Published online Sep 20, 2023. doi: 10.5493/wjem.v13.i4.59
Peer-review started: June 17, 2023
First decision: July 4, 2023
Revised: July 16, 2023
Accepted: August 11, 2023
Article in press: August 11, 2023
Published online: September 20, 2023
Processing time: 89 Days and 15 Hours
Orphan diseases are rare diseases that affect less than 200000 individuals within the United States. Most orphan diseases are of neurologic and genetic origin. With the current advances in technology, more funding has been devoted to de
Core Tip: Neurologic orphan diseases are rare conditions that impact a small percentage of the population. Through new advances in technology and research, the use of genetic treatment for these conditions is increasing. Recent advances in clustered regularly interspaced palindromic repeats/Cas9, adeno-associated viral vectors, antisense oligonucleotides, and mammalian target of rapamycin inhibitors have shown improvements in the care of patients and their quality of life.
- Citation: Kioutchoukova IP, Foster DT, Thakkar RN, Foreman MA, Burgess BJ, Toms RM, Molina Valero EE, Lucke-Wold B. Neurologic orphan diseases: Emerging innovations and role for genetic treatments. World J Exp Med 2023; 13(4): 59-74
- URL: https://www.wjgnet.com/2220-315X/full/v13/i4/59.htm
- DOI: https://dx.doi.org/10.5493/wjem.v13.i4.59
Orphan diseases are rare diseases that affect less than 200000 individuals within the United States[1]. Despite being rare, these diseases affect over 300 million individuals globally[2]. Of the 7000 diseases listed on the National Institute of Health's Office of Rare Diseases site, most are neurological and of genetic origin[3]. 90% of orphan diseases have serious neurological effects[2]. Neurologic orphan diseases are fatal, drastically decrease quality of life, and are defined by long periods of disability[4]. However, diagnosis and treatment of rare central nervous system (CNS) disorders pose a challenge[2]. Lack of access to diagnostic genomic sequencing, screening tests, and specialists contributes to the difficulty of diagnosing and managing neurologic orphan diseases[2]. Most neurologic orphan diseases don’t have treatments that prevent disease progression[4]. Additionally, clinical trials investigating neurologic orphan disease therapeutics have the lowest success rate[5]. Global Genes, a rare disease advocacy organization, states that since 2021, a total of $22.9 billion has been invested in research on neurologic orphan diseases, which offers a promising future for patients with these conditions[6].
One study found that the use of whole genome sequencing within clinical practice increases the diagnosis of neurologic orphan diseases[7]. Genetic conditions such as Huntington's disease and Friedreich's ataxia are caused by repeat expansions which can effectively be detected by whole genome sequencing due to its high sensitivity and specificity for repeat expansions[7].
The Patient Identification and Engagement for Rare CNS Disorders initiative is designed to investigate and improve barriers to diagnosis and clinical research trials[2]. This initiative plans to address the underrepresentation of certain groups from clinical trials and to improve access for those individuals in the participation of trials investigating new gene therapy approaches[2]. The future for the diagnosis and management of neurologic orphan diseases looks promising and hopes to improve the efficacy of therapeutic agents by gene targeting methods. Some forms of gene therapy include the use of clustered regularly interspaced palindromic repeats (CRISPR)/Cas9, antisense oligonucleotides (ASO), adeno-associated viruses (AAV), and mammalian target of rapamycin (mTOR) inhibitors.
CRISPR/Cas9 is a form of gene-editing where a guide RNA binds to a target sequence of genomic DNA, followed by the endonuclease, Cas9, binding to the guide RNA. Cas9 then creates a double strand break in the genomic DNA which is then repaired. This process allows for the inclusion or exclusion of desired genes to create a desired mutation[8].
ASO are small molecules that can modify gene expression preventing or altering protein production. If a certain protein is undesired, an ASO can be designed to cause the protein to be terminated or partially expressed and modify it so that it is not harmful[9].
AAV are used as a vector for gene therapy by using a non-enveloped virus engineered to deliver deoxyribonucleic acid (DNA) to targeted cells[10]. This gene therapy has shown preclinical and clinical access in gene replacement, gene silencing, and gene editing[11].
mTOR is a kinase closely correlated with the occurrence of neurodegenerative diseases and tumors in humans. The goal of mTOR inhibition therapies is to block the mTOR signaling pathway that may be contributing to abnormal signal transduction to block the occurrence and development of disease[12].
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disease caused by deletions or mutations in the survival motor neuron (SMN1) gene[13]. Specifically, diagnostic testing commonly demonstrates the absence of SMN1 exon 7 on chromosome 5[14,15]. SMA is characterized by progressive muscle weakness and atrophy resulting from progressive degeneration and irreversible loss of anterior horn cells in the spinal cord[16]. The severity of SMA can range from more mild cases where the onset occurs in adulthood and progresses at a slow rate to more severe cases where the onset can occur in the first months of life and result in respiratory failure[17].
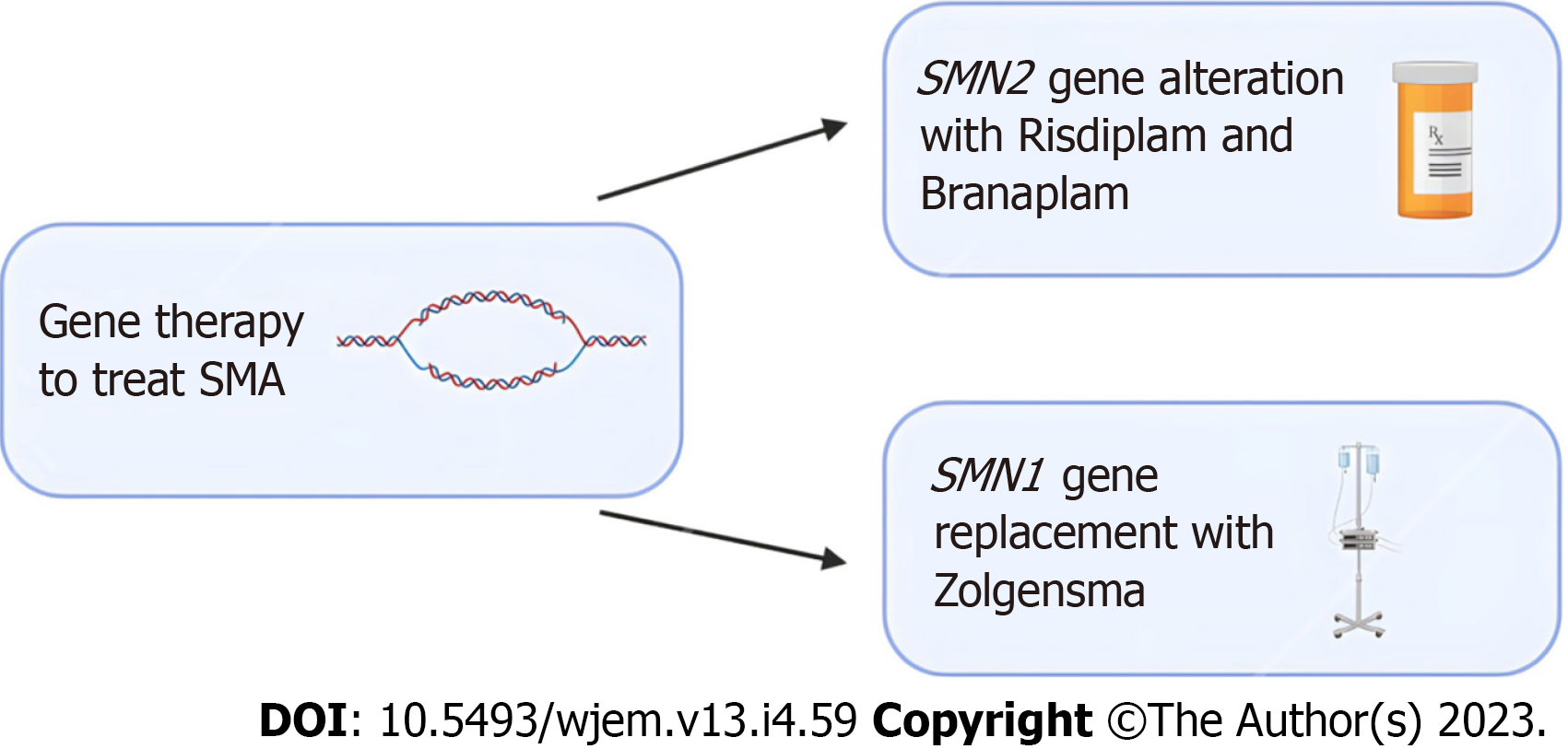
In recent years, new treatment options like gene therapy involving splicing modulation of SMN2 and SMN1 genes and the development of the first approved drugs for SMA treatment have shown promise in treating SMA. When therapy is initiated early, it can significantly alter the natural course of the disease. Current evidence in these treatments is limited to a small scope of patients and more research is needed for conclusive results[17].
Nusinersen was the first drug that received approval for the treatment of SMA. Nusinersen enhances the inclusion of exon 7 in mRNA transcripts of SMN2 by suppressing the binding of certain splicing factors which results in an increase in functional SMN2-mRNA with included exon 7[18-20]. Various studies in infants and young children have displayed improvements in prolonged time of death and improved motor functions[21-23]. Furthering the potential of SMN2 gene alteration, Risdiplam, and Branaplam are oral medications that have been shown to cross the blood-brain barrier and increase the number of full-length SMN proteins[24].
Gene therapy has also shown promise with SMN1-gene replacement. Studies in mice have shown prolonged survival following successful vector delivery of intact SMN1-gene across the blood-brain barrier[25,26]. Zolgensma is a gene therapy medicine that is administered as an intravenous infusion and uses adeno-associated virus vectors to deliver a functional copy of SMN to motor neuron cells[27]. Clinical trials in children treated with Zolgensma have shown improved survival, motor function, and developmental milestones following treatment[28,29]. A summary of emerging treatment options for SMA can be found in Figure 1 below.
Duchenne muscular dystrophy (DMD) is the most common hereditary neuromuscular disease and is one of the most severe forms of inherited muscular dystrophy. Symptoms of DMD include severe and progressive muscle wasting, muscle weakness, and difficulty with movement[30]. Late stages of DMD often require the need for assisted ventilation and the use of a wheelchair to perform daily activities and lead to premature death in the mid-twenties due to respiratory muscle weakness or cardiomyopathy[31].
DMD is an X-linked inherited disorder that predominantly affects males. The onset of DMD occurs due to mutations in the dystrophin gene on chromosome Xp21. This results in a ceased production of dystrophin in cardiac and skeletal muscle. The absence of dystrophin results in a loss of myofibril membrane integrity through cycles of necrosis and regeneration. Fibrous connective tissue and fat then replace muscle over time, resulting in the progression of expressed clinical symptoms[30,32].
In recent years, there has been progress in the development of diagnosis and therapeutics for DMD, but the current treatments given do not cure DMD[33]. Daily prednisone treatment is commonly used to increase muscle strength and function, improve pulmonary function, and significantly slow the progression of weakness[34,35]. While this treatment does not cure the disease, it does improve the overall quality of life for patients.
The most advanced therapy is antisense oligonucleotides -mediated exon skipping which has shown promise in clinical application. For this technique, the administration of 20-30 bp long antisense oligonucleotide hybridizes to splice motifs necessary for pre-mRNA processing and mask RNA splicing signals. This leads to the exclusion of the intron and adjacent exon, creating an in-frame mRNA without the targeted exon. This mRNA can then be translated into a truncated and partially functional dystrophin protein[33,36].
CRISPR/Cas technology has been used therapeutically to treat DMD by upregulating a dystrophin homolog, utrophin, to compensate for the lack of dystrophin protein which has been successfully demonstrated in patient cells. In these patients, full-length dystrophin was restored in patient cells carrying duplication mutations[37]. Manipulations resulting from CRISPR-Cas9 were shown to restore the expression of truncated but partially functional dystrophin, improve skeletal and cardiac muscle function, and increase the survival of mdx mice significantly[38].
Emerging vector-mediated gene therapy in mice has been shown to deliver a functional DMD gene to cells lacking dystrophin protein and has been shown to increase exercise capacity[39]. While promising, this technique is challenging due to the very large size of the dystrophin gene and the widespread distribution of muscles[40].
Dravet syndrome (DS), first described by Charlotte Dravet in 1978, is an early-onset epileptic syndrome characterized by a variety of refractory seizures and neurodevelopmental impairment that often persist into adulthood[41,42]. The clinical features of DS typically progress over time, most commonly first presenting as a bilateral tonic-clonic in the first year of life, with half of the patients being febrile[43]. The disease progresses to multiple types of seizures, often leading to poor therapeutic control and the majority of patients (93%) experiencing status epilepticus[43,44]. Additionally, patients with DS will develop neurodevelopmental delay, as well as motor and cognitive impairment that will persist into adulthood. It should be noted, however, the diagnosis is highly clinical as both magnetic resonance imaging (MRI) and electroencephalogram (EEG) studies may be nonspecific[45].
Previously named severe myoclonic epilepsy of infancy, the molecular basis of DS arises from a de novo mutation on chromosome 2q24 on the sodium voltage-gated channel alpha subunit (SCN1A) gene[45,46]. Although the role of the SCN1A variant in the pathogenesis of DS has not been fully elucidated, some studies have suggested that the diffuse neuronal hyperactivity in DS patients is correlated with a loss of inhibitory GABAergic interneurons which have an SCN1A mutant in non-coding regions[47,48].
Currently, DS is managed symptomatically with a series of anti-seizure medications (valproic acid being the first line of treatment), though this regimen has variable efficacy[48]. Gene-specific therapies for the causes of DS continue to gradually emerge. One study used Targeted Augmentation of Nuclear Gene Output technology, utilizing ASO, to successfully increase the expression of the SCN1A protein in mice models. In addition to a higher expression of this SCN1A product, the incidence of seizures in these DS mice was significantly reduced[49]. The use of viral vectors to target genes has shown success in some diseases but poses a challenge in DS[50]. The SCN1A coding sequence is 6kb long, which exceeds the carrying capacity of adeno-associated viruses[50]. However, the use of a different viral vector, such as lentiviruses, poses another challenge as it demonstrates a limited spread in neural tissue therefore it cannot effectively treat large brain areas[50]. Another approach to upregulating the expression of SCN1A is through the use of dCas9-based gene activation systems. One study found that the use of dCas9 resulted in the upregulation of SCN1A in brain tissue and cultured neurons[51]. Overall, there have been significant developments in viral vectors and molecular techniques that demonstrate promising results.
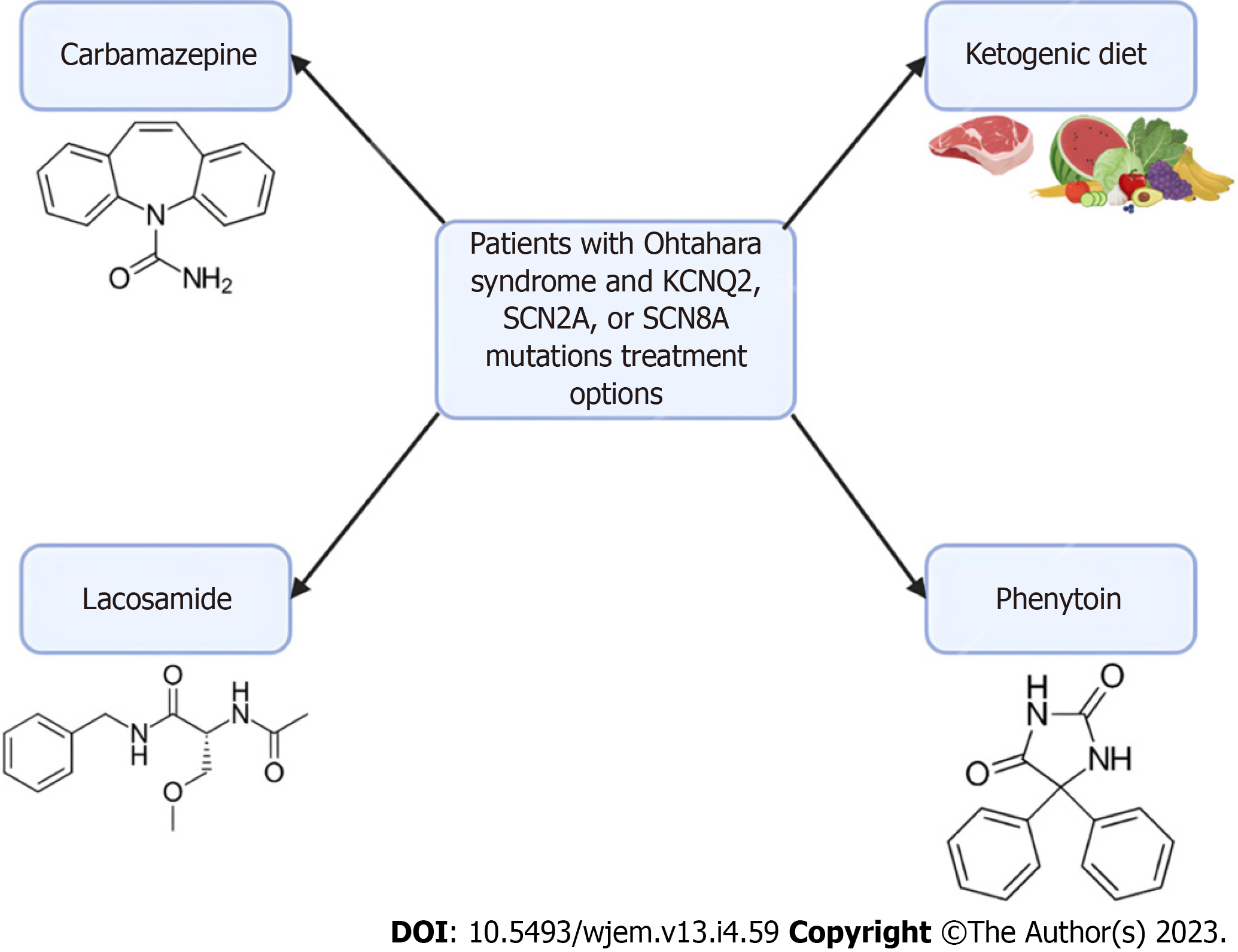
Ohtahara syndrome, also referred to as Early Infantile Developmental and Epileptic Encephalopathy (EIDEE) syndrome, is a group of devastating pediatric conditions characterized by frequent spasms in neonates and infants leading to severe cognitive and physical disabilities and even death[52,53]. There are numerous causes of Ohtahara syndrome, including structural brain defects, metabolic derangements, and genetic variants. Multiple studies have identified the most common variants associated with Ohtahara syndrome are PRRT2, SCN1A, KCNQ2, and SLC2A1[54]. The extensive distribution of these channelopathies and their various penetrance help explain the evolution of focal seizures to status epilepticus seen in EIDEE[54,55].
Seizure episodes in Ohtahara syndrome are initially treated with anti-seizure medications but are usually only in managing the frequency of seizures. Patients with Ohtahara syndrome with KCNQ2, SCN2A, or SCN8A mutations have been found to respond to sodium channel anti-seizure medications, such as carbamazepine, lacosamide, or phenytoin[56,57]. Additionally, implementing a ketogenic diet has been shown to provide some improvement in many infants[58]. Currently, the rise in genetic testing has allowed identifying the monogenic etiology of various EIDEE. An AAV-based gene replacement therapy has been proposed for Ohtahara syndrome particularly for the transmembrane sodium channel SLD13A5, though these principles could provide insight into future therapeutic targets of Ohtahara syndrome[59]. However, challenges exist for gene therapy due to the limited knowledge of the disease mechanism and progression of Ohtahara syndrome. Further research is needed in patients with Ohtahara syndrome to evaluate the efficacy of gene therapies such as viral vectors. A summary can be found in Figure 2 below.
Lennox-gastaut syndrome (LGS) is another severe pediatric epileptic and encephalopathic disorder characterized by severe pediatric seizures, treatment-resistant epilepsy, and cognitive impairments[60,61]. As in various other childhood epilepsy disorders, LGS is caused by several etiologies, including genetic predispositions, anatomical brain abnormalities, hypoxic-reperfusion encephalopathies, meningitis, and head trauma[61]. The majority of children affected by LGS have underlying genetic disorders, often chromosomal syndromes, or de novo pathogenic variants[61,62].
As LGS is characterized by seizures resistant to pharmacologic therapy, management often includes a multidisciplinary approach, often including dietary and surgical interventions. In one case series, 50% of children received a greater than 50% reduction in seizure frequency, and almost a quarter of the children achieved a greater than 90% reduction[63]. Additionally, the serotonergic agent fenfluramine is commonly used in LGS and has been shown to have significant benefits for the reduction of generalized tonic-clonic seizures and drop seizures[64]. Fenfluramine increases the level of serotonin in the extracellular compartment and acts as a serotonergic 5-HT2 receptor agonist and an alpha-1 receptor antagonist to decrease anti-epileptic activity. Since LGS seizures evolve over a patient’s life, an LGS algorithm detailing several anti-epileptics medications as well as adjunctive therapy including a ketogenic diet, possible surgical resection, and close EEG monitoring[65].
In the discourse of neurologic orphan diseases, one important consideration in the context of neurodegenerative movement disorders is Friedreich Ataxia (FRDA). In the second half of the 19th century, its original description by German professor of medicine at Heidelberg, Nikolaus Friedreich, remarked FRDA was a degenerative atrophy of the posterior spinal cord columns[66]. Further, between the years of 1863-1877, Friedreich published the earliest and most extensive works on “Friedreich’s ataxia”, which provided insightful descriptions of this neurological disorder as being marked with the principal abnormality of axonal thinning without axonal loss of the dorsal spinal roots[66]. Unfortunately, Friedreich received little recognition for his academic effort during his lifetime, however, an anonymous obituary of “perhaps his most important work” details Friedreich’s rapid scientific progress and 59 publications[67]. Nonetheless, the full extent of FRDA’s etiology couldn’t be fully appreciated until direct genetic testing became available in the late 1960s[68]. Thus, the proceeding discussion will review current knowledge on this debilitating condition and evaluate the recent developments in treatment approaches.
As the most common inherited ataxia—defined as the compromised coordination of voluntary muscle move
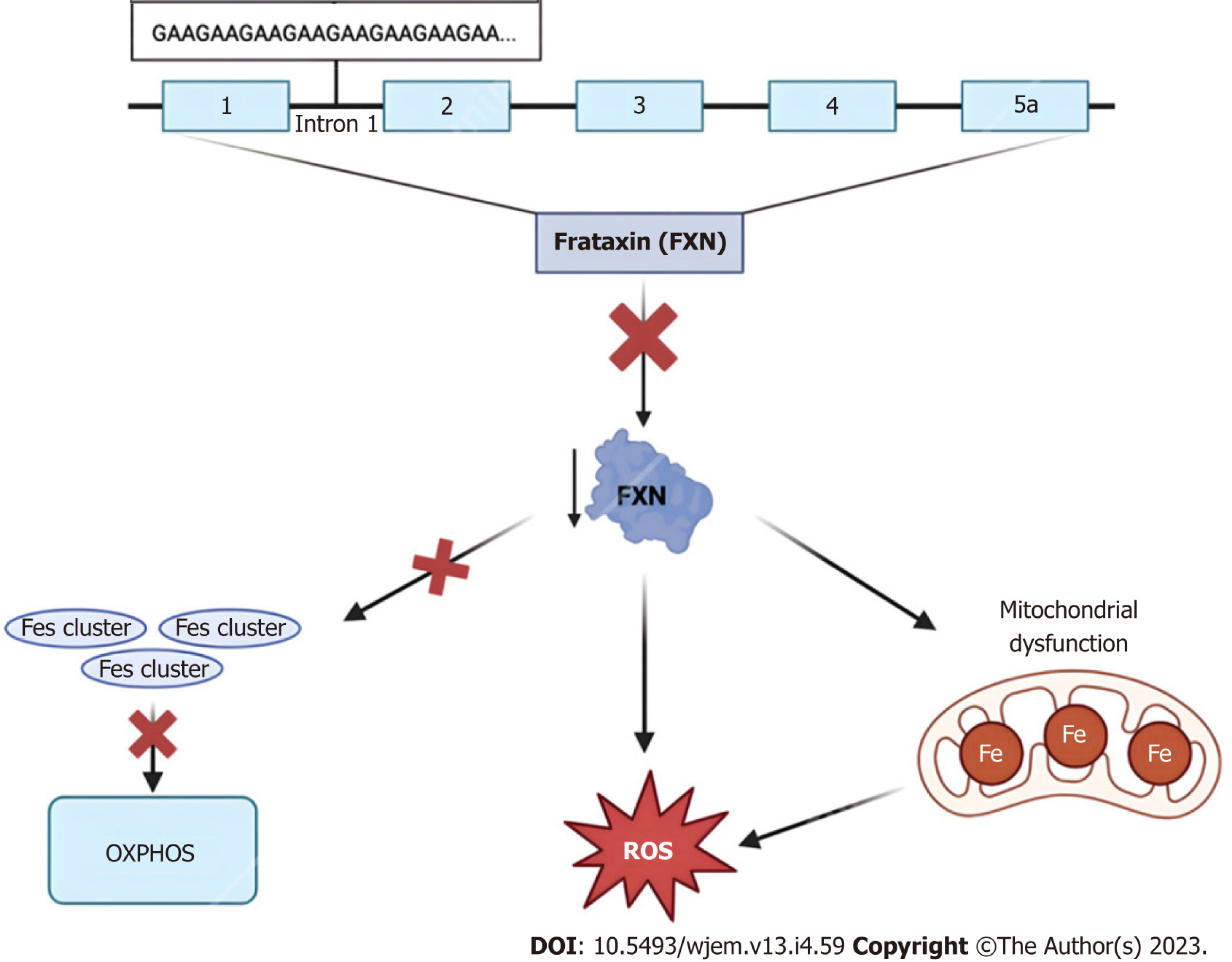
Clinically, due to frataxin’s ubiquitous biochemical role in numerous cellular pathways, humans with frataxin deficiency manifest with dysfunction in the central and peripheral nervous system, heart, skeleton, and even endocrine organs including the pancreas[71]. Regarding FRDA in particular, many of these patients classically present with neuropathologic disabilities including progressive ataxia, peripheral sensory loss, and muscle weakness beginning between the ages of 5 and 15 years old[76]. The aforementioned manifestations are chiefly secondary to neuropathy in the dorsal root ganglia, accompanied by degeneration of both peripheral sensory nerve fibers and posterior columns of the spinal cord[76]. Moreover, non-neurological areas of morbidity include the heart, typically in the form of left ventricular hypertrophy due to mitochondrial proliferation, and the pancreas, with approximately 10% of all FRDA patients developing diabetes mellitus[76,77].
Before the dawn of genetic testing, the constellation of the above-mentioned signs and symptoms as well as MRI was employed in the diagnosis of FRDA[78]. Presently, however, this neurologic disorder is more precisely diagnosed using modern genetic testing such as Southern blot and conventional polymerase chain reaction (PCR) techniques[79]. The accurate diagnosis of FRDA helps in differentiating it from other ataxias and provides a guide for physicians to appropriately tackle the treatment and management of this progressive disorder. Despite the heterogeneity of symptomatic presentation, most patients will lose the ability to independently walk, stand, and/or sit within 10-15 years of initial disease onset[76]. Therefore, there is a persisting call for advancements in techniques and approaches for treating FRDA.
In the past, there were no effective protective pharmacological agents for FRDA, and thus treatment was aimed at symptomatic management and physical therapy for impaired motor function[80]. Further, aggressive surveillance was and still is used in managing progressive cardiomyopathy, arrhythmias, and diabetes mellitus to improve the quality of life for these patients[80,81]. More recently, treatment approaches to address the mitochondrial dysfunction caused by a mutation to the frataxin protein include mitochondrial function enhancers, free radical scavengers, and iron chelators such as coenzyme Q10 and its synthetic analog idebenone, as well as vitamin E[82]. Although the employment of such molecules is mechanistically sound, the pre-clinical and clinical trial data on their use has demonstrated little to no therapeutic effect across multiple studies and even worsened enzymatic activity[80]. As of February 2023, however, the first FDA-approved treatment for FRDA became available within the United States. Developed by Reata Pharmaceuticals, Inc., the small molecule Omaveloxolone, or SKYCLARYS™, is an orally active drug aimed at alleviating oxidative stress, mitochondrial damage and dysfunction as a direct result of Nrf2 pathway suppression found in Friedreich patients[83]. As a novel pharmaceutical with early proven safety and effectiveness, Omaveloxolone is a promising emerging option for FRDA treatment.
In addition to symptomatic treatment and management, newer therapeutics for neurological disorders are being developed, evidenced by the rapid increase in research surrounding gene and cell therapy for FRDA between the years 2000 and the present day. Considering the severity of neurophysiological abnormalities is strongly correlated with the size of the GAA repeat expansion implicated in the genetic inheritance of FRDA, many of these novel techniques are aimed at altering this pattern. Specifically, the use of the CRISPR-Cas9 system in GAA expansion-based animal models such as YG8-derived cells and mouse models demonstrated promising results in the successful editing of GAA expansion both in vitro and in vivo[84]. Contrastingly, instead of altering the primary genetic mutation, other more recent methods of ataxia reversal are aimed at inducing the expression of the frataxin protein itself[85]. Particularly, Piguet et al[86] demonstrated the complete reversal of sensory ataxia and cardiomyopathy via viral vector-based introduction of AAV-expressing frataxin (AAV-FXN) in cells and mouse models. Finally, other prospective therapeutics look to stimulate the direct transcription of FXN and/or increase FXN mRNA stability via interferon administration, or indirectly via the stimulation of NRF2—a transcription factor whose levels are intimately associated with FXN mRNA expression[87]. Altogether, these animal-based models hold promising treatment options for future application to clinical medicine, for now, more research is necessary before translation into FRDA human-based clinical trials.
