia-Gimeno MA, Waelkens E, Sanz P, Bollen M. A complex of catalytically inactive protein phosphatase-1 sandwiched between Sds22 and inhibitor-3. Biochemistry. 2007;46:8909-8919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 239. | Helps NR, Barker HM, Elledge SJ, Cohen PT. Protein phosphatase 1 interacts with p53BP2, a protein which binds to the tumour suppressor p53. FEBS Lett. 1995;377:295-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 104] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 240. | Garcia A, Cayla X, Guergnon J, Dessauge F, Hospital V, Rebollo MP, Fleischer A, Rebollo A. Serine/threonine protein phosphatases PP1 and PP2A are key players in apoptosis. Biochimie. 2003;85:721-726. [PubMed] |
| 241. | Parrish AB. Regulation of Apoptosis Following Mitochondrial Cytochrome c Release. Department of Pharmacology and Molecular Cancer Biology. USA: Duke University 2010; . |
| 242. | Ayllón V, Martínez-A C, García A, Cayla X, Rebollo A. Protein phosphatase 1alpha is a Ras-activated Bad phosphatase that regulates interleukin-2 deprivation-induced apoptosis. EMBO J. 2000;19:2237-2246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 111] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 243. | Ayllón V, Cayla X, García A, Roncal F, Fernández R, Albar JP, Martínez C, Rebollo A. Bcl-2 targets protein phosphatase 1 alpha to Bad. J Immunol. 2001;166:7345-7352. [PubMed] |
| 244. | Luo W, Peterson A, Garcia BA, Coombs G, Kofahl B, Heinrich R, Shabanowitz J, Hunt DF, Yost HJ, Virshup DM. Protein phosphatase 1 regulates assembly and function of the beta-catenin degradation complex. EMBO J. 2007;26:1511-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 107] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 245. | Yu YM, Pace SM, Allen SR, Deng CX, Hsu LC. A PP1-binding motif present in BRCA1 plays a role in its DNA repair function. Int J Biol Sci. 2008;4:352-361. [PubMed] |
| 246. | Hsu LC. Identification and functional characterization of a PP1-binding site in BRCA1. Biochem Biophys Res Commun. 2007;360:507-512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 247. | Szatmari E, Habas A, Yang P, Zheng JJ, Hagg T, Hetman M. A positive feedback loop between glycogen synthase kinase 3beta and protein phosphatase 1 after stimulation of NR2B NMDA receptors in forebrain neurons. J Biol Chem. 2005;280:37526-37535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 248. | Canettieri G, Morantte I, Guzmán E, Asahara H, Herzig S, Anderson SD, Yates JR, Montminy M. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat Struct Biol. 2003;10:175-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 160] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 249. | Zhao S, Brandt NR, Caswell AH, Lee EY. Binding of the catalytic subunit of protein phosphatase-1 to the ryanodine-sensitive calcium release channel protein. Biochemistry. 1998;37:18102-18109. [PubMed] |
| 250. | Sonnleitner A, Fleischer S, Schindler H. Gating of the skeletal calcium release channel by ATP is inhibited by protein phosphatase 1 but not by Mg2+. Cell Calcium. 1997;21:283-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 251. | Xiao B, Tian X, Xie W, Jones PP, Cai S, Wang X, Jiang D, Kong H, Zhang L, Chen K. Functional consequence of protein kinase A-dependent phosphorylation of the cardiac ryanodine receptor: sensitization of store overload-induced Ca2+ release. J Biol Chem. 2007;282:30256-30264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 67] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 252. | Lu Z, Wan G, Guo H, Zhang X, Lu X. Protein phosphatase 1 inhibits p53 signaling by dephosphorylating and stabilizing Mdmx. Cell Signal. 2013;25:796-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 253. | Fresu M, Bianchi M, Parsons JT, Villa-Moruzzi E. Cell-cycle-dependent association of protein phosphatase 1 and focal adhesion kinase. Biochem J. 2001;358:407-414. [PubMed] |
| 254. | Brichese L, Valette A. PP1 phosphatase is involved in Bcl-2 dephosphorylation after prolonged mitotic arrest induced by paclitaxel. Biochem Biophys Res Commun. 2002;294:504-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 255. | Li X, Lin HH, Chen H, Xu X, Shih HM, Ann DK. SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci Signal. 2010;3:ra32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 256. | Chuang MJ, Wu ST, Tang SH, Lai XM, Lai HC, Hsu KH, Sun KH, Sun GH, Chang SY, Yu DS. The HDAC inhibitor LBH589 induces ERK-dependent prometaphase arrest in prostate cancer via HDAC6 inactivation and down-regulation. PLoS One. 2013;8:e73401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 257. | Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1094] [Cited by in RCA: 1203] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 258. | Chen CS, Weng SC, Tseng PH, Lin HP, Chen CS. Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J Biol Chem. 2005;280:38879-38887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 259. | Guergnon J, Dessauge F, Dominguez V, Viallet J, Bonnefoy S, Yuste VJ, Mercereau-Puijalon O, Cayla X, Rebollo A, Susin SA. Use of penetrating peptides interacting with PP1/PP2A proteins as a general approach for a drug phosphatase technology. Mol Pharmacol. 2006;69:1115-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 41] [Reference Citation Analysis (0)] |
| 260. | Godet AN, Guergnon J, Maire V, Croset A, Garcia A. The combinatorial PP1-binding consensus Motif (R/K)x( (0,1))V/IxFxx(R/K)x(R/K) is a new apoptotic signature. PLoS One. 2010;5:e9981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |