Copyright
©The Author(s) 2022.
World J Clin Infect Dis. Apr 26, 2022; 12(1): 20-32
Published online Apr 26, 2022. doi: 10.5495/wjcid.v12.i1.20
Published online Apr 26, 2022. doi: 10.5495/wjcid.v12.i1.20
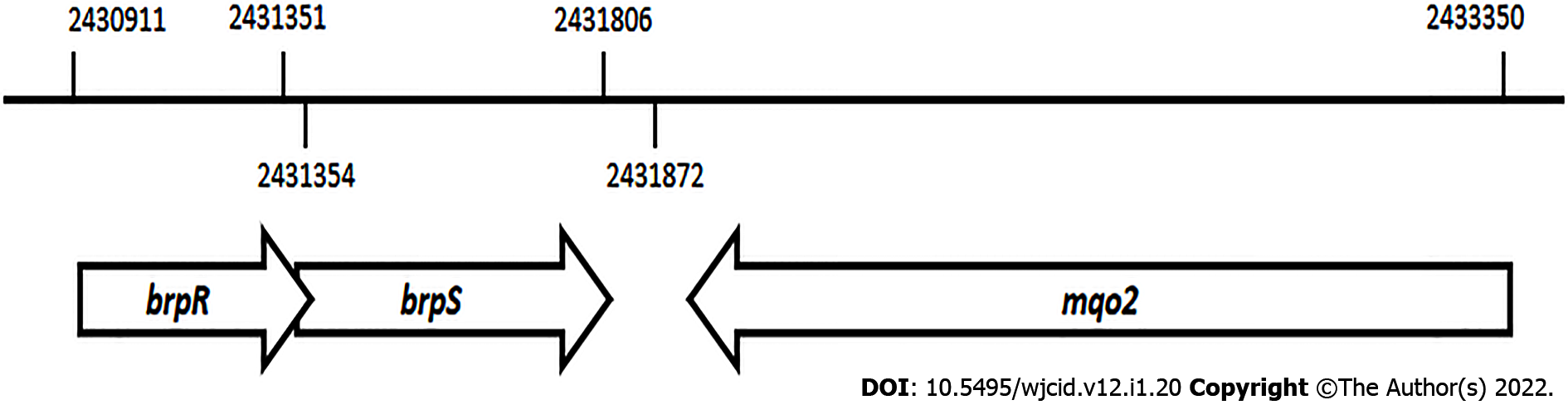
Figure 1 Schematic representation of the chromosomal position and organization of the brpR, brpS, and mqo2 genes in the Staphylococcus aureus strain MW2 genome.
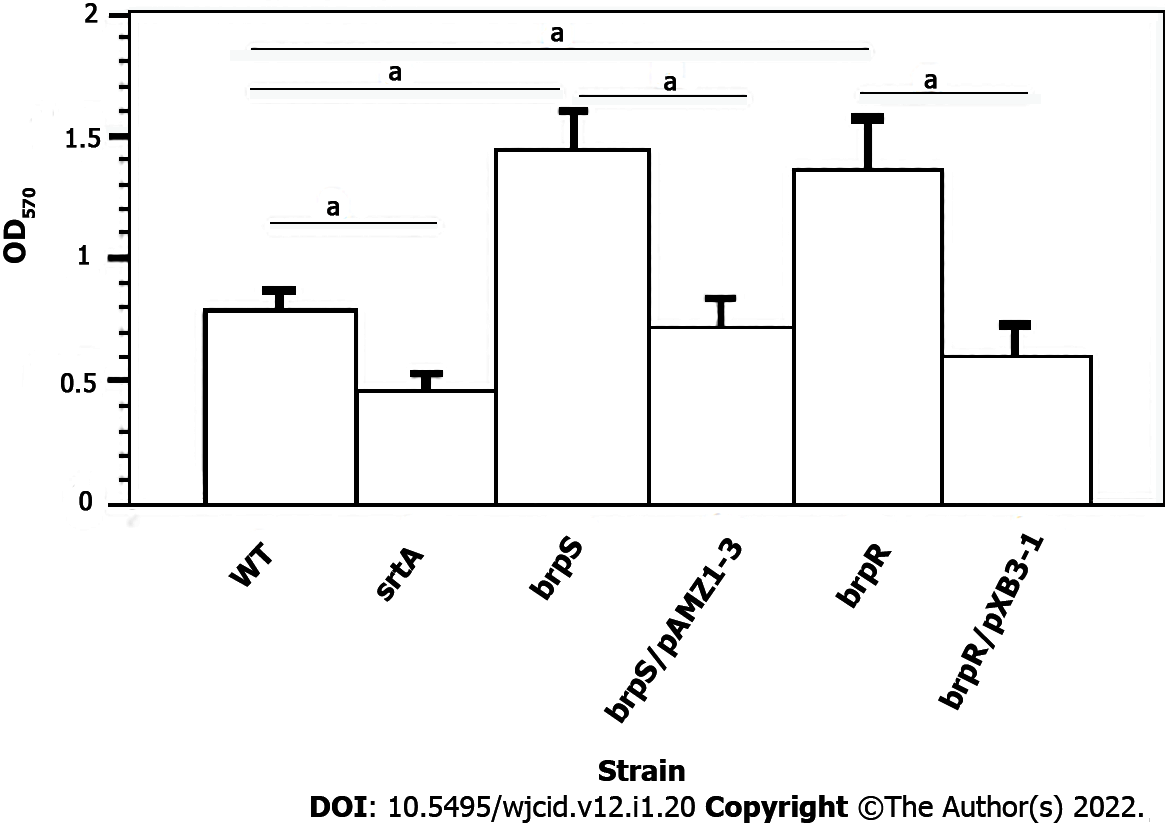
Figure 2 Effect of brpR and brpS mutations and complementation on Staphylococcus aureus biofilm formation.