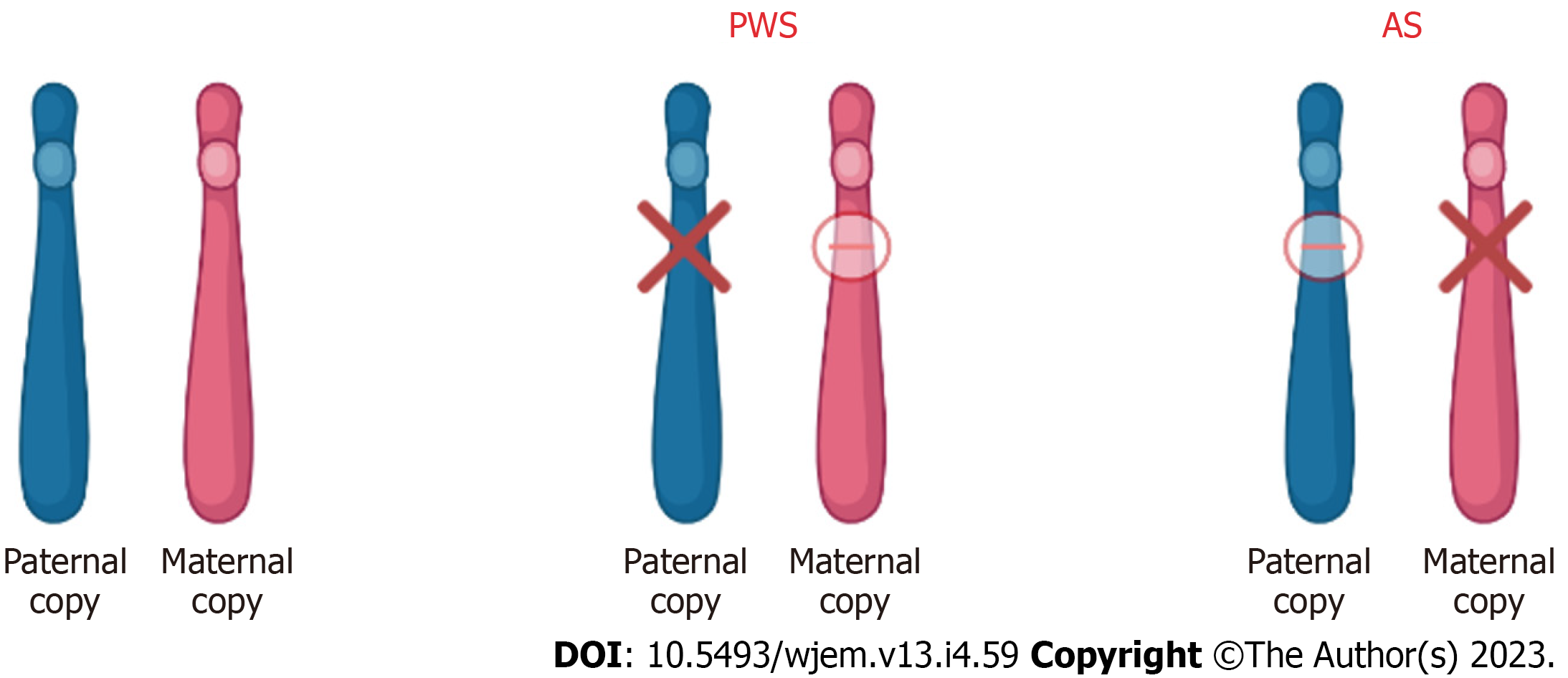
Prader-Willi syndrome (PWS) and Angelman syndrome (AS) are the first known examples of diseases relative to imprinted genes. These diseases have their own clinically distinct behavioral, cognitive, and neurologic phenotypes. However, they are still considered “sister disorders” due to the common origin of the disease from the imprint gene abnormalities in the 15q11-q13 region[88]. The difference in the diseases is that with PWS, the contribution is paternal whereas with AS, the contribution is maternal. This parent-of-origin difference is the reason for the variable expression of the disease depending on the sex of the parent from whom the disease is inherited[88,89].
PWS is suspected in individuals that present with clinical features of hypotonia (during the first few years) that lead to hyperphagia, hypogonadism, short stature, and mild mental retardation[90]. However, regarding the 15q11-q13 region, the abnormality is suggestive of PWS, but not diagnostic. The diagnosis of PWS is established by identifying the abnormal DNA-methylation and maternal-allele imprinting in the Prader-Willi critical region. This can be due to either the deletion of the paternal allele, maternal disomy, and/or imprinting defect causing the absence of the paternal allele[91]. Moreover, point mutation does not cause PWS because it is a factor of multiple gene products. An exception to this rule is when the point mutation is seen in the imprinting control region[92]. In these cases, although very rare, the loss of function has been seen to contribute to many different aspects of the PWS phenotype[93].
The underlying molecular defect in PWS further opens a great scope of molecular treatment and epigenetic therapy. An increasing number of studies are pointing toward gene SNORD116 being the main causative player in PWS[94-96]. Deactivation of the SNORD116 gene is associated with a complex of a zinc finger protein ZNF274 and SETDB1 a histone H3 methyltransferase[97-99]. Thus, CRISPR-Cas systems can be employed to deactivate ZNF274 and/or SETDB1 and increase the expression of SNORD116. One of the drawbacks to this approach is that the SETDB1 is not specific and thus, targeting it can have many off-label systemic effects.
Another approach explored in many studies was to inhibit G9a, another histone H3 methyltransferase, which restored the targeted genes from the silenced maternal chromosome[100-102]. Based on these studies, the use of a G9a inhibitor improved perinatal lethality and poor growth, which ultimately can improve the life span[100-102]. But again, one of the drawbacks of these research studies is that they are all performed with either mouse models or PWS patient-derived cells. Thus, the efficacy of gene therapy in PWS patients is yet to be explored and proven to be beneficial. Moreover, during the development of new drug therapies, focus should be put on off-target effects. Epigenetic drugs are known to have complex and broad effects, and thus, the development of specific molecular therapy for PWS patients is necessary.
AS clinically presents as severe developmental delay, minimal or no speech, difficulty walking, and a unique behavioral phenotype that includes frequent smiling and excessive laughter[103]. As mentioned above, defects in the 15q11.2-q13 chromosome region are the genetic basis of both PWS and AS. In terms of AS however, the maternally expressed allele does not produce functional gene product, and the paternal allele is imprinted. The gene of interest in terms of AS is the UBE3A sequence which is mutated in certain individuals with AS[103].
In terms of epigenetic therapy, restoration of the mutated UBE3A gene expression is more favorable and plausible rather than targeting known activities of the molecule. However, there haven’t been any successful ways researched to do so. Thus, expressing the silenced paternal UBE3A has been an adequate alternative. The most common way proposed is by administering topoisomerase-I inhibitors and reactivating the inhibition of paternal UBE3A.
Topoisomerase-I inhibitors like topotecan and irinotecan are FDA-approved chemotherapeutic drugs. Moreover, these drugs are also shown to promote the expression of paternal UBE3A in a dose-dependent manner. The drugs work by reactivating the gene via reduced transcription of an imprinted antisense RNA[104-106]. Despite proving their potential, topoisomerase inhibitors are not currently used as a first-line treatment for AS due to their side effects. According to one study, topoisomerase inhibitors also reduced the levels of other genes that are linked to autism, thus increasing the potential of autism characteristics in individuals[107].
Thus, as discussed, all the approved therapies for PWS and AS target the management of symptoms rather than the actual disease. Thus, there is currently no cure for either AS or PWS. Gene and molecular therapies offer a promising way into precision medicine and the development of novel therapies however a lot more research needs to be done regarding offsetting the systemic side effects. A summary of the chromosome 15 abnormalities in PWS and AS is shown in Figure 4 below.
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder affecting both the upper and lower motor neurons and is the most common motor neuron disease in adults[108-110]. The onset of ALS symptoms typically occurs between 50-70 years old and is more common in males than females[111-113]. While some individuals can survive over 10 years with ALS, most survive less than 3 years due to the rapid disease progression[114]. Current treatments focus on the treatment of symptoms; however, their effect is often minimal by extending a patient’s survival by only three months[112]. Although the etiology of ALS is currently unknown, there does appear to be a genetic component as nearly 10 percent of diagnosed individuals reported having a family member with ALS and are said to have familial ALS (fALS), while the remaining individuals are classified as having sporadic ALS (sALS)[115].
In recent years, the number of genetic mutations linked to ALS has increased. Superoxide dismutase-1 (SOD1) was the first gene discovered to be associated with ALS, and mutations of the SOD1 gene account for 10%-15% of fALS and 1%-2% of sALS[110,115-117]. More recently, mutations to the C90RF72 gene have been associated with 30%-40% of fALS and nearly 6% of sALS[115,116]. Mutations to other genes such as TDP-43, FUS, OPTN, TBK, GRN, NEK1, and C21ORF2 have also been linked to ALS[115,116]. Current evidence suggests that many of these mutations result in the aggregation of misfolded proteins, which then causes ALS[110,112]. Furthermore, knockout models tested on mice have shown that the removal of these genes typically does not result in symptoms of ALS, suggesting that these mutations result in a gain of function[112,116,117].
Current ALS treatments in clinical trials focus on gene therapy as opposed to treatment of the symptoms of ALS. These treatments attempt to replace the mutated gene, inactivate the mutated gene, or introduce a new gene to fight ALS[117]. Tofersen, an ASO, is currently in clinical trials for individuals suffering from ALS because of a SOD1 mutation. Tofersen causes the degradation of SOD1 mRNA, decreasing the amount of SOD1 protein[118]. However, ASOs such as Tofersen are unable to cross the blood-brain barrier and must be injected into the cerebrospinal fluid[117,118]. This can cause many unwanted side effects, such as inflammation, infection, and long and repeated injections[117]. As a result, nanoparticles, such as calcium phosphate lipid nanoparticles, are being developed to reduce the need for direct injection of ASOs into the CSF[117]. Additional research is being conducted on AAV vectors for the modification of genes. The major advantage of AAV vectors is that only one injection is necessary[116]. AAV vectors have the potential to deliver gene-silencing material, such as antisense sequences, to mutated genes such as the SOD1 gene[116]. However, immunoreactivity to AAV has been documented in both animal and human studies presenting future challenges to the use of AAV vectors for ALS treatment[116].
Huntington’s disease (HD) is a neurodegenerative disorder resulting from the expansion of a CAG trinucleotide sequence on the HTT gene which encodes the huntingtin protein[119-121]. For an individual to be at risk for HD, they must have greater than 36 glutamine repeats, and as the number of repeats increases the age of onset typically decreases[120]. HD is inherited in an autosomal dominant manner, with symptoms first appearing in mid-life[121,122]. Individuals with HD experience cognitive, motor, and psychiatric symptoms that are progressive over time[119]. Current treatments for HD focus on treating symptoms but often have limited benefits, and there is no available disease-modifying treatment[121,123].
The HTT gene encodes the huntingtin protein, which is involved in a variety of cellular functions including cell division, vesicle transportation, and transcription regulation[120]. Repeat CAG trinucleotide sequences found on exon 1 of the HTT gene cause the formation of mutant huntingtin protein aggregates within neurons[124,125]. Individuals who possess 60 or more CAG repeats will develop HD before the age of 20[124,125]. However, the CAG repeat length explains less than 50% of the HD age of onset[124]. Other factors such as glutamic acid polymorphisms and glutamate and N-methyl-D-aspartate receptor polymorphisms also play a role in the onset of HD[124]. These genetic modifiers all present potential targets for genetic treatment for HD.
As a result of the many functions of the huntingtin protein, the embryonic knockout of the HTT gene in mice was lethal[120,121]. Furthermore, the inactivation of the HTT gene after birth has been linked to neurological deterioration in mice[121]. Because of this, many HD disease-modifying treatments being researched focus on gene editing as opposed to gene silencing[125]. In a phase 1-2a clinical trial conducted between 2015-2017, an ASO known as IONIS-HTTRx was studied for its safety, ability to remain in the CSF, and ability to reduce mutant HTT in the CSF[126]. Upon completion of this trial, it was found that IONIS-HTTRx did not increase the number of adverse effects and resulted in a dose-dependent reduction of the mutant HTT concentration[126]. However, phase three trials show that IONIS-HTTRx may cause worse motor and cognitive function[127]. Along with ASOs, RNA interference (RNAi) technology is being developed to treat HD. RNAi technology attempts to decrease the amount of mutant HTT being translated using small non-coding RNAs[127]. This RNAi technology is typically delivered through an AAV. However, this technology is in the early stages of development, with significant testing still needed.
Neurofibromatosis-1 (NF-1), also known as Von Recklinghausen disease, is an autosomal dominant disorder characterized by changes in the nervous system, bones, and skin[128]. Risks associated with NF-1 include bone abnormalities, vasculopathy, and cognitive impairment[128]. NF-1 is also the common type of hamartoma neoplastic syndrome, which is the formation of benign tumorlike malformations consisting of abnormal cells and tissues[128]. Neurofibromatosis also consists of neurofibromatosis type 2 (NF-2) and Schwannmatosis[128]. NF-2 presents with similar cutaneous manifestations as NF-1 but mainly exhibits schwannomas (tumors of the nervous system), meningiomas (tumors of the meninges), and ependymomas (brain and spinal cord tumors)[129]. Of the three kinds of NF, NF-1 accounts for 96% of all cases, NF-2 accounts for 3% and Schwannomatosis accounts for less than 1%[130].
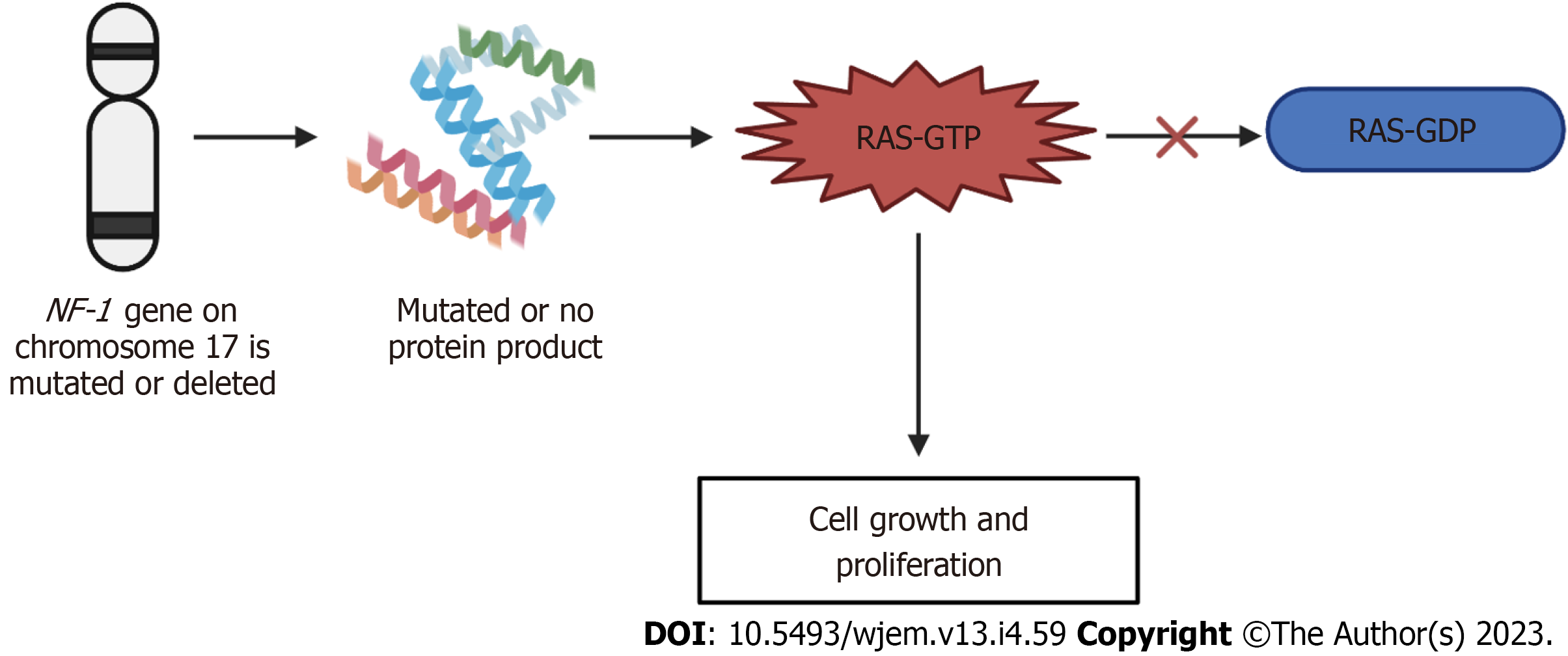
NF-1 is known as an autosomal dominant disorder that affects 1 in 2600 to 3000 individuals. All generations are included. The expression of this disease differs between individuals of the same family and from one affected family to another[128]. The NF-1 gene is located on chromosome 17 and is a tumor suppressor gene. The NF-1 gene produces a product called neurofibromin, which catalyzes the hydrolysis of guanosine triphosphate (GTP)-bound Ras to guanosine diphosphate-bound Ras, ultimately inactivating Ras GTPase[131]. Neurofibromin is expressed in various tissues throughout the body and the function of NF-1 is to modulate the activity of the RAS pathway[132]. This pathway in turn delivers signals from the granulocyte-monocyte colony-stimulating factor to proliferating cells[132]. In turn, the NF-1 protein promotes the conversion of the activated complex, Ras-GTP, to Ras-GDP, the inactivated form[133]. When the NF-1 gene is mutated or deleted, the typical phenotype of NF-1 results due to activation of the Ras-GTP, ultimately resulting in cell growth and prolferation[128]. With this disorder, penetrance is complete[134]. NF-1 has a high degree of variability in clinical presentation, which may include cutaneous, bony, vascular, and cognitive features along with multiple neoplasms. Due to these manifestations overlapping with other genetic conditions, accurate diagnosis of NF1 is important for clinical care and genetic counseling[135]. The mechanism of neurofibromatosis-1 is summarized in Figure 5 below.
New gene techniques were shown to help with NF-1-related pain. A study conducted on this rare autosomal disease suggested that collapsing response mediator protein 2 (CRMP2) is a key target for therapeutic intervention[136]. Moutal et al[136] highlighted there is a direct connection between the amount of neurofibromin expressed and pain. CRMP2 regulates the activity of calcium channels and increases the Ca2+ current and release in sensory neurons[137]. Additionally, CRMP2 binds to the C-terminus of neurofibromin, suggesting a possible correlation between CRMP2 and the pain experienced by patients with NF-1[136]. In the study, they used clustered regularly interspaced short palindromic repeats of the CRISPR-associated 9 (CAS9 genome), a commonly used DNA editing device[136]. With this, the researchers created a novel rat model of NF-1-related pain[136]. The delivery guide of Cas9 nuclease plasmid was used to generate allele-specific C-terminal truncation of neurofibromin[136]. Additionally, researchers used (S)-LCM, an inactive enantiomer of the drug Vimpat to inhibit CRMP2 phosphorylation, uncoupling CRMP2 from the NF-1 protein[136]. The rats with truncation of neurofibromin showed increases in voltage-gated calcium and sodium resulting in increased nociceptor excitability and behavioral hyperalgesia[136]. As the protein CRMP2 regulates these channels and binds to the C-terminus of neurofibromin, this indicated a possible mechanism underlying NF1 pain[136]. Targeting CRMP2 phosphorylation offers a new therapeutic way to manage the pain associated with NF-1. However, future research is needed in human trials to further investigate the link between CRMP2 and the NF-1 protein.
In children with neurofibromatosis 1, low-grade gliomas are the most common tumor and often result in significant visual loss due to tumor progression along with motor deficits[138]. The current therapeutical agent used for the management of tumor progression is vincristine/carboplatin, or vinblastine, a chemotherapeutic agent[139]. Although this agent prevents tumor progression, restoration of vision or motor function is rare[140]. A new type of therapeutic approach involves the use of mTOR inhibitors[140]. When mTOR is activated, abnormal cell growth results leading to cell proliferation and the formation of new blood vessels through angiogenesis[140]. Everolimus is derived from rapamycin and inhibits angiogenesis, hypoxia-inducible factor 1, and vascular endothelial growth factor production, and ultimately prevents the proliferation of cells[141]. When Everolimus was given to patients with NF-1, the tumor progression was significantly halted and demonstrated sufficient penetration of the blood-brain barrier[141]. This offers a promising therapeutic for children with low-grade gliomas as it is available orally and has minimal toxicities. However, further researcher is needed on Everolimus in combination with other agents to determine if it is the superior therapeutic agent or if a combination offers better results.
Tuberous sclerosis is an inherited autosomal dominant neurocutaneous genetic disorder[142]. This genetic disorder affects multiple systems and often presents in children with skin lesions, seizures, and hamartomas of the brain, kidney, and heart[142]. Additionally, tuberous sclerosis impacts 1 in 5500 individuals. Affected individuals may also present with developmental delays[142]. Cardiac rhabdomyomas of cortical tubers may also be present prenatally[142]. Adulthood signs show osseous, renal, or pulmonary lesions[142]. Skin lesions are found in 90% of patients of all ages whilst, hypopigmented macules are found in early childhood[142]. Ungual fibromas appear during puberty and facial angiofibromas are more commonly found towards adolescence[143].
Tuberous sclerosis disorder arises from four mutations in tuberous sclerosis complex 1 (TSC1) (9q34) and tuberous sclerosis complex 2 (TSC2) (16p13.3) genes which respectively encode hamartin and tuberin[144,145]. This disorder has been known to have a broad spectrum of mutations in both genes[144]. No regions seem more liable to mutations and the frequency is consistently higher in TSC2 rather than in TSC1[144]. 15% of patients admitted who meet clinical criteria for tuberous sclerosis demonstrated no identifiable genetic mutations[143]. The disease itself is caused by a mutation of these genes and causes dysfunction of proteins hamartin or tuberin[143]. Hamartin’s role helps control the proliferation of cell growth, division, and cell size[143]. Tuberin functions to regulate cell growth and protein synthesis through the downstream inhibition of mTOR[146]. Therefore, the loss of either of these proteins can lead to the overgrowth of lesions in many vital organs[146].
Gene therapy for tuberous sclerosis type 2 was conducted on a mouse model by delivering an adeno-associated virus (AAv9) which encoded a condensed form of tuberin[147]. A mouse model of TSC2 was generated by AAV-Cre recombinase disruption of Tsc2-floxed alleles at birth, leading to a shortened lifespan (mean 58 d) and brain pathology consistent with TSC[147]. When these mice were injected intravenously on day 21 with AAV9-cTuberin, the mean survival was extended to 462 d with a reduction in brain pathology[147]. This study demonstrated the potential of treating life-threatening TSC2 Lesions with a single intravenous injection of AAV9-cTuberin[147]. Preventions for this disease will vary depending on the developmental stage of the specific individual as tuberous sclerosis has a highly variable clinical course and the prognosis may be uncertain[147]. Options pertaining to tuberous sclerosis are limited to surgery for treating symptoms of tuberous sclerosis related to the growth of hamartomas[148]. Therefore, to better understand the genetic causation for this disease, clinical trials of mammalian target of rapamycin inhibitors, including sirolimus and Everolimus have been conducted[149]. mTOR is an evolutionary conserved serine-threonine kinase that regulates cell growth and cell survival[149]. The connection between TSC and mTOR led to the clinical use of mTOR allosteric inhibitors, Sirolimus and Everolimus[149]. Both sirolimus and Everolimus inhibit mTOR selectively with similar molecular mechanisms but distinct clinical profiles[149]. Everolimus has been approved for subependymal giant cell astrocytomas and renal angiomyolipomas in TSC patients[149]. However, sirolimus is not approved for TSC and has undergone considerable investigation to treat various aspects of TSC. However, the use of sirolimus has been studied in older children and adults with TSC[150-153]. One study published in 2023 investigated the use of sirolimus in children under the age of 2 with tuberous sclerosis complex and found that sirolimus was safe to use in children[154]. The study reported that the most common adverse events due to sirolimus use included anemia, hyperlipidemia, and thrombocytosis, which were able to be managed well[154]. Despite proving its safety through this study, further research is needed into sirolimus to demonstrate safety and efficacy through larger studies and clinical trials, but it offers a promising therapeutic option for patients with tuberous sclerosis.
Neurologic orphan diseases impact less than 200000 individuals within the United States and because they’re rare, diagnosis and treatment are often difficult. However, in recent years, data has suggested that the use of whole genome sequencing has increased the diagnosis of neurologic orphan diseases, allowing patients who have these conditions to be more easily identified. With new advances in technology, more research is being devoted to developing therapeutic options for patients with neurologic orphan diseases. Such advances in research include the use of AAV vectors which have shown positive results in SMA, ALS, Friedreich Ataxia, and NF-1. Additionally, the use of CRISPR/Cas has demonstrated promising results in DMD, Dravet syndrome, Friedreich Ataxia, NF-1, and Tuberous Sclerosis. The use of therapeutics that can cross the blood-brain barrier, such as Risdiplam and Branaplam, has increased the number of treatment options available for patients with SMA. Additionally, research into mTOR inhibitors offers a promising option for patients with NF-1 and tuberous sclerosis. Although further research is needed, these treatment options can significantly impact the quality of life and survival of patients with neurologic orphan diseases.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Neurosciences
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bagheri-Mohammadi S, Iran; Jabbarpour Z, United Kingdom; Liu Y, China S-Editor: Liu JH L-Editor: A P-Editor: Xu ZH
| 1. | Griggs RC, Batshaw M, Dunkle M, Gopal-Srivastava R, Kaye E, Krischer J, Nguyen T, Paulus K, Merkel PA; Rare Diseases Clinical Research Network. Clinical research for rare disease: opportunities, challenges, and solutions. Mol Genet Metab. 2009;96:20-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 259] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 2. | The Lancet Neurology. Rare diseases: maintaining momentum. Lancet Neurol. 2022;21:203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 3. | Murphy SM, Puwanant A, Griggs RC; Consortium for Clinical Investigations of Neurological Channelopathies (CINCH) and Inherited Neuropathies Consortium (INC) Consortia of the Rare Disease Clinical Research Network. Unintended effects of orphan product designation for rare neurological diseases. Ann Neurol. 2012;72:481-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Stephenson D, Ollivier C, Brinton R, Barrett J. Can Innovative Trial Designs in Orphan Diseases Drive Advancement of Treatments for Common Neurological Diseases? Clin Pharmacol Ther. 2022;111:799-806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Dowden H, Munro J. Trends in clinical success rates and therapeutic focus. Nat Rev Drug Discov. 2019;18:495-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 351] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 6. | Vinci L. New Report Shows 28% Increase in YoY Funding for Rare Disease Drug Development in 2021. January 13, 2022. Available from: https://www.businesswire.com/news/home/20220112005794/en/New-Report-Shows-28-Increase-in-YoY-Funding-for-Rare-Disease-Drug-Development-in-2021. |
| 7. | Ibañez K, Polke J, Hagelstrom RT, Dolzhenko E, Pasko D, Thomas ERA, Daugherty LC, Kasperaviciute D, Smith KR; WGS for Neurological Diseases Group, Deans ZC, Hill S, Fowler T, Scott RH, Hardy J, Chinnery PF, Houlden H, Rendon A, Caulfield MJ, Eberle MA, Taft RJ, Tucci A; Genomics England Research Consortium. Whole genome sequencing for the diagnosis of neurological repeat expansion disorders in the UK: a retrospective diagnostic accuracy and prospective clinical validation study. Lancet Neurol. 2022;21:234-245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 118] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 8. | Redman M, King A, Watson C, King D. What is CRISPR/Cas9? Arch Dis Child Educ Pract Ed. 2016;101:213-215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 93] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 9. | Hill SF, Meisler MH. Antisense Oligonucleotide Therapy for Neurodevelopmental Disorders. Dev Neurosci. 2021;43:247-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 10. | Naso MF, Tomkowicz B, Perry WL 3rd, Strohl WR. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs. 2017;31:317-334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 510] [Cited by in RCA: 867] [Article Influence: 123.9] [Reference Citation Analysis (0)] |
| 11. | Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18:358-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1440] [Cited by in RCA: 1564] [Article Influence: 260.7] [Reference Citation Analysis (0)] |
| 12. | Xu T, Sun D, Chen Y, Ouyang L. Targeting mTOR for fighting diseases: A revisited review of mTOR inhibitors. Eur J Med Chem. 2020;199:112391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 13. | Ross LF, Kwon JM. Spinal Muscular Atrophy: Past, Present, and Future. Neoreviews. 2019;20:e437-e451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 14. | Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2629] [Cited by in RCA: 2895] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 15. | D'Amico A, Mercuri E, Tiziano FD, Bertini E. Spinal muscular atrophy. Orphanet J Rare Dis. 2011;6:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 354] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 16. | Prior TW, Leach ME, Finanger E. Spinal Muscular Atrophy. In: Adam MP, Mirzaa GM, Pagon RA, Wallace SE, Bean LJ, Gripp KW, Amemiya A, editors. GeneReviews®. Seattle (WA): University of Washington, Seattle, 1993. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1352/. |
| 17. | Schorling DC, Pechmann A, Kirschner J. Advances in Treatment of Spinal Muscular Atrophy - New Phenotypes, New Challenges, New Implications for Care. J Neuromuscul Dis. 2020;7:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 151] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 18. | Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26:1333-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 346] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 19. | Rigo F, Hua Y, Krainer AR, Bennett CF. Antisense-based therapy for the treatment of spinal muscular atrophy. J Cell Biol. 2012;199:21-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 20. | Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, Hua Y, Rigo F, Matson J, Hung G, Kaye EM, Shihabuddin LS, Krainer AR, Bennett CF, Cheng SH. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med. 2011;3:72ra18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 439] [Cited by in RCA: 419] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 21. | Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, Norris DA, Xia S, Bennett CF, Bishop KM. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017-3026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 668] [Cited by in RCA: 742] [Article Influence: 82.4] [Reference Citation Analysis (0)] |
| 22. | Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, Tizzano E, Topaloglu H, Tulinius M, Montes J, Glanzman AM, Bishop K, Zhong ZJ, Gheuens S, Bennett CF, Schneider E, Farwell W, De Vivo DC; ENDEAR Study Group. Nusinersen vs Sham Control in Infantile-Onset Spinal Muscular Atrophy. N Engl J Med. 2017;377:1723-1732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1537] [Article Influence: 192.1] [Reference Citation Analysis (0)] |
| 23. | Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, Iannaccone ST, Kirschner J, Kuntz NL, Saito K, Shieh PB, Tulinius M, Mazzone ES, Montes J, Bishop KM, Yang Q, Foster R, Gheuens S, Bennett CF, Farwell W, Schneider E, De Vivo DC, Finkel RS; CHERISH Study Group. Nusinersen vs Sham Control in Later-Onset Spinal Muscular Atrophy. N Engl J Med. 2018;378:625-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 780] [Cited by in RCA: 1003] [Article Influence: 143.3] [Reference Citation Analysis (0)] |
| 24. | Poirier A, Weetall M, Heinig K, Bucheli F, Schoenlein K, Alsenz J, Bassett S, Ullah M, Senn C, Ratni H, Naryshkin N, Paushkin S, Mueller L. Risdiplam distributes and increases SMN protein in both the central nervous system and peripheral organs. Pharmacol Res Perspect. 2018;6:e00447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 25. | Dayton RD, Wang DB, Klein RL. The advent of AAV9 expands applications for brain and spinal cord gene delivery. Expert Opin Biol Ther. 2012;12:757-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 26. | Valori CF, Ning K, Wyles M, Mead RJ, Grierson AJ, Shaw PJ, Azzouz M. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med. 2010;2:35ra42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 219] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 27. | Blair HA. Onasemnogene Abeparvovec: A Review in Spinal Muscular Atrophy. CNS Drugs. 2022;36:995-1005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 28. | Al-Zaidy SA, Kolb SJ, Lowes L, Alfano LN, Shell R, Church KR, Nagendran S, Sproule DM, Feltner DE, Wells C, Ogrinc F, Menier M, L'Italien J, Arnold WD, Kissel JT, Kaspar BK, Mendell JR. AVXS-101 (Onasemnogene Abeparvovec) for SMA1: Comparative Study with a Prospective Natural History Cohort. J Neuromuscul Dis. 2019;6:307-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 29. | Mercuri E, Sumner CJ, Muntoni F, Darras BT, Finkel RS. Spinal muscular atrophy. Nat Rev Dis Primers. 2022;8:52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 199] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 30. | Duan D, Goemans N, Takeda S, Mercuri E, Aartsma-Rus A. Duchenne muscular dystrophy. Nat Rev Dis Primers. 2021;7:13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 696] [Article Influence: 174.0] [Reference Citation Analysis (0)] |
| 31. | Venugopal V, Pavlakis S. Duchenne Muscular Dystrophy. In: StatPearls. Treasure Island (FL): StatPearls Publishing, 2023. Available from: http://www.ncbi.nlm.nih.gov/books/NBK482346/. |
| 32. | Falzarano MS, Scotton C, Passarelli C, Ferlini A. Duchenne Muscular Dystrophy: From Diagnosis to Therapy. Molecules. 2015;20:18168-18184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 183] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 33. | Sun C, Shen L, Zhang Z, Xie X. Therapeutic Strategies for Duchenne Muscular Dystrophy: An Update. Genes (Basel). 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 34. | Ciafaloni E, Moxley RT. Treatment options for Duchenne muscular dystrophy. Curr Treat Options Neurol. 2008;10:86-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 35. | Ortez C, Natera de Benito D, Carrera García L, Expósito J, Nolasco G, Nascimento A. [Advances in the treatment of Duchenne muscular dystrophy]. Medicina (B Aires). 2019;79 Suppl 3:77-81. [PubMed] |
| 36. | Aartsma-Rus A, Straub V, Hemmings R, Haas M, Schlosser-Weber G, Stoyanova-Beninska V, Mercuri E, Muntoni F, Sepodes B, Vroom E, Balabanov P. Development of Exon Skipping Therapies for Duchenne Muscular Dystrophy: A Critical Review and a Perspective on the Outstanding Issues. Nucleic Acid Ther. 2017;27:251-259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 135] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 37. | Wong TWY, Cohn RD. Therapeutic Applications of CRISPR/Cas for Duchenne Muscular Dystrophy. Curr Gene Ther. 2017;17:301-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 38. | Wang JZ, Wu P, Shi ZM, Xu YL, Liu ZJ. The AAV-mediated and RNA-guided CRISPR/Cas9 system for gene therapy of DMD and BMD. Brain Dev. 2017;39:547-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 39. | Rodgers BD, Bishaw Y, Kagel D, Ramos JN, Maricelli JW. Micro-dystrophin Gene Therapy Partially Enhances Exercise Capacity in Older Adult mdx Mice. Mol Ther Methods Clin Dev. 2020;17:122-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Duan D. Systemic AAV Micro-dystrophin Gene Therapy for Duchenne Muscular Dystrophy. Mol Ther. 2018;26:2337-2356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 324] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 41. | Lazaridis D, Eraikhuemen N, Williams K, Lovince J. Treatment of Seizures Associated with Lennox-Gastaut and Dravet Syndromes: A Focus on Cannabidiol Oral Solution. P T. 2019;44:255-266. [PubMed] |
| 42. | Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, Hirsch E, Jain S, Mathern GW, Moshé SL, Nordli DR, Perucca E, Tomson T, Wiebe S, Zhang YH, Zuberi SM. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58:512-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2280] [Cited by in RCA: 3403] [Article Influence: 425.4] [Reference Citation Analysis (0)] |
| 43. | Li W, Schneider AL, Scheffer IE. Defining Dravet syndrome: An essential pre-requisite for precision medicine trials. Epilepsia. 2021;62:2205-2217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 70] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 44. | Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W, Engel J, French J, Glauser TA, Mathern GW, Moshé SL, Nordli D, Plouin P, Scheffer IE. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia. 2010;51:676-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2956] [Cited by in RCA: 2872] [Article Influence: 191.5] [Reference Citation Analysis (0)] |
| 45. | Wirrell EC, Laux L, Donner E, Jette N, Knupp K, Meskis MA, Miller I, Sullivan J, Welborn M, Berg AT. Optimizing the Diagnosis and Management of Dravet Syndrome: Recommendations From a North American Consensus Panel. Pediatr Neurol. 2017;68:18-34.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 198] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 46. | Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1001] [Cited by in RCA: 884] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 47. | Tahara M, Higurashi N, Hata J, Nishikawa M, Ito K, Hirose S, Kaneko T, Mashimo T, Sakuma T, Yamamoto T, Okano HJ. Developmental changes in brain activity of heterozygous Scn1a knockout rats. Front Neurol. 2023;14:1125089. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 48. | He Z, Li Y, Zhao X, Li B. Dravet syndrome: Advances in etiology, clinical presentation, and treatment. Epilepsy Res. 2022;188:107041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 49. | Han Z, Chen C, Christiansen A, Ji S, Lin Q, Anumonwo C, Liu C, Leiser SC, Meena, Aznarez I, Liau G, Isom LL. Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome. Sci Transl Med. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 223] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 50. | Higurashi N, Broccoli V, Hirose S. Genetics and gene therapy in Dravet syndrome. Epilepsy Behav. 2022;131:108043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 51. | Colasante G, Lignani G, Brusco S, Di Berardino C, Carpenter J, Giannelli S, Valassina N, Bido S, Ricci R, Castoldi V, Marenna S, Church T, Massimino L, Morabito G, Benfenati F, Schorge S, Leocani L, Kullmann DM, Broccoli V. dCas9-Based Scn1a Gene Activation Restores Inhibitory Interneuron Excitability and Attenuates Seizures in Dravet Syndrome Mice. Mol Ther. 2020;28:235-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 148] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 52. | Ohtahara S, Ohtsuka Y, Yamatogi Y, Oka E. The early-infantile epileptic encephalopathy with suppression-burst: developmental aspects. Brain Dev. 1987;9:371-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 85] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 53. | Radaelli G, de Souza Santos F, Borelli WV, Pisani L, Nunes ML, Scorza FA, da Costa JC. Causes of mortality in early infantile epileptic encephalopathy: A systematic review. Epilepsy Behav. 2018;85:32-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 54. | Symonds JD, Elliott KS, Shetty J, Armstrong M, Brunklaus A, Cutcutache I, Diver LA, Dorris L, Gardiner S, Jollands A, Joss S, Kirkpatrick M, McLellan A, MacLeod S, O'Regan M, Page M, Pilley E, Pilz DT, Stephen E, Stewart K, Ashrafian H, Knight JC, Zuberi SM. Early childhood epilepsies: epidemiology, classification, aetiology, and socio-economic determinants. Brain. 