Published online Feb 28, 2023. doi: 10.5495/wjcid.v13.i1.1
Peer-review started: August 28, 2022
First decision: December 13, 2022
Revised: December 28, 2022
Accepted: February 1, 2023
Article in press: February 1, 2023
Published online: February 28, 2023
Processing time: 182 Days and 11.8 Hours
Leprosy is a disease caused by Mycobacterium leprae (M. leprae), an intracellular pathogen that has tropism and affects skin and nervous system cells. The disease has two forms of presentation: Paucibacillary and multibacillary, with different clinical and immunological manifestations. Unlike what occurs in the multibacillary form , the diagnostic tests for the paucibacillary form are nonspecific and not very sensitive, allowing the existence of infected individuals without treatment, which contributes to the spread of the pathogen in the population. To mitigate this contamination, more sensitive diagnostic tests capable of detecting paucibacillary patients are needed.
To predict the three-dimensional structure models of M. leprae antigens with serodiagnostic potential for leprosy.
In this in silico study, satisfactory templates were selected in the Protein Data Bank (PDB) using Basic Local Alignment Search Tool to predict the structural templates of ML2038, ML0286, ML0050, and 85B antigens by comparative modeling. The templates were selected according to general criteria such as sequence identity, coverage, X-ray resolution, Global Model Quality Estimate value and phylo
The three-dimensional structure models of ML2038, ML0286, ML0050, and 85B antigens of M. leprae were predicted using the templates PDB: 3UOI (90.51% identity), PDB: 3EKL (87.46% identity), PDB: 3FAV (40.00% identity), and PDB: 1F0N (85.21% identity), respectively. The QMEAN and Z-score values indicated the good quality of the structure models. These data refer to the monomeric units of antigens, since some of these antigens have quaternary structure. The validation of the models was performed with the final three-dimensional structure - monomer (ML0050 and 85B antigens) and quaternary structures (ML2038 and ML0286). The majority of amino acid residues were observed in favorable and allowed regions in the Ramachandran plot, indicating correct positioning of the side chain and absence of steric impediment. The MolProbity score value and Verify 3D results of all models indicated a satisfactory prediction.
The polarized immune response against M. leprae creates a problem in leprosy detection. The selection of immunodominant epitopes is essential for the development of more sensitive serodiagnostic tests, for this it is important to know the three-dimensional structure of the antigens, which can be predicted with bioinformatics tools.
Core Tip: Leprosy is a disease with high clinical and epidemiological impact, because it causes irreversible and disfiguring sequelae and has a high incidence in endemic countries. Its variability of manifestations, with different immune responses and the difficulty of cultivating Mycobacterium leprae (M. leprae) in the laboratory, makes it difficult to develop sensitive and specific tests for the diagnosis of the disease, thus emphasizing the importance of in silico studies to solve this problem. In this sense, this study aimed to obtain three-dimensional models of M. leprae antigens, which have stood out in previous studies as candidates for the serological diagnosis of leprosy.
- Citation: Melo de Assis BL, Viana Vieira R, Rudenco Gomes Palma IT, Bertolini Coutinho M, de Moura J, Peiter GC, Teixeira KN. Three-dimensional models of antigens with serodiagnostic potential for leprosy: An in silico study. World J Clin Infect Dis 2023; 13(1): 1-10
- URL: https://www.wjgnet.com/2220-3176/full/v13/i1/1.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v13.i1.1
Leprosy is a chronic transmissible and infectious disease caused by the bacillus Mycobacterium leprae (M. leprae), which persists as a major public health problem in Brazil. The etiological agent of this disease mainly affects the skin, peripheral nerves, and eyes, presents slow evolution, and causes deformity and physical disability, when not correctly diagnosed and treated[1].
According to the World Health Organization (WHO)[1], in 2019, there were 202 185 cases of M. leprae infection worldwide, 93% of which were reported in the Americas. In addition, according to the WHO, Brazil contributed 27 864 new cases of the disease in 2019, placing it in second place among the countries with the highest number of leprosy cases, behind only India.
Clinically, leprosy may manifest in two different forms, which differ mainly by the immune response developed by the host against the pathogen. The current classification of leprosy came into being in 1982, from the WHO Committee, which proposed a simplified and operational classification of paucibacillary and multibacillary individuals based on the likely relationship between clinical form and smear[2].
The manifestations of leprosy are distinguished both by clinical presentations and characteristics of the immune response. Such presentations are considered antagonistic, which guide the understanding of the pattern of dual response observed in Th1 and Th2 Lymphocytes. In leprosy, the paucibacillary form is characterized by the Th1 response, which confers some resistance of the organism to the pathogen, thereby this is a milder form of the infection; the multibacillary form, a more severe infection, is characterized by the development of the Th2 pattern, which does not have an effective mechanism against the pathogen[3].
Due to the variety of clinical manifestations of leprosy, the WHO[4] recommends that the diagnosis be based on loss of sensitivity of a hypopigmented or reddish skin area, visualization of thickening or augmentation of a peripheral nerve, accompanied by weakness of muscle tissue supplied by the nerve, which can only be observed or visualization of acid-alcohol fast bacilli by means of the intradermal sample smear technique. In addition, enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR) tests can be used to complement the diagnosis, especially in suspected multibacillary form. Thus, it is noted that the recognition of the disease can be based both on factors arising from the clinical examination (anamnesis and physical examination) and complementary tests, requiring the expertise of the professionals who will perform and interpret the procedure, especially with regard to histological techniques, and reliability of the tests available.
However, the tests used to diagnose the multibacillary form, such as serological tests by ELISA have become ineffective since the paucibacillary patient presents low titers of antibodies. Due to the high production of pro-inflammatory cytokines, there is little bacterial proliferation, which makes it difficult to find bacilli at the site of the lesion by the intradermal smear technique. Consequently, when applied to patients with paucibacillary leprosy, the recommended tests have low sensitivity and specificity, which may lead to false-negative results, contributing to the spread of the disease[5]. Thus, tests aimed at improving these parameters and diagnosing both severe and mild forms of leprosy are necessary.
To support the development of diagnostic tests with greater sensitivity for any type of clinical manifestation of leprosy, Santana[6] mapped epitopes of M. leprae, constructed by the spot synthesis technique, using serum from leprosy patients, based on a previous study[7]. Of 12 selected proteins, seven were promising candidates for immunodominant antigens. Bioinformatics tools were used to verify molecular mass, isoelectric point, hydrophobicity, and acid-basic characteristics[5]; however, none of the proteins has experimentally resolved three-dimensional structure, which hinders a complete biochemical characterization.
The three-dimensional structure of a protein is fundamental to determine the selection of the best candidates in order to obtain a functional epitope given that the peptides hidden in the protein core are not of interest, since the antibodies do not have access to this region. In turn, antigenic determinants actively participate in antigen-antibody binding, which, when established, generate conformational and structural changes, which are important in the process of antigenic neutralization [6].
In this study, the three-dimensional structures of M. leprae antigens selected by Santana[6] were predicted with bioinformatics tools in order to provide support for the elaboration of possible serological diagnostic tests for both multibacillary and paucibacillary clinical forms of leprosy.
The FASTA sequences of antigens ML2038, ML0286, ML0050, and 85B (GenBank WP_010908683.1, WP_010907650, WP_010907488.1, and CAA43269.1, respectively) available at The National Center for Biotechnology Information (ncbi.nlm.nih.gov) were used to select templates for molecular modeling at Protein Data Bank (PDB) (rcsb.org), through local alignment using the BLAST (Basic Local Alignment Search Tool)[8], aiming to find templates with (1) three-dimensional structures resolved; (2) high sequence identity and coverage values as much as possible; (3) high X-ray resolution; and (4) phylogenetic proximity to the target proteins. The choice of templates also took into account the Global Model Quality Estimate (GMQE) value, which predicts the overall quality of the model and ranges from 0 to 1, with higher numbers indicating higher quality. The identity between primary sequence and template, as well as sequence coverage, were verified through global alignment using ClustalX 2.1 software[9].
The antigens whose template was satisfactory were modeled by the comparative modeling methodology using the Modeller 9v13[10] software. The selection of the models released by Modeller was performed using the Discrete Optimized Protein Energy (DOPE) method, which predicts the lowest energy models, therefore the most stable ones. The selected three-dimensional models were visualized and analyzed with the ViewerLite 4.2 (Accelrys Inc.) and PyMol (Schrödinger Inc.) software, and the quality was verified using the QMEAN score[11] and Z-score. The validation of the models was performed using the MolProbity[12] and Verify 3D platforms[13].
Templates were selected for target proteins ML2038, ML0286, ML0050, and 85B antigens - 3UOI (90.51% identity and 1.90 Å resolution), 3EKL (87.46% identity and resolution of 1.51 Å), 3FAV (40.00% identity and 2.15 Å resolution), and 1F0N (85.21% identity and 1.80 Å resolution), respectively. In addition, a 100.00% coverage was obtained for ML2038 and ML0050 proteins, 99.00% for ML0286, and 87.00% for 85B antigen (Table 1).
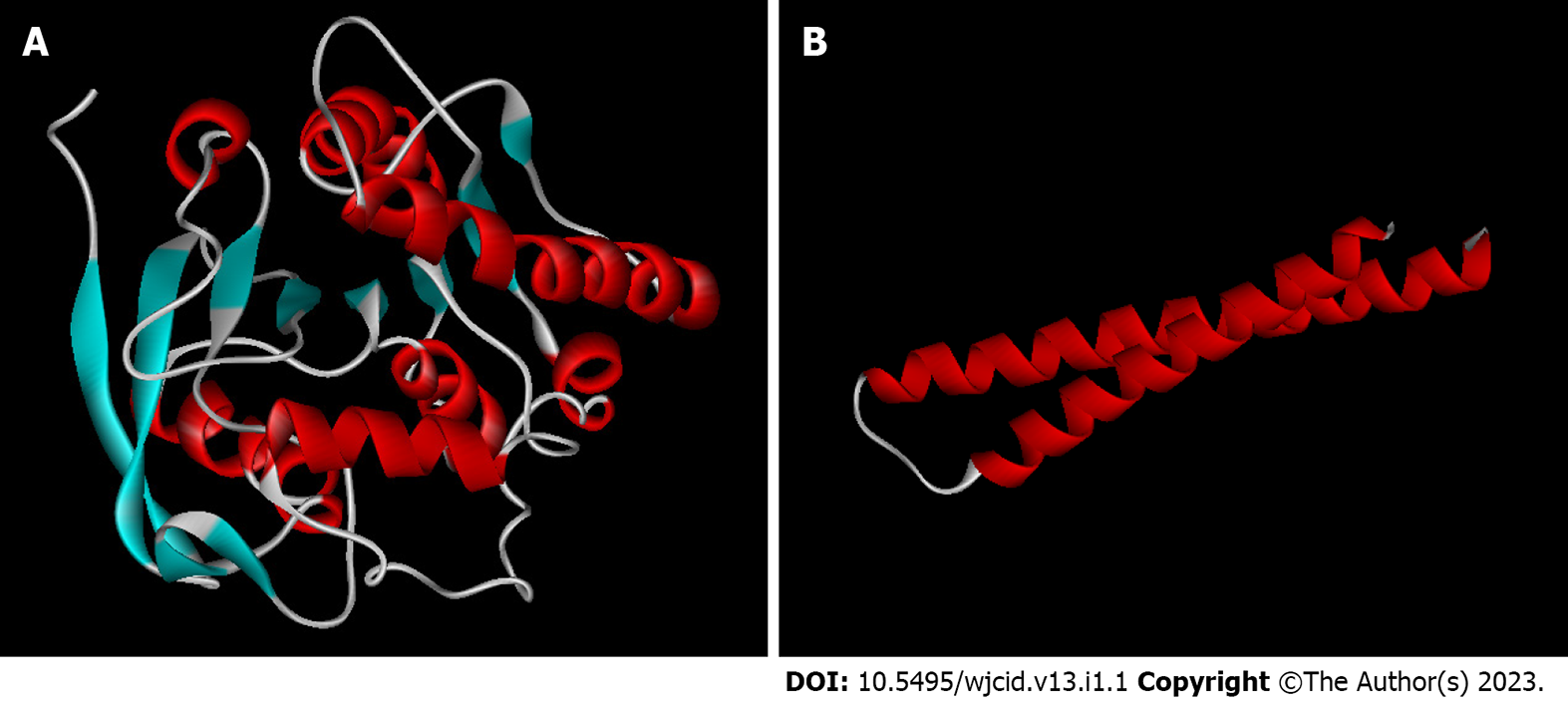
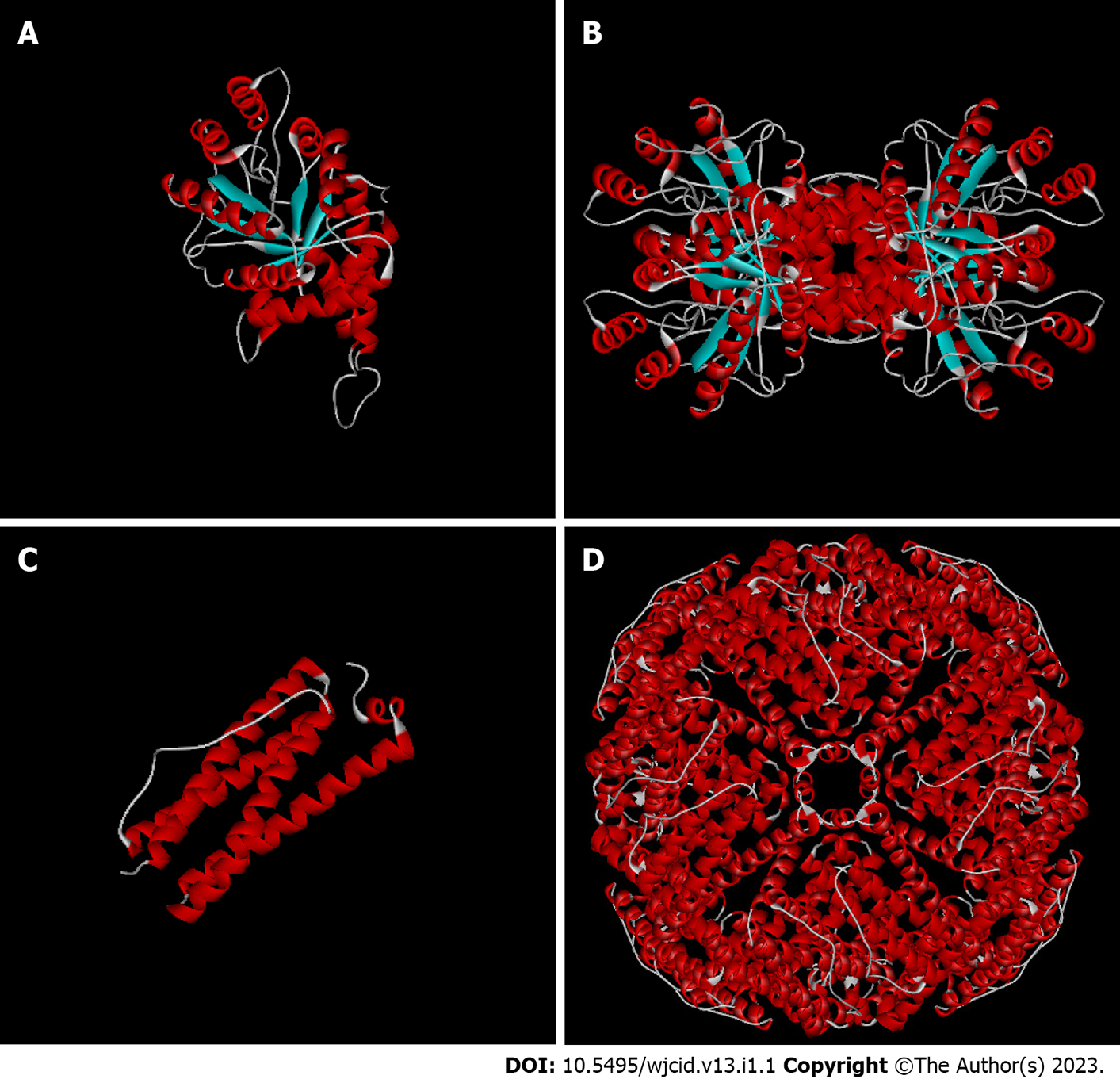
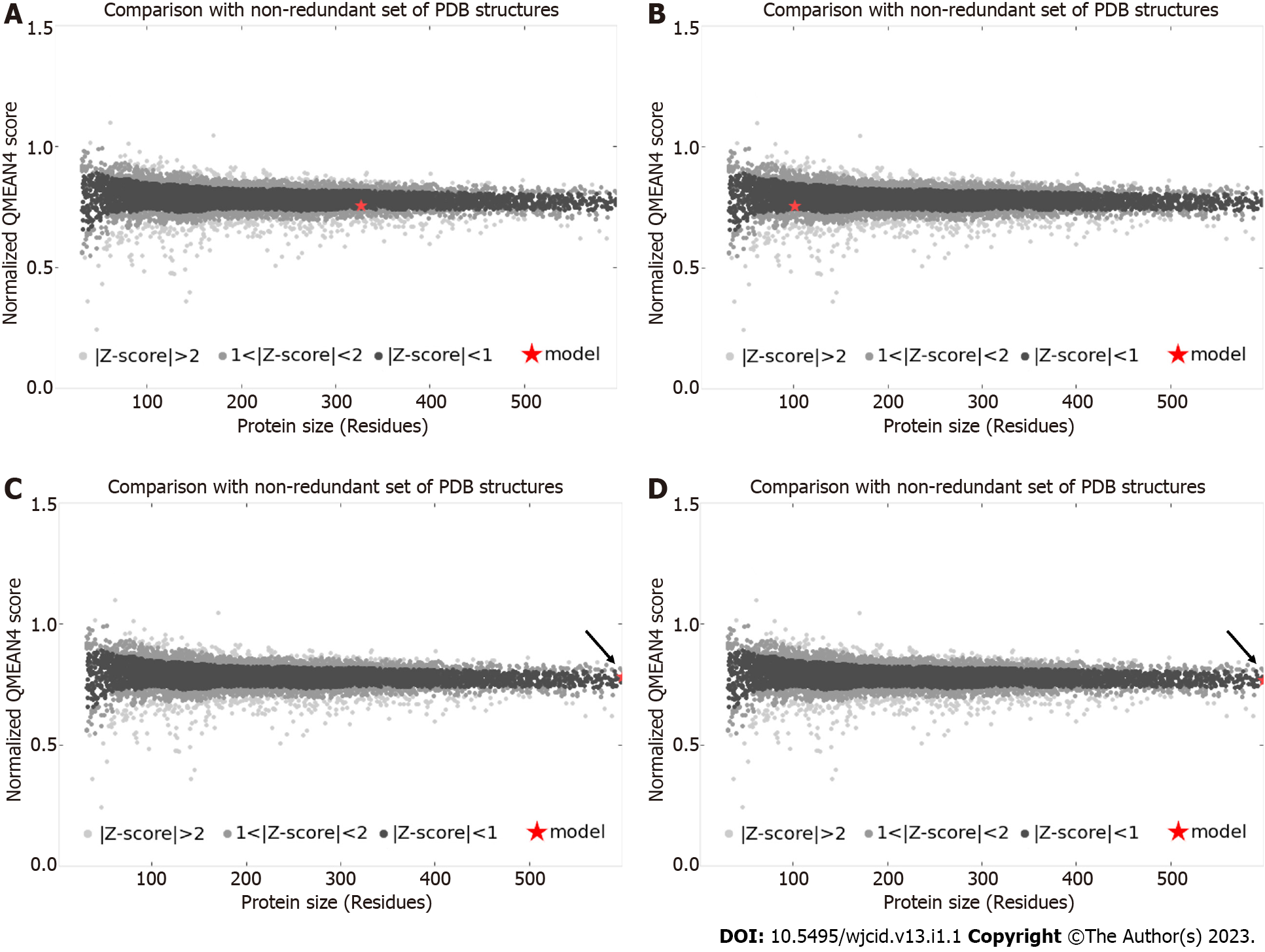
The primary sequences of the target proteins were submitted to comparative molecular modeling using the Modeller 9v13 software which generated the three-dimensional structures as shown in Figures 1 and 2. The comparative modeling of ML0050 was performed with a pipeline designed for low identity templates[14]. QMEAN scores for the structural models were 0.13 (ML2038), 0.56 (ML0286), -0.62 (ML0050), and -0.39 (85B antigen); this score typically ranges from 0 to 4.0, and values closer to 0 indicate a good model. QMEAN score refers to the monomeric subunits that were modeled; however, the ML2038 and ML0286 antigens are quaternary proteins. ML0050 can dimerize, but functionally, the protein is a monomer. Validation of the predicted structural models was performed with the functional structure of each antigen, i.e., ML0050 and 85B antigens in monomeric form and ML2038 and ML0286 in quaternary form. The Z-scores of all structure models were > 0.5 but < 1.0. Figure 3 shows the Z-score plot which indicates the quality of the three-dimensional models obtained.
Validation was performed using MolProbity software; parameters related to the geometry of the angles of the chemical groups of amino acid residues were evaluated in the models obtained by molecular modeling. The MolProbity score, outlier residues, and the percentage of residues in favorable regions in the Ramachandran plot were considered (Table 2). On Verify3D software, 3D/1D score for all models was more than 80%; it means that the models have good quality.
| Antigen | Identification | Functional structure | MolProbity Score | Ramachandran plota (%) | Outliers (%) |
| ML2038 | Bacterioferritin | Homopolymer | 1.31 | 98.48 | 0.08 |
| ML0050 | ESAT-6-like protein esxB | Monomer | 0.77 | 98.61 | 0.00 |
| ML0286 | Fructose-bisphosphate aldolase | Homotetramer | 1.32 | 96.47 | 0.59 |
| 85B Ag | 85B antigen | Monomer | 1.85 | 93.95 | 0.00 |
The leprosy infection caused by the M. leprae induces marked lesions on the skin and peripheral nerves and, when not properly treated, leads to irreversible and disfiguring sequelae in the patient, such as destruction of cartilage and leonine facies[15]. In addition, the great social stigma related to the disease is added, which contributes even more to the worsening of the psychological issue of these patients[16]. It is also worth noting the high prevalence of this pathology in the Americas, especially in Brazil, which in 2017 was responsible for 92.3% of the cases reported on the continent, evidencing the clinical and epidemiological importance of the pathology for the country[15]. Thus, there is a need for tests to identify the disease early, to prevent its spread and enable the early institution of the recommended therapy, avoiding or reducing the associated sequelae.
However, there are variations in the clinical manifestation of leprosy. In the multibacilla
Moreover, as there is little presence of initial signs and symptoms, and there may be asymptomatic presentation and similarity with other diseases, making the pure clinical diagnosis of leprosy difficult[5,15].
In addition, there is also the difficulty of growing M. leprae in artificial or cellular media, constituting one of the major obstacles to leprosy research. The available forms of Hansen's bacillus cultivation are inoculation in captive armadillo and in the paw pads of immunocompetent mice (Shepard method) and immunodeficient mice (Prabhakaran method). The use of armadillos for leprosy research is limited by the difficult management of the animal, while research with different types of mice is extensive, taking up to 8 mo for adequate immunological manifestation[17-19].
Therefore, in silico studies aimed at the identification and mapping of B-cell epitopes[20] which predict the type of interaction and the affinity energy between the epitope and the Fab region of the antibodies, are valid for the development of serological tests for the diagnosis of leprosy. This kind of study allows the recognition of immunodominant protein domains and their selection for the preparation of immunohistochemical tests, as well as for the manufacture of synthetic peptides, which are less expensive and easy to manipulate[15,21]. Thus, there is a decreased possibility of cross-reactivity with antigens from other pathogens, increasing the specificity and sensitivity of the test.
This study aimed to predict the three-dimensional structure models of M. leprae antigens that, according to previous studies[6,7], have the potential to be used in serodiagnostic tests for both paucibacillary and multibacillary leprosy. One of these antigens, ML2038, is the Bacterioferritin protein or Major Membrane Protein II (MMP-II), which is encoded by the BFRA gene. This protein is present in the cell membrane of M. leprae and is responsible for iron storage in restricted situations. In addition, MMP-II has a high identity with its homologue found in M. tuberculosis and has the ability to activate CD4+ and CD8+ T cells[6,15,22].
Due to the high primary sequence identity of the template (91.14%), it was possible to predict the three-dimensional structure of the ML2038 antigen by comparative modeling. The quality of the model was evaluated by the QMEAN score, a parameter used to analyze the structure obtained and compare it with others already known, in relation to physicochemical and evolutionary characteristics, in order to identify problematic regions for subsequent correction[23,24].
The QMEAN score of the three-dimensional structure of ML2038 was 0.56. This value was applied to the Z-score plot, a tool that combines the QMEAN score with those of proteins with structure deposited in the PDB, demonstrating that the model obtained by the study is similar to what is expected for native proteins with molecular mass similarity [24,25,26].
Using these data, it was possible to correct the errors found in the configuration of amino acid residues using tools of the MolProbity platform, for subsequent validation of the three-dimensional models. Thus, correcting the outliers, the MolProbity score equal to 1.31 was obtained. This score evaluates the log of the clashscore, percentage of amino acids in unfavorable regions in the Ramachandran chart, and bad rotations of the lateral chains; MolProbity score values lower than the crystallographic resolution of the template (1.90 Å) indicate three-dimensional model quality[27].
The results of the Ramachandran plot also contribute to the reliability of the three-dimensional organization of the ML2038 model, which predicts that 98.48% of residues are in permitted regions, indicating the stability of the predicted structure.
In a study conducted by Maeda et al[22], in which sera from paucibacillary and multibacillary patients, sera from patients with tuberculosis, and sera from healthy individuals vaccinated with Bacillus of Calmette-Guérin (BCG) were tested, and a better sensitivity of the serological test was observed for individuals with the multibacillary form (82.4%, 95.0% confidence interval [CI]: 71.8-90.3) than those with the paucibacillary form of leprosy (39.0%, 95.0%CI: 28.8-50.1), when screened for antibodies against MMP-II. Furthermore, healthy individuals or individuals with tuberculosis showed high specificity, indicating the low influence of homologous M. bovis antigens, and ancestry between M. leprae and M. tuberculosis on the diagnostic test result.
Similar results were obtained in a study by Kai et al[28], in which paucibacillary and multibacillary patients tested positive, 47.6% and 85.1%, respectively when investigated for antibodies against ML2038, and 20.2% and 57.0% when investigated for antibodies against PGL-I (Phenolic Glycolipid-I), one of the first isolated M. leprae-specific antigens currently instituted in serological tests for leprosy[28,29].
In this way, Tsukamoto et al[30] conducted clinical research associated with the search for anti-MMP-II and anti-MMP-I antibodies. This association allowed the increase in the sensitivity of the test and the rescue of patients with false-negative results obtained when researched for antigens purely against ML2038. Thus, it is perceived that the literature converges in relation to the possibility and advantage of the use of Bacterioferritin in the diagnostic test of leprosy patients.
In the face of these studies, Santana[6] points out in a study aimed to identify antigen-antibody recognition for some antigens of the M. leprae that the ML2038, as well as ML0286 and ML0050, have no reactivity to the sera from leprosy patients. The author reports that the possible reasons for this non-recognition would be the presence of conformal immunodominant protein domains, which depend on the novel structure of the protein to act as epitopes, and the variability of sera from patients of different ethnicities and regions.
The ML0286 antigen (Putative Fructose Bisphosphate Aldolase), encoded by the FBA gene, is a cytosolic enzyme, soluble and integral to the energy metabolism of M. leprae. This enzyme catalyzes the conversion of fructose 1-6-diphosphate into glyceraldehyde-3-phosphate and dihydroxyacetone-phosphate by metabolizing this sugar. Puckett et al[31] have suggested that this enzyme regulates the glycolytic and gluconeogenic metabolism of carbohydrates in mycobacteria, indicating participation in aminoglycoside resistance in strains resistant to these antibiotics. The overexpression of Rv0363c, another classification for this aldolase, can maintain the energy supply for resistant M. leprae strains, providing adenosine triphosphate (ATP) at the state of antimicrobial resistance. Homology modeling has revealed that ML0286 is a homotetramer, with 96.47% of residues in favorable regions, and is therefore feasible for the elaboration of serological tests for recognition of the protein epitope[7,32,33].
The ML0050 antigen, also called ESAT-6-like protein esxB or culture filtrate protein (CFP-10), acts on the virulence and pathogenicity of M. leprae together with ESAT-6 protein, which is secreted by the ESX-1 secretion system and improves the stability of other externalized antigens[15,19,34]. In addition, ML0050 is encoded by the esxB gene, having recognized its expression only in the Mycobacterium genus[19]. For such evolutionary conservation, it is understandable that there is homology among such proteins in distinct species of mycobacteria, such as between M. leprae and M. tuberculosis, increasing the chances of cross-reactivity in serological tests.
According to Geluk et al[35], although the specificity of ML0050 is high for the Mycobacterium genus, the absence of differentiation between both paucibacillary and multibacillary leprosy is a problem with the use of serological tests based on this antigen. Moreover, even if the authors point out the properties of this antigen as a measure of disease progression and effectiveness of leprosy treatment, it is noted that there are obstacles to its use.
The 85B antigen (MPT59), also called ML2028 or diacylglycerol acetyltransferase, is encoded by the FBPB gene, having function in cell wall synthesis through its mycolyl-transferase activity[15,36]. Studies have shown that the 85B antigen induces the proliferation and release of high levels of IFN-gamma in T lymphocyte cultures of immunized mice. According to Spencer et al[7], ML2028 may represent a biomarker of disease progression because a patient who developed leprosy had the strongest response to the antigen about 19 mo before clinical diagnosis; due to this incubation time and delay in epitope reactivity, the authors report that this characteristic is dependent on bacterial burden.
The molecular modeling of the 85B antigen was possible due to the similarity between homologous proteins of M. leprae and M. tuberculosis. Thus, the identity rate between the two reaches 85.21%, with the antigen having 11 beta-sheet and 6 alpha-helix regions. According to Santana[6], the external location of reactive peptides facilitates the action of antibodies in the recognition of the antigenic target.
According to Serafín-López et al[37], the specificity of 85B antigen reaches 100%, so that control patients or tuberculosis patients showed no reactivity against the antigen. The results also showed a high degree of antigenicity in leprosy patients, regardless of the clinical classification, and it is therefore an important candidate for serological markers.
Regarding the effector mechanisms of the immune system for mycobacteria and other pathogens, it would be feasible that surface antigens and those secreted would be better candidates for serodiagnostic tests, since these would be more exposed to the components of the immune system. However, this placement needs to be better evaluated, at least in relation to M. leprae, in which a polarity of response is observed. Therefore, it is important to know the biological functionality of the antigen and also its three-dimensional structure in order to be able to evaluate the localization of epitopes within the structure, and thus try to understand the behavior of the antibody response against M. leprae, which is exploited by serological tests. In this context, the prediction of satisfactory structural models is able to collaborate with the development of these tests.