All experiments represent the mean ± SD from five different runs done in triplicate for each strain. Biofilm formation was done on wild-type Staphylococcus aureus (S. aureus) strain Newman (WT, open column), S. aureus Newman srtA mutant (black column), S. aureus Newman brpS mutant (right striped column), S. aureus Newman brpS mutant/pAMZ1-3 (left striped column), S. aureus Newman brpR mutant (dark right striped column), and S. aureus Newman brpR mutant/pXB31 (dark left striped column). Differences were statistically compared by analysis of variance where aP < 0.001.
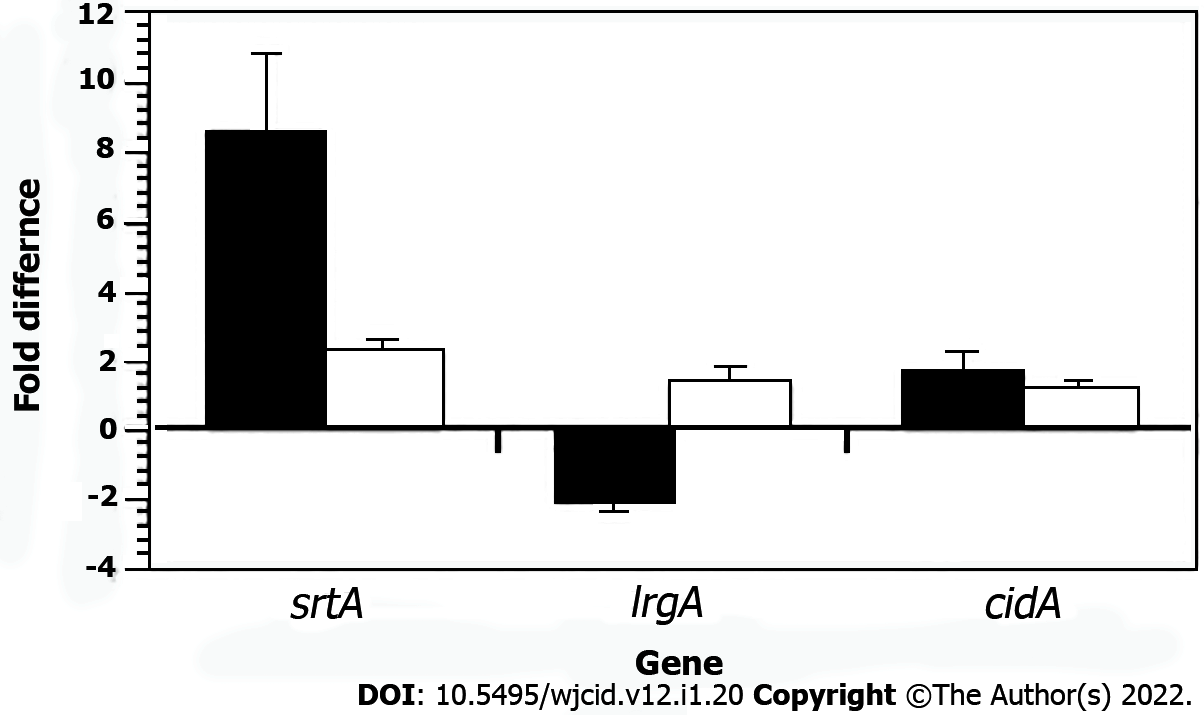
Figure 3 Quantitative reverse transcribed-polymerase chain reaction results of Staphylococcus aureus Newman cidA, lrgA, and srtA transcription in wild-type bacteria (standardized to 0) compared to a Staphylococcus aureus Newman brpS mutant (black column) and the complemented brpS mutant (white column).
The data represents the mean ± SD from three separate runs.
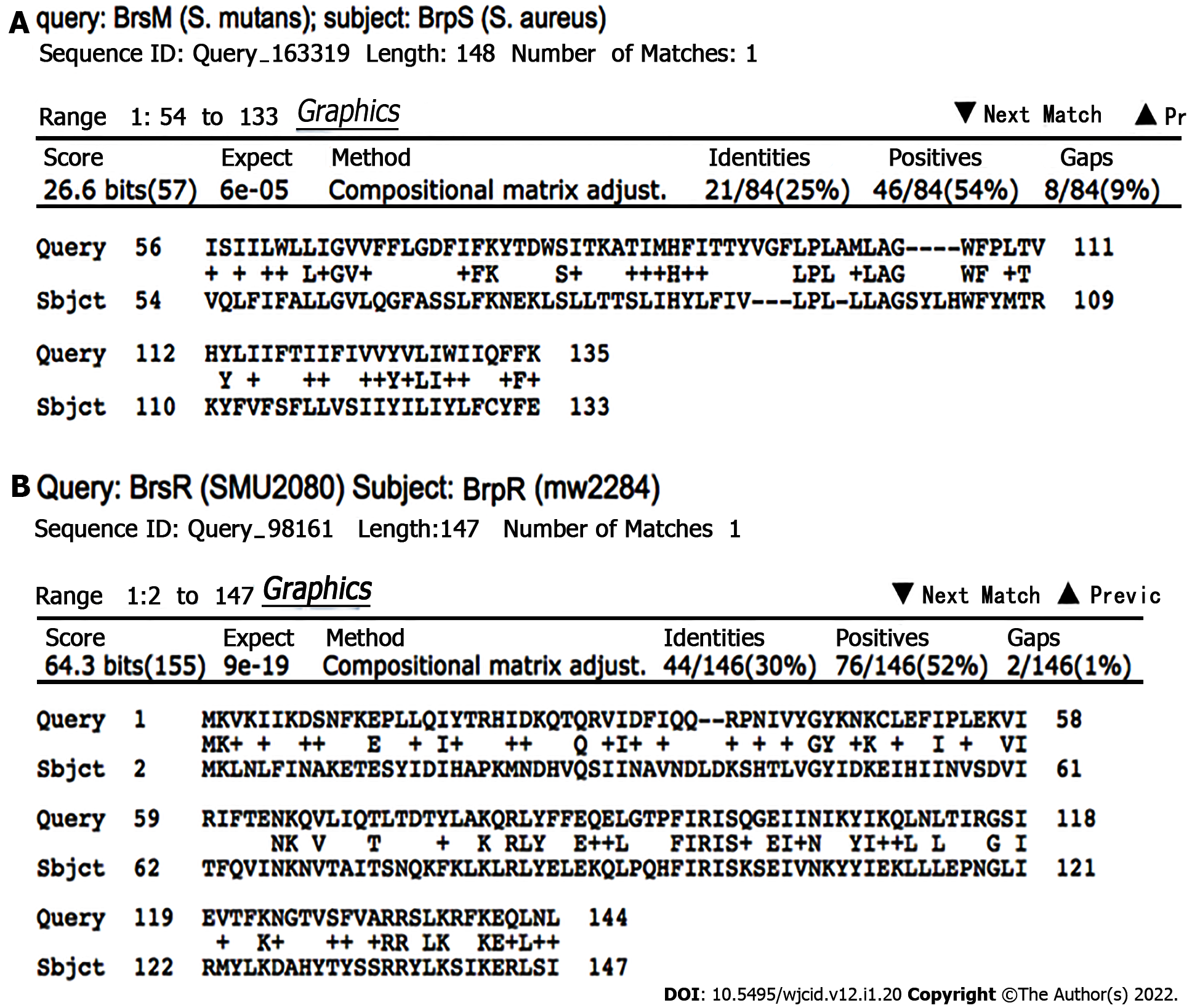
Figure 4 Bioinformatic comparison of the BrsRM proteins of Streptococcus mutans with the BrpRS proteins of Staphylococcus aureus.
