Published online Aug 27, 2014. doi: 10.5496/wjmg.v4.i3.69
Revised: May 13, 2014
Accepted: May 16, 2014
Published online: August 27, 2014
Processing time: 265 Days and 11.4 Hours
Recently, an epoch-making genome engineering technology using clustered regularly at interspaced short palindromic repeats (CRISPR) and CRISPR associated (Cas) nucleases, was developed. Previous technologies for genome manipulation require the time-consuming design and construction of genome-engineered nucleases for each target and have, therefore, not been widely used in mouse research where standard techniques based on homologous recombination are commonly used. The CRISPR/Cas system only requires the design of sequences complementary to a target locus, making this technology fast and straightforward. In addition, CRISPR/Cas can be used to generate mice carrying mutations in multiple genes in a single step, an achievement not possible using other methods. Here, we review the uses of this technology in genetic analysis and manipulation, including achievements made possible to date and the prospects for future therapeutic applications.
Core tip: This review introduces the latest information about the genome manipulation technology of the clustered regularly at interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) system to readers. We focus particularly on the application of CRISPR/Cas in mammalian cultured cells and mice. The problems of off-target effects and the prospects for therapeutic applications of CRISPR/Cas in the future are also discussed.
- Citation: Horii T, Hatada I. Genome engineering using the CRISPR/Cas system. World J Med Genet 2014; 4(3): 69-76
- URL: https://www.wjgnet.com/2220-3184/full/v4/i3/69.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v4.i3.69
The recent development of site-specific endonuclease technologies for selective genome cleavage has been an important advance in mammalian genome engineering. Zinc-finger nucleases (ZFNs) consist of specific DNA-binding zinc-finger proteins and a nuclease domain of the Fok I endonuclease[1-3]. Cleavage with Fok I requires dimerization of the protein; therefore, fusion to a pair of zinc-finger proteins provides target specificity, and allows cleavage of the target DNA locus, generating double-strand breaks (DSBs).
On the other hand, transcription activator-like effector (TALE) nucleases (TALENs) are fusions of DNA-binding domain TALE repeats with the cleavage domain of the Fok I restriction enzyme. TALE repeats are highly conserved 33-35 amino acid sequences found in naturally occurring TALEs encoded by Xanthomonas bacteria[4]. Each TALE repeat binds to a single base pair of DNA and the identities of the amino acids at two positions have been associated with specificities for different nucleotides[5,6].
These chimeric nucleases enable genome editing by inducing targeted DNA DSBs that are repaired by error-prone, non-homologous end joining (NHEJ) or homology-directed repair (HDR)[7-10]. NHEJ-mediated repair induces small insertions or deletions (indels) at the cleavage site, and results in disruption of gene function by frame-shift mutations. In the presence of a single- or double-stranded DNA template containing homology to the sequences flanking the DSB, mutant alleles with precise-point mutations or DNA inserts can be produced by HDR. However, both ZFNs and TALENs require the design of DNA-binding proteins and the construction of complicated plasmids for expression of these, making these methods time-consuming and laborius.
Recently, a new efficient genome manipulation technology, clustered regularly at interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) system, which uses the RNA-guided nuclease, Cas9, and is derived from the immune system of bacteria and archaea, has been developed. CRISPR/Cas technology has the advantages of a highly efficienct mutation rate and simple-to-design target-specific RNA molecules, compared to the complex ZFN and TALEN systems. Therefore, CRISPR/Cas has been rapidly adopted and applied to many species in a short period of time[11-40].
Several reviews about CRISPR/Cas have already been published[41-44]; however, this technology is progressing rapidly, with new reports published weekly. Here, we introduce recent research made possible by CRISPR/Cas technologies and discuss the application of these reagents for genetic analysis and manipulation. We also show the therapeutic potential of CRISPR/Cas and make discussion of future prospects for the field.
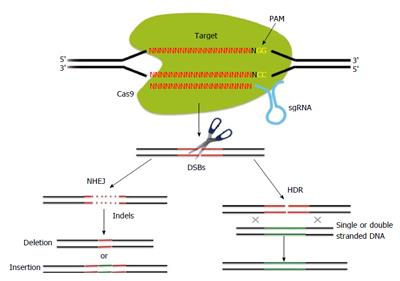
CRISPR/Cas is the RNA-based acquired immunity system in bacteria and archaea[45,46]. CRISPR RNA-guided Cas9 nucleases use short RNAs to target and cleave DNA elements captured from foreign invaders (termed “spacers”) in a sequence-specific manner. In the type II CRISPR/Cas system, a single gene encoding the Cas9 protein and two RNAs, a mature CRISPR RNA (crRNA) which is transcribed from spacers, and a partially complementary transacting RNA (tracrRNA) are sufficient for RNA-guided cleavage of foreign DNAs. For maturation of crRNA, RNase III and tracrRNA are necessary[47]; however, this process can be simplified by an engineered small guide RNA (sgRNA) containing a hairpin that mimics the tracrRNA-crRNA complex and short guide sequence[48] with a protospacer-adjacent motif (with the sequence NGG, Figure 1)[49]. Thus, the Cas9 endonuclease can generate sequence-specific DSBs of target DNAs bound to sgRNAs (Figure 1). DSBs generated by the Cas9 endonuclease are repaired by NHEJ or HDR[7-10]. NHEJ-mediated repair leads to the generation of small indels at the targeted site, which results in disruption of gene function via frame-shift mutations. In the presence of a single- or double-stranded DNA template with homology to the sequences flanking the DSB, mutant alleles with precise-point mutations or DNA inserts can be produced by HDR.
When the CRISPR/Cas system in bacteria and archaea was elucidated, many researchers expected that it functions in the cells of eukaryotic organisms such as yeast, plants, and even mammals. In January 2013, several papers using the CRISPR/Cas system in human cells were published in succession[50-53]. Cho et al[52] showed that combination of Cas9 protein and artificial sgRNAs efficiently cleaved two genomic sites and induced indels with approximately 33% frequencies using human embryonic kidney (HEK) 293T-cells. Two papers published in Science used other cell types or targeting loci[50,51]. For the endogenous AAVS1 safe harbor genomic locus, Mali et al[50] suceeded in gene targeting using 293T-cells (10%-25%), human chronic myelogenous leukemia K562 cells (8%-13%), and human induced pluripotent stem (iPS) cells (2%-4%). In addition, they also used HDR to integrate either a double-stranded DNA donor construct (SA-2A-Puro-pA + CAG-GFP-pA) or an oligo donor into the native AAVS1locus, and obtained 293T or iPS clones showing HDR-mediated integration.
CRISPR/Cas also enables NHEJ- and HDR-mediated genome editing in mouse ES cells[54,55]. The high efficiency of the CRISPR/Cas system coupled with the ability to easily create synthetic sgRNAs make it possible to target multiple genes simultaneously, which is not possible using previous methods[54]. Wang et al[54] transformed embryonic stem cells using CRISPR/Cas system for three different genes (Tet1, Tet2, and Tet3), and found that > 20% (20/96) of ES cell clones had mutations in all six alleles. To further test the potential of multiplexed gene targeting using the CRISPR/Cas system, sgRNAs targeting five genes (Tet1, Tet2, Tet3, Sry, and Uty) were ,mixed and co-transfected with a Cas9-expressing vector into ES cells; of 96 clones screened using an restriction fragment length polymorphism assay, 10% carried mutations at all five loci.
The use of the CRISPR/Cas system in combination with haploid ES cells[56-58] provides a powerful platform to manipulate the mammalian genome, because disruption of only one allele can cause loss-of-function phenotypes in haploid ES cells. We have recently reported that co-transfection of mouse haploid ES cells with vectors expressing Cas9 nuclease and sgRNAs targeting Tet1, Tet2, and Tet3 results in the complete disruption of all three genes, causing a loss-of-function phenotype with higher efficiency (50%)[59] than that previously reported using diploid ES cells[54]. Thus, the CRISPR/Cas system used in the context of haploid cells will be useful for the efficient disruption of multiple genes.
Homologous recombination in mouse ES cells is the most popular method for targeted modifications of the mouse genome; however, generating gene-modified mice through germline chimeras is both time consuming and expensive. Therefore, alternative methods have been developed to accelerate the process of genome modification by the introduction of site-specific nucleases into fertilized embryos to generate DNA DSBs at a target locus in various species. ZFNs and TALENs have been used to produce several gene-modified rodents[60-62]. Although these technologies are widely used in other animals, their use in mice has been limited, principally because the ZFN and TALEN systems are labor-intensive and expensive techniques that do not perform substantially better than ordinary gene knockout technology. On the other hand, CRISPR/Cas-mediated genome editing has successfully demonstrated one-step generation of gene-modified mice, and this technology became widely used within only one year[54,55,63-65]. To understand the functions of genes in families of two or more members, animals carrying multiple mutated genes are required; however, ZFNs or TALENs cannot be multiplexed to generate animals with several targeted loci. In contrast, the CRISPR/Cas system can be used to generate mice carrying mutations in multiple genes in one step[54]. Co-injection of Cas9 mRNA and sgRNAs for Tet1 and Tet2 into fertilized embryos led to the generation of mice with biallelic mutations in both genes with an efficiency of 78% (22/28). Wang et al[54] also showed that co-injection of Cas9 mRNA and sgRNAs with mutant oligos generated precise-point mutations simultaneously in two target genes with an efficiency of 20% (2/10). Using this “one-step” procedure, Yang et al[55] produced mice carrying a tag or a fluorescent reporter construct in the Oct4, Sox2, and Nanog genes. In addition, Mecp2 conditional mutant mice with two loxP sites were generated[55]. These results show that a single step by CRISPR/Cas-mediated genome editing can generate mice having NHEJ- or HDR-mediated mutations in multiple genes.
Compared to ZFNs and TALENs, CRISPR/Cas technology has the advantages of a highly efficient mutation rate and the simplicity of the design of target-specific sgRNAs. It is difficult to compare the off-target effect risk among ZFN, TALEN, and CRISPR/Cas. Although the cleavage of off-target sites has also been observed in ZFN and TALEN systems[66,67], it appears to be less likely because they require two adjacent recognition sites, while the CRISPR/Cas system requires only one. Therefore, it is important to pay careful attention to the specificity of CRISPR/Cas target sequences, because off-target mutations are detrimental to experimental results.
When genome-edited mice are produced using the CRISPR/Cas system, they are rarely influenced by off-target effects. For example, of seven double-mutant mice produced by injection with high RNA concentrations, none showed effects at potential off-target loci using the Surveyor assay[54]. Mashiko et al[65] found only one off-target mutation in a total of 144 sites examined. In addition, Fujii et al[64] proposed that off-target effects are mostly avoided by the careful control of Cas9 mRNA concentration. Surprisingly, the optimized CRISPR/Cas system has a higher gene targeting rate and a lower occurrence of off-target effects compared to ZFN[64]. Mutant mouse ES cells generated by the CRISPR/Cas system also showed a very low Cas9-mediated cleavage rate in off-target loci[55]. These reports suggest that the CRISPR/Cas system is highly specific in the “one-step generation” of mutant mice and mouse ES cells.
By contrast, study of the CRISPR/Cas system in human cancer cell lines indicated a widespread occurrence of off-target mutations[68,69]. Cas9-mediated cleavage can be abolished by single mismatches at the sgRNA-target site interface, particularly in the last 10-12 nucleotides located at the 3’ end of the 20-nt sgRNA-targeting sequence[48,51]. Using human cell lines (U2OS.EGFP, HEK293, and K562), Fu et al[68] found that one or two mismatches are tolerated to varying degrees, depending on their position along the sgRNA-DNA interface. In addition, they easily detected off-target alterations induced by 66% (4/6) of CRISPR/Cas experiments targeting endogenous loci by examination of partially mismatched sites. However, these mismatches were mainly located in the 5’ region, with only one base mismatch detected in the last 12 nucleotides at the 3’ end of one off-target locus.
Yang et al[55] considered several possibilities to explain the lower off-target cleavage rate observed in animals derived from manipulated zygotes compared to the results reported for CRISPR/Cas-treated human cell lines including the following: (1) the cells analyzed in mice and humans are clonal and heterogenous populations, respectively; (2) the transformed human cell lines may have different DNA damage responses, resulting in a different mutagenesis rate compared to normal one-cell embryos; and (3) introduced nucleotides are short-lived RNA or long-lived DNA plasmids in mouse and human systems, respectively, which lead to more extensive cleavage in human cells; however, a definitive explanation has not yet been found.
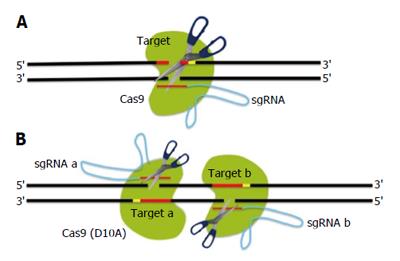
Several measures to improve the specificity of Cas9-mediated genome editing have been assessed. Firstly, it was hypothesized that cleavage specificity may be improved by increasing the length of the region of base pairing between the sgRNA and its target locus. To test this, Ran et al[70] generated sgRNAs with 20 or 30 nucleotides guide sequences; however, they found that extension of the guide sequence did not improve Cas9 targeting specificity. Next, Ran et al[70] developed a strategy that combines the D10A mutant nickase version of Cas9[48,51,71] with a pair of offset sgRNAs complementary to opposite strands of the target site (Figure 2A: DSBs using wild-type Cas9 endonuclease; B: DSBs using a pair of sgRNAs guiding Cas9 D10A). Whereas nicking of both DNA strands by a pair of Cas9 nickases leads to site-specific DSBs and NHEJ, individual nicks are predominantly repaired by the high-fidelity base excision repair pathway[72]. As a result, this double nicking method can reduce off-target activity by 50- to 1500-fold and assisted gene knockout without reduction of on-target cleavage efficiency[70,73]. Double nicking allows not only NHEJ-mediated indels but also insertion into the genome via HDR in human cells.
In the case of mutant animals produced by CRISPR/Cas, off-target mutations will be eliminated by backcrossing to wild-type animals. Therefore, if researchers do not use F0 pups obtained by CRISPR/Cas for experiments, off-target effects should not be a concern. RNA interference (RNAi) experiment to induce sequence-specific gene silencing is now a standard method for the functional analysis of genes. However, designed small RNA frequently repress translation from unexpected loci[74-76]. To remove this off-target effect, two or more independent small RNAs are generally used in RNAi experiments. In CRISPR/Cas experiment, use of two or more independent sgRNAs for a gene will be also an effective control to remove off-target noise and improve the reliability of the obtained phenotype. Nevertheless, more detailed work will be necessary to determine the frequency of off-target mutations, and improve the specificity in CRISPR/Cas systems.
Precise genome modifications by CRISPR/Cas system excite the interest of scientists working in both basic science and applied fields, including gene therapy. Undoubtedly, the CRISPR/Cas system is a strong candidate for application in human gene therapy. Several human iPS cell lines have been generated from patients for stem cell-based gene therapy by correction of gene mutations. But, gene targeting in human pluripotent stem cells including ES and iPS cells has been very difficult historically[77]. Nevertheless, ZFNs and TALENs are capable of correcting gene mutations mediated by HDR repair mechanisms in human iPS cells[78-82] and the CRISPR/Cas system has also recently been applied to the gene therapy model[50,83]. Of course, this application will require a highly efficienct gene editing rate and no off-target mutations.
CRISPR/Cas is thought to be applicable for genome editing based only on NHEJ or HDR; however, nuclease-null Cas9 (Cas9N) can work as a transcriptional activator or silencer without changing DNA sequences[84,85]. Mali et al[84] produced a Cas9N directly fused with the VP64 activation domain to generate a Cas9N-fusion protein capable of transcriptional activation. This Cas9N-VP64 protein robustly activated transcription of reporter constructs and endogeneous REX1, OCT4, SOX2, and NANOG genes when this fusion protein is combined with sgRNA-targeting sequences near the promoter[84]. This is the example of RNA-guided transcriptional activation. By contrast, a Cas9N-sgRNA complex is specifically able to interfere with transcriptional elongation, transcription factor binding, or RNA polymerase binding[85]. This technology could be applied to genome-wide screens for gene function.
Prior genome-editing technologies, ZFNs and TALENs, suggest new applications for CRISPR/Cas. For example, Konermann et al[86] developed a light-inducible genome-editing system, using transcriptional effectors and the customizable TALE DNA-binding domain. They suceeded in transcriptional activation and epigenetic modification of endogenous genes using primary neurons as well as brain of living mice.
Bacterial DNA methyltransferases[87-91] and human DNA metyltransferase 3a and 3b subunits[92-94] have been fused to zinc-finger proteins and successfully demonstrated to perform targeted DNA methylation. Efficient targeting of DNA demethylation was also demonstrated using fusions of TALE repeat arrays and the TET1 hydroxylase catalytic domain (TALE-TET1)[95]. These targeted methylation and demethylation technologies will be applicable for gene therapy of cancer and other epigenetic diseases such as Beckwith-Wiedemann and Angelman syndromes mediated by abnormal DNA methylation, or of Huntington disease, which is caused by by extra repetitive DNA sequences. In addition, Jiang et al[96] inserted an inducible XIST transgene into chromosome 21 using ZFN in iPS cells derived from a Down’s syndrome patient. In this system, chromosome 21 are coated with XIST non-coding RNA, followed by stable heterochromatin modifications, DNA methylation and chromosome-wide transcriptional silencing. This successful silencing of trisomy is the first step for chromosome therapy using genome engineering. These applications developed using ZFN and TALEN systems will be also applicable using the CRISPR/Cas technique.
In the future, CRISPR/Cas may be used to target the viral DNA that becomes integrated into the chromosomes of people with lifetime infections (e.g., HIV). If this viral genetic material can be disrupted using the CRISPR/Cas system, this could negate the need for patients to continue taking antiviral drugs throughout their lives.
CRISPR/Cas has already been applied to many species in which genome engineering has been difficult, because this technology has the advantages of a highly efficient mutation rate and a simple system for design of target-specific sgRNA. Although improvements in the specificity of CRISPR/Cas will be necessary to eliminate off-target effects, the technique will be indispensable for researchers in both basic and applied science.
P- Reviewer: Guo ZS, Vargas FR, Wei L, Xuei X S- Editor: Wen LL L- Editor: A E- Editor: Liu SQ
| 1. | Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156-1160. [PubMed] |
| 2. | Smith J, Berg JM, Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27:674-681. [PubMed] |
| 3. | Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 429] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 4. | Bonas U, Stall RE, Staskawicz B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol Gen Genet. 1989;218:127-136. [PubMed] |
| 5. | Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509-1512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2048] [Cited by in RCA: 1812] [Article Influence: 113.3] [Reference Citation Analysis (0)] |
| 6. | Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1451] [Cited by in RCA: 1370] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 7. | Barnes DE. Non-homologous end joining as a mechanism of DNA repair. Curr Biol. 2001;11:R455-R457. [PubMed] |
| 8. | Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2264] [Cited by in RCA: 2064] [Article Influence: 137.6] [Reference Citation Analysis (0)] |
| 9. | van den Bosch M, Lohman PH, Pastink A. DNA double-strand break repair by homologous recombination. Biol Chem. 2002;383:873-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 10. | Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 572] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 11. | Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741-743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 749] [Cited by in RCA: 685] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 12. | Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220-228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 621] [Cited by in RCA: 632] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 13. | Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 273] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 14. | Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One. 2013;8:e68708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 275] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 15. | Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904-13909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 950] [Cited by in RCA: 968] [Article Influence: 80.7] [Reference Citation Analysis (0)] |
| 16. | Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31:681-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 507] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 17. | Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31:684-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 343] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 18. | Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1249] [Cited by in RCA: 1155] [Article Influence: 105.0] [Reference Citation Analysis (0)] |
| 19. | Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant. 2013;6:1975-1983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 438] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 20. | Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229-1232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 662] [Cited by in RCA: 643] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 21. | Chiu H, Schwartz HT, Antoshechkin I, Sternberg PW. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics. 2013;195:1167-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 22. | Katic I, Großhans H. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics. 2013;195:1173-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 23. | Tzur YB, Friedland AE, Nadarajan S, Church GM, Calarco JA, Colaiácovo MP. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics. 2013;195:1181-1185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 24. | Waaijers S, Portegijs V, Kerver J, Lemmens BB, Tijsterman M, van den Heuvel S, Boxem M. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics. 2013;195:1187-1191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 25. | Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028-1034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 681] [Cited by in RCA: 712] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 26. | Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 933] [Cited by in RCA: 708] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 27. | Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23:1233-1236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 567] [Cited by in RCA: 583] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 28. | Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 437] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 29. | Seth A, Stemple DL, Barroso I. The emerging use of zebrafish to model metabolic disease. Dis Model Mech. 2013;6:1080-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 198] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 30. | Hisano Y, Ota S, Kawahara A. Genome editing using artificial site-specific nucleases in zebrafish. Dev Growth Differ. 2014;56:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Upadhyay SK, Kumar J, Alok A, Tuli R. RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda). 2013;3:2233-2238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 339] [Cited by in RCA: 212] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 32. | Blitz IL, Biesinger J, Xie X, Cho KW. Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis. 2013;51:827-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 33. | Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, Grainger RM. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 220] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 34. | Sebo ZL, Lee HB, Peng Y, Guo Y. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly (Austin). 2013;8:52-57. [PubMed] |
| 35. | Wang Y, Li Z, Xu J, Zeng B, Ling L, You L, Chen Y, Huang Y, Tan A. The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Res. 2013;23:1414-1416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 222] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 36. | Daimon T, Kiuchi T, Takasu Y. Recent progress in genome engineering techniques in the silkworm, Bombyx mori. Dev Growth Differ. 2014;56:14-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 37. | Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 495] [Cited by in RCA: 468] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 38. | Frøkjær-Jensen C. Exciting prospects for precise engineering of Caenorhabditis elegans genomes with CRISPR/Cas9. Genetics. 2013;195:635-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 39. | Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014;24:125-131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 229] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 40. | Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982-4987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 362] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 41. | Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2477] [Cited by in RCA: 2480] [Article Influence: 206.7] [Reference Citation Analysis (0)] |
| 42. | Menke DB. Engineering subtle targeted mutations into the mouse genome. Genesis. 2013;51:605-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Wei C, Liu J, Yu Z, Zhang B, Gao G, Jiao R. TALEN or Cas9-rapid, efficient and specific choices for genome modifications. J Genet Genomics. 2013;40:281-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 44. | Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029-1035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 774] [Cited by in RCA: 821] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 45. | Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1269] [Cited by in RCA: 1384] [Article Influence: 106.5] [Reference Citation Analysis (0)] |
| 46. | Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1581] [Cited by in RCA: 1655] [Article Influence: 110.3] [Reference Citation Analysis (0)] |
| 47. | Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602-607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2124] [Cited by in RCA: 1758] [Article Influence: 125.6] [Reference Citation Analysis (0)] |
| 48. | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11979] [Cited by in RCA: 10993] [Article Influence: 845.6] [Reference Citation Analysis (0)] |
| 49. | Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568-571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 459] [Cited by in RCA: 462] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 50. | Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6555] [Cited by in RCA: 7001] [Article Influence: 583.4] [Reference Citation Analysis (0)] |
| 51. | Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10336] [Cited by in RCA: 11160] [Article Influence: 930.0] [Reference Citation Analysis (0)] |
| 52. | Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1480] [Cited by in RCA: 1461] [Article Influence: 121.8] [Reference Citation Analysis (0)] |
| 53. | Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1499] [Cited by in RCA: 1639] [Article Influence: 136.6] [Reference Citation Analysis (0)] |
| 54. | Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2799] [Cited by in RCA: 2789] [Article Influence: 232.4] [Reference Citation Analysis (0)] |
| 55. | Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1203] [Cited by in RCA: 1242] [Article Influence: 103.5] [Reference Citation Analysis (0)] |
| 56. | Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 204] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 57. | Yang H, Shi L, Wang BA, Liang D, Zhong C, Liu W, Nie Y, Liu J, Zhao J, Gao X. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell. 2012;149:605-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 58. | Elling U, Taubenschmid J, Wirnsberger G, O’Malley R, Demers SP, Vanhaelen Q, Shukalyuk AI, Schmauss G, Schramek D, Schnuetgen F. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 197] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 59. | Horii T, Morita S, Kimura M, Kobayashi R, Tamura D, Takahashi RU, Kimura H, Suetake I, Ohata H, Okamoto K. Genome engineering of mammalian haploid embryonic stem cells using the Cas9/RNA system. PeerJ. 2013;1:e230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 60. | Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 211] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 61. | Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 778] [Cited by in RCA: 713] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 62. | Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31:23-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 288] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 63. | Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 491] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 64. | Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013;41:e187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 176] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 65. | Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 352] [Cited by in RCA: 348] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 66. | Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 394] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 67. | Grau J, Boch J, Posch S. TALENoffer: genome-wide TALEN off-target prediction. Bioinformatics. 2013;29:2931-2932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 68. | Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822-826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2216] [Cited by in RCA: 2424] [Article Influence: 202.0] [Reference Citation Analysis (0)] |
| 69. | Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3576] [Cited by in RCA: 3444] [Article Influence: 287.0] [Reference Citation Analysis (0)] |
| 70. | Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2581] [Cited by in RCA: 2525] [Article Influence: 210.4] [Reference Citation Analysis (0)] |
| 71. | Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579-E2586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1687] [Cited by in RCA: 1890] [Article Influence: 145.4] [Reference Citation Analysis (0)] |
| 72. | Dianov GL, Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483-3490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 275] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 73. | Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132-141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1098] [Cited by in RCA: 1050] [Article Influence: 95.5] [Reference Citation Analysis (0)] |
| 74. | Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1709] [Cited by in RCA: 1691] [Article Influence: 76.9] [Reference Citation Analysis (0)] |
| 75. | Saxena S, Jónsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312-44319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 300] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 76. | Snøve O, Holen T. Many commonly used siRNAs risk off-target activity. Biochem Biophys Res Commun. 2004;319:256-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 103] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 77. | Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 532] [Cited by in RCA: 467] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 78. | Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 996] [Cited by in RCA: 958] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 79. | Choi SM, Kim Y, Shim JS, Park JT, Wang RH, Leach SD, Liu JO, Deng C, Ye Z, Jang YY. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013;57:2458-2468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 181] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 80. | Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 825] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 81. | Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 438] [Cited by in RCA: 416] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 82. | Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 631] [Cited by in RCA: 584] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 83. | Horii T, Tamura D, Morita S, Kimura M, Hatada I. Generation of an ICF syndrome model by efficient genome editing of human induced pluripotent stem cells using the CRISPR system. Int J Mol Sci. 2013;14:19774-19781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 84. | Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833-838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1434] [Cited by in RCA: 1369] [Article Influence: 114.1] [Reference Citation Analysis (0)] |
| 85. | Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3647] [Cited by in RCA: 3560] [Article Influence: 296.7] [Reference Citation Analysis (0)] |
| 86. | Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 624] [Cited by in RCA: 639] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 87. | Xu GL, Bestor TH. Cytosine methylation targetted to pre-determined sequences. Nat Genet. 1997;17:376-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 122] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 88. | McNamara AR, Hurd PJ, Smith AE, Ford KG. Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships. Nucleic Acids Res. 2002;30:3818-3830. [PubMed] |
| 89. | Carvin CD, Parr RD, Kladde MP. Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Res. 2003;31:6493-6501. [PubMed] |
| 90. | Smith AE, Ford KG. Specific targeting of cytosine methylation to DNA sequences in vivo. Nucleic Acids Res. 2007;35:740-754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 91. | Nomura W, Barbas CF. In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J Am Chem Soc. 2007;129:8676-8677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 92. | Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, Jeltsch A. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007;35:100-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 116] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 93. | Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, Blancafort P. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2012;7:350-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 166] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 94. | Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol. 2013;425:479-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 128] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 95. | Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013;31:1137-1142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 374] [Cited by in RCA: 370] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 96. | Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY. Translating dosage compensation to trisomy 21. Nature. 2013;500:296-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 249] [Article Influence: 20.8] [Reference Citation Analysis (0)] |