Our research group has been trying to use bioinformatics tools as allies of experimental research in order to corroborate and confirm data. So far the results have been very satisfactory and have saved research time and financial costs.
This study was motivated by our group's previous studies on diagnostic tests for leprosy. Promising and relevant data have been achieved in experimental research with patient serum and have been confirmed by bioinformatics analyses. This study predicting Mycobacterium leprae (M. leprae) antigen models is only one step towards future research on the development of more sensitive diagnostic tests for leprosy.
The aim of this study was to provide reliable three-dimensional structure models for the analysis of immunodominant epitopes that can be tested later, in the form of synthetic peptides, as possible candidates for the development of diagnostic tests that can detect patients with paucibacillary leprosy. The structure and location of the epitope within the antigen structure is important to understand the behavior of the humoral response of patients.
The methods used were classic methods of bioinformatics, which were well established and had proven reliability. Comparative modeling is the simplest methodology of molecular modeling, which was used in this study due to antigen conditions. Once the input data is well filtered, the results are very satisfactory, which can be proven by the similarity of the structures with the homologous ones.
The results obtained in this study were considered of good quality; no important parameters, such as steric impediment and lack of stability, were observed. Therefore, the structure models of M. leprae antigens are satisfactory for the research of immunodominant epitopes.
The structural models of M. leprae antigens are considered high-quality models by validation parameters and can be used for the mapping of epitope candidates for serodiagnostic tests.
The research perspective is to continue the study and map the epitopes and evaluate them through experimental studies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Immunology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Liu S, China S-Editor: Liu JH L-Editor: Ma JY-MedE P-Editor: Liu JH
| 1. | Brasil, Ministério da saúde. Boletim epidemiológico: hanseníase. Boletim Epidemiológico Especial 2021; 1: 9-51. Available from: https://bvsms.saude.gov.br/bvs/periodicos/boletim_epidemiologico_SVS_numero_especial_jan_2021.pdf. |
| 2. | Froes LAR Jr, Trindade MAB, Sotto MN. Immunology of leprosy. Int Rev Immunol. 2022;41:72-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 3. | de Sousa JR, Sotto MN, Simões Quaresma JA. Leprosy As a Complex Infection: Breakdown of the Th1 and Th2 Immune Paradigm in the Immunopathogenesis of the Disease. Front Immunol. 2017;8:1635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 4. | World Health Organization (WHO). Guidelines for the diagnosis, treatment and prevention of leprosy. World Health Organization 2019; 1-30. Available from: https://apps.who.int/iris/bitstream/handle/10665/274127/9789290226383-eng.pdf. |
| 5. | Torres RT, Fachi MM, Böger B, Marson BM, Ferreira VL, Pontarolo R, Guimarães TM. Sensitivity and specificity of multibacillary and paucibacillary leprosy laboratory tests: A systematic review and meta-analysis. Diagn Microbiol Infect Dis. 2021;100:115337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. |
Santana JF.
Mapeamento de epítopos imunodominantes de antígenos de Mycobacterium leprae: caracterização in vitro, |
| 7. | Spencer JS, Duthie MS, Geluk A, Balagon MF, Kim HJ, Wheat WH, Chatterjee D, Jackson M, Li W, Kurihara JN, Maghanoy A, Mallari I, Saunderson P, Brennan PJ, Dockrell HM. Identification of serological biomarkers of infection, disease progression and treatment efficacy for leprosy. Mem Inst Oswaldo Cruz. 2012;107 Suppl 1:79-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 8. | Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57523] [Cited by in RCA: 61997] [Article Influence: 1771.3] [Reference Citation Analysis (0)] |
| 9. | Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947-2948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21085] [Cited by in RCA: 20902] [Article Influence: 1161.2] [Reference Citation Analysis (0)] |
| 10. | Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10124] [Cited by in RCA: 10508] [Article Influence: 328.4] [Reference Citation Analysis (0)] |
| 11. | Benkert P, Tosatto SC, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins. 2008;71:261-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 700] [Cited by in RCA: 758] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 12. | Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB 3rd, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2746] [Cited by in RCA: 3016] [Article Influence: 430.9] [Reference Citation Analysis (0)] |
| 13. | Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1572] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 14. | Tramontano A. Homology modeling with low sequence identity. Methods. 1998;14:293-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Soares BA, Scandelari JPS, Bottolo BM Wagatuma, de Moura J. Engineered biomarkers for immunodiagnosis of leprosy. Editor(s): Joel Faintuch, Salomao Faintuch. Precision Medicine for Investigators,Precision Medicine for Investigators, Practitioners and Providers. Academic Press, 2020; 309-317. [DOI] [Full Text] |
| 16. | Silva WC da S, Costa NL, Argentino S, Oliveira NP, Rodrigues D da S. A estigmatização da Hanseníase: Vivências dos pacientes tratados em uma unidade básica de saúde. BJD. 2020;6:15824-15833. [DOI] [Full Text] |