A: BrsM, a two-component system membrane protein responsible for activating competence in response to sensing competitor organisms within a niche, shares sequence similarity with BrpS; B: BrsR, which is the cognate response regulator to BrsM, shares sequence similarity with BrpR. BLASTp NCBI. Algorithm parameters: Max target sequences = 100, automatically adjusted parameters for short input sequences, expect threshold = 10, word size = 3, max matches in a query range = 0, matrix = BLOSUM62, gap costs = 11 existence and 1 extension, and a conditional compositional score matrix adjustment.
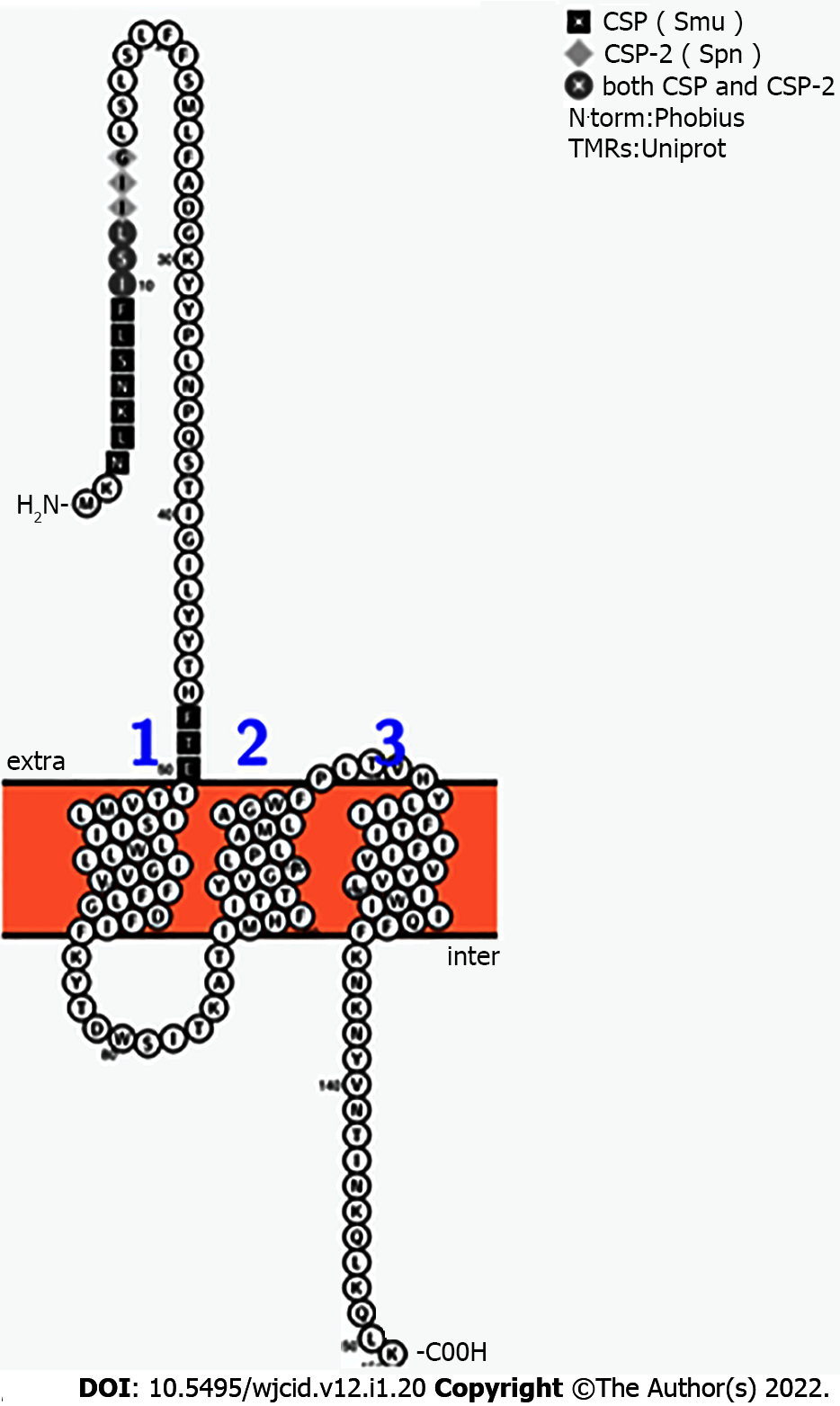
Figure 5 Highlight of amino acid residues shared by competence stimulating pheromone, competence stimulating pheromone-2, and BrpS.
The predicted exterior segment of BrpS, which spans N’-1-MKNLKNSLFISLIIGLSLSLFFSMLFADGKYYPLNPQSTIGILYYTHFT-50-C’, was compared to competence stimulating pheromone (CSP) and CSP-2 by BLASTp. The competence stimulating peptides of Streptococcus mutans (1-SGSLSTFFRLFNRSFTQA-18, CSP) and Streptococcus pneumoniae (1-EMRISRIILDFLFLRKK-17, CSP-2) has 56% similarity to CSP and 30% similarity to CSP-2. CSP: Competence stimulating pheromone.