| 17. | Prabhakaran K, Harris EB, Kirchheimer WF. Hairless mice, human leprosy and thymus-derived-lymphocytes. Experientia. 1975;31:784-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 18. | Shepard CC. The experimental disease that follows the injection of human leprosy bacilli into foot-pads of mice. J Exp Med. 1960;112:445-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 382] [Cited by in RCA: 285] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 19. | Akama T, Tanigawa K, Kawashima A, Wu H, Ishii N, Suzuki K. Analysis of Mycobacterium leprae gene expression using DNA microarray. Microb Pathog. 2010;49:181-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Soares BA, Teixeira KN, de Santana JF, de Assis BLM, Zocatelli-Ribeiro C, Scandelari JPS, Thomaz-Soccol V, Machado-de-Ávila RA, Alvarenga LM, de Moura J. Epitope mapping from Mycobacterium leprae proteins: Convergent data from in silico and in vitro approaches for serodiagnosis of leprosy. Mol Immunol. 2021;138:48-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Potocnakova L, Bhide M, Pulzova LB. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J Immunol Res. 2016;2016:6760830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 231] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 22. | Maeda Y, Mukai T, Kai M, Fukutomi Y, Nomaguchi H, Abe C, Kobayashi K, Kitada S, Maekura R, Yano I, Ishii N, Mori T, Makino M. Evaluation of major membrane protein-II as a tool for serodiagnosis of leprosy. FEMS Microbiol Lett. 2007;272:202-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Benkert P, Künzli M, Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510-W514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 627] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 24. | Biozentrum, University of Basel’s. QMEAN: Qualitative Model Energy Analysis [citado em 11 de julho de 2022]. Em: Swiss-model [internet]. Available from: https://swissmodel.expasy.org/qmean/help#references. |
| 25. | Wiederstein M. ProSA-web Help Page [citado em 11 de julho de 2022]. Em: ProSA-web: Protein Structure Analysis [internet]. Available from: https://prosa.services.came.sbg.ac.at/prosa_help.html. |
| 26. | Silva LX, Bastos LL, Santos LH. Modelagem computacional de proteínas. BIOINFO. 2021;1:1-38. [DOI] [Full Text] |
| 27. | Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10115] [Cited by in RCA: 11821] [Article Influence: 738.8] [Reference Citation Analysis (0)] |
| 28. | Kai M, Nguyen Phuc NH, Hoang Thi TH, Nguyen AH, Fukutomi Y, Maeda Y, Miyamoto Y, Mukai T, Fujiwara T, Nguyen TT, Makino M. Serological diagnosis of leprosy in patients in vietnam by enzyme-linked immunosorbent assay with Mycobacterium leprae-derived major membrane protein II. Clin Vaccine Immunol. 2008;15:1755-1759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 29. | Schlesinger LS, Horwitz MA. Phenolic glycolipid-1 of Mycobacterium leprae binds complement component C3 in serum and mediates phagocytosis by human monocytes. J Exp Med. 1991;174:1031-1038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 62] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Tsukamoto Y, Maeda Y, Makino M. Evaluation of major membrane protein-I as a serodiagnostic tool of pauci-bacillary leprosy. Diagn Microbiol Infect Dis. 2014;80:62-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 31. | Puckett S, Trujillo C, Eoh H, Marrero J, Spencer J, Jackson M, Schnappinger D, Rhee K, Ehrt S. Inactivation of fructose-1,6-bisphosphate aldolase prevents optimal co-catabolism of glycolytic and gluconeogenic carbon substrates in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Williams DL, Torrero M, Wheeler PR, Truman RW, Yoder M, Morrison N, Bishai WR, Gillis TP. Biological implications of Mycobacterium leprae gene expression during infection. J Mol Microbiol Biotechnol. 2004;8:58-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 33. | Sharma D, Lata M, Singh R, Deo N, Venkatesan K, Bisht D. Cytosolic Proteome Profiling of Aminoglycosides Resistant Mycobacterium tuberculosis Clinical Isolates Using MALDI-TOF/MS. Front Microbiol. 2016;7:1816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 34. | Agarwal S, Nguyen DT, Lew JD, Teeter LD, Yamal JM, Restrepo BI, Brown EL, Dorman SE, Graviss EA. Differential positive TSPOT assay responses to ESAT-6 and CFP-10 in health care workers. Tuberculosis (Edinb). 2016;101S:S83-S91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 35. | Geluk A, van den Eeden SJ, Dijkman K, Wilson L, Kim HJ, Franken KL, Spencer JS, Pessolani MC, Pereira GM, Ottenhoff TH. ML1419c peptide immunization induces Mycobacterium leprae-specific HLA-A*0201-restricted CTL in vivo with potential to kill live mycobacteria. J Immunol. 2011;187:1393-1402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 36. | Mattos AMM. Detecção de anticorpos IgG específicos para os antígenos ESAT-6, CPF-10, 16kDa e HBHA em pacientes com tuberculose ativa: importância do diagnóstico e efeito do tratamento quimioterápico. Masters dissertation, Universidade Federal de Juiz de Fora. 2009. Available from: https://repositorio.ufjf.br/jspui/handle/ufjf/9792. |
| 37. | Serafín-López J, Talavera-Paulin M, Amador-Molina JC, Alvarado-Riverón M, Vilchis-Landeros MM, Méndez-Ortega P, Fafutis-Morris M, Paredes-Cervantes V, López-Santiago R, León CI, Guerrero MI, Ribas-Aparicio RM, Mendoza-Hernández G, Carreño-Martínez C, Estrada-Parra S, Estrada-García I. Enoyl-coenzyme A hydratase and antigen 85B of Mycobacterium habana are specifically recognized by antibodies in sera from leprosy patients. Clin Vaccine Immunol. 2011;18:1097-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (1)] |