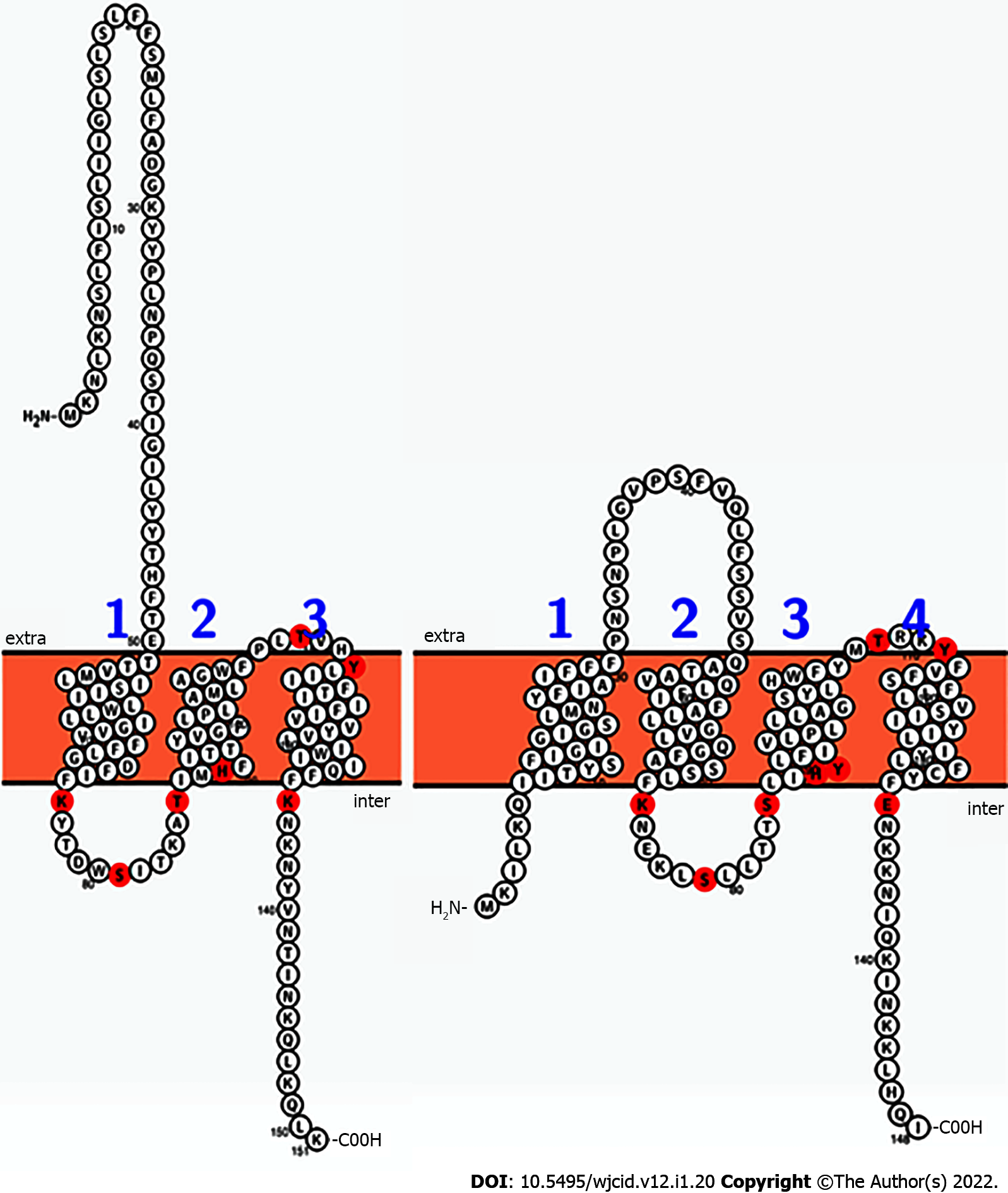
Figure 6 Comparison of the sequences and predicted topologies of the putative two-component system membrane sensor BrpS (left, Staphylococcus aureus) and BrsM (right, Streptococcus mutans).
According to this prediction, residues likely to be reactive (red) are topologically arranged in similar loci among both proteins. Intra is proposed to correspond with the cytoplasmic space, and extra is proposed to correspond with the extracellular milieu of the cell. Figures generated by Protter.
- Citation: Zank A, Schulte L, Brandon X, Carstensen L, Wescott A, Schwan WR. Mutations of the brpR and brpS genes affect biofilm formation in Staphylococcus aureus. World J Clin Infect Dis 2022; 12(1): 20-32
- URL: https://www.wjgnet.com/2220-3176/full/v12/i1/20.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v12.i1.20