2021;144:2879-2891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 128] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 55. | Cossu A, Santos JL, Galati G, Nikanorova M, Costa P, Mang Y, Silahtaroglu A, Rubboli G, Tommerup N, Dalla Bernardina B, Møller RS, Cantalupo G, Gardella E. PRRT2 benign familial infantile seizures (BFIS) with atypical evolution to encephalopathy related to status epilepticus during sleep (ESES). Neurol Sci. 2023;44:2173-2176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 56. | Pisano T, Numis AL, Heavin SB, Weckhuysen S, Angriman M, Suls A, Podesta B, Thibert RL, Shapiro KA, Guerrini R, Scheffer IE, Marini C, Cilio MR. Early and effective treatment of KCNQ2 encephalopathy. Epilepsia. 2015;56:685-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 189] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 57. | Wolff M, Johannesen KM, Hedrich UBS, Masnada S, Rubboli G, Gardella E, Lesca G, Ville D, Milh M, Villard L, Afenjar A, Chantot-Bastaraud S, Mignot C, Lardennois C, Nava C, Schwarz N, Gérard M, Perrin L, Doummar D, Auvin S, Miranda MJ, Hempel M, Brilstra E, Knoers N, Verbeek N, van Kempen M, Braun KP, Mancini G, Biskup S, Hörtnagel K, Döcker M, Bast T, Loddenkemper T, Wong-Kisiel L, Baumeister FM, Fazeli W, Striano P, Dilena R, Fontana E, Zara F, Kurlemann G, Klepper J, Thoene JG, Arndt DH, Deconinck N, Schmitt-Mechelke T, Maier O, Muhle H, Wical B, Finetti C, Brückner R, Pietz J, Golla G, Jillella D, Linnet KM, Charles P, Moog U, Õiglane-Shlik E, Mantovani JF, Park K, Deprez M, Lederer D, Mary S, Scalais E, Selim L, Van Coster R, Lagae L, Nikanorova M, Hjalgrim H, Korenke GC, Trivisano M, Specchio N, Ceulemans B, Dorn T, Helbig KL, Hardies K, Stamberger H, de Jonghe P, Weckhuysen S, Lemke JR, Krägeloh-Mann I, Helbig I, Kluger G, Lerche H, Møller RS. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140:1316-1336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 311] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 58. | van der Louw E, van den Hurk D, Neal E, Leiendecker B, Fitzsimmon G, Dority L, Thompson L, Marchió M, Dudzińska M, Dressler A, Klepper J, Auvin S, Cross JH. Ketogenic diet guidelines for infants with refractory epilepsy. Eur J Paediatr Neurol. 2016;20:798-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 138] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 59. | Goodspeed K, Liu JS, Nye KL, Prasad S, Sadhu C, Tavakkoli F, Bilder DA, Minassian BA, Bailey RM. SLC13A5 Deficiency Disorder: From Genetics to Gene Therapy. Genes (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 60. | Mastrangelo M. Lennox-Gastaut Syndrome: A State of the Art Review. Neuropediatrics. 2017;48:143-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 61. | Arzimanoglou A, French J, Blume WT, Cross JH, Ernst JP, Feucht M, Genton P, Guerrini R, Kluger G, Pellock JM, Perucca E, Wheless JW. Lennox-Gastaut syndrome: a consensus approach on diagnosis, assessment, management, and trial methodology. Lancet Neurol. 2009;8:82-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 339] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 62. | Asadi-Pooya AA. Lennox-Gastaut syndrome: a comprehensive review. Neurol Sci. 2018;39:403-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 63. | Lemmon ME, Terao NN, Ng YT, Reisig W, Rubenstein JE, Kossoff EH. Efficacy of the ketogenic diet in Lennox-Gastaut syndrome: a retrospective review of one institution's experience and summary of the literature. Dev Med Child Neurol. 2012;54:464-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 64. | Knupp KG, Scheffer IE, Ceulemans B, Sullivan JE, Nickels KC, Lagae L, Guerrini R, Zuberi SM, Nabbout R, Riney K, Shore S, Agarwal A, Lock M, Farfel GM, Galer BS, Gammaitoni AR, Davis R, Gil-Nagel A. Efficacy and Safety of Fenfluramine for the Treatment of Seizures Associated With Lennox-Gastaut Syndrome: A Randomized Clinical Trial. JAMA Neurol. 2022;79:554-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 70] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 65. | Cross JH, Auvin S, Falip M, Striano P, Arzimanoglou A. Expert Opinion on the Management of Lennox-Gastaut Syndrome: Treatment Algorithms and Practical Considerations. Front Neurol. 2017;8:505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 66. | Pandolfo M. Friedreich ataxia. Arch Neurol. 2008;65:1296-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 67. | Koeppen AH. Nikolaus Friedreich and degenerative atrophy of the dorsal columns of the spinal cord. J Neurochem. 2013;126 Suppl 1:4-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 68. | Dürr A, Cossee M, Agid Y, Campuzano V, Mignard C, Penet C, Mandel JL, Brice A, Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N Engl J Med. 1996;335:1169-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 705] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 69. | Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Cañizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423-1427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2041] [Cited by in RCA: 1945] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 70. | Pandolfo M. Molecular pathogenesis of Friedreich ataxia. Arch Neurol. 1999;56:1201-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 73] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 71. | Koeppen AH. Friedreich's ataxia: pathology, pathogenesis, and molecular genetics. J Neurol Sci. 2011;303:1-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 357] [Cited by in RCA: 316] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 72. | Pastore A, Puccio H. Frataxin: a protein in search for a function. J Neurochem. 2013;126 Suppl 1:43-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 73. | Czechowska E. [Parasitic diseases of the visual system (author’s transl)]. Klin Oczna. 1977;47:407-409. [PubMed] |
| 74. | Bencze KZ, Kondapalli KC, Cook JD, McMahon S, Millán-Pacheco C, Pastor N, Stemmler TL. The structure and function of frataxin. Crit Rev Biochem Mol Biol. 2006;41:269-291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 134] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 75. | Sandi C, Sandi M, Anjomani Virmouni S, Al-Mahdawi S, Pook MA. Epigenetic-based therapies for Friedreich ataxia. Front Genet. 2014;5:165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 76. | Delatycki MB, Corben LA. Clinical features of Friedreich ataxia. J Child Neurol. 2012;27:1133-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 77. | Hanson E, Sheldon M, Pacheco B, Alkubeysi M, Raizada V. Heart disease in Friedreich's ataxia. World J Cardiol. 2019;11:1-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 78. | Wessel K, Schroth G, Diener HC, Müller-Forell W, Dichgans J. Significance of MRI-confirmed atrophy of the cranial spinal cord in Friedreich's ataxia. Eur Arch Psychiatry Neurol Sci. 1989;238:225-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 29] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 79. | Muthuswamy S, Agarwal S, Dalal A. Diagnosis and Genetic Counseling for Friedreich’s Ataxia: A time for consideration of TP-PCR in an Indian Setup. Hippokratia 2013; 17: 38–41. [PubMed] |
| 80. | Lynch DR, Farmer JM, Balcer LJ, Wilson RB. Friedreich ataxia: effects of genetic understanding on clinical evaluation and therapy. Arch Neurol. 2002;59:743-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 92] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 81. | Schulz JB, Boesch S, Bürk K, Dürr A, Giunti P, Mariotti C, Pousset F, Schöls L, Vankan P, Pandolfo M. Diagnosis and treatment of Friedreich ataxia: a European perspective. Nat Rev Neurol. 2009;5:222-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 176] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 82. | Montenegro L, Turnaturi R, Parenti C, Pasquinucci L. Idebenone: Novel Strategies to Improve Its Systemic and Local Efficacy. Nanomaterials (Basel). 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 83. | Lee A. Omaveloxolone: First Approval. Drugs. 2023;83:725-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 84. | Ouellet DL, Cherif K, Rousseau J, Tremblay JP. Deletion of the GAA repeats from the human frataxin gene using the CRISPR-Cas9 system in YG8R-derived cells and mouse models of Friedreich ataxia. Gene Ther. 2017;24:265-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 85. | Evans-Galea MV, Pébay A, Dottori M, Corben LA, Ong SH, Lockhart PJ, Delatycki MB. Cell and gene therapy for Friedreich ataxia: progress to date. Hum Gene Ther. 2014;25:684-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 86. | Piguet F, de Montigny C, Vaucamps N, Reutenauer L, Eisenmann A, Puccio H. Rapid and Complete Reversal of Sensory Ataxia by Gene Therapy in a Novel Model of Friedreich Ataxia. Mol Ther. 2018;26:1940-1952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 87. | Zhang S, Napierala M, Napierala JS. Therapeutic Prospects for Friedreich's Ataxia. Trends Pharmacol Sci. 2019;40:229-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 88. | Cassidy SB, Dykens E, Williams CA. Prader-Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet. 2000;97:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 89. | Ma VK, Mao R, Toth JN, Fulmer ML, Egense AS, Shankar SP. Prader-Willi and Angelman Syndromes: Mechanisms and Management. Appl Clin Genet. 2023;16:41-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 90. | Vogels A, De Hert M, Descheemaeker MJ, Govers V, Devriendt K, Legius E, Prinzie P, Fryns JP. Psychotic disorders in Prader-Willi syndrome. Am J Med Genet A. 2004;127A:238-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 91. | Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genet Med. 2012;14:10-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 916] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 92. | Brøndum-Nielsen K. The genetic basis for Prader-Willi syndrome: the importance of imprinted genes. Acta Paediatr Suppl. 1997;423:55-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 93. | Cheon CK. Genetics of Prader-Willi syndrome and Prader-Will-Like syndrome. Ann Pediatr Endocrinol Metab. 2016;21:126-135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 94. | Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, Rosenfeld JA, Lamb AN, Sahoo T. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18:1196-1201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 252] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 95. | Wang SE, Jiang YH. Potential of Epigenetic Therapy for Prader-Willi Syndrome. Trends Pharmacol Sci. 2019;40:605-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 96. | Bieth E, Eddiry S, Gaston V, Lorenzini F, Buffet A, Conte Auriol F, Molinas C, Cailley D, Rooryck C, Arveiler B, Cavaillé J, Salles JP, Tauber M. Highly restricted deletion of the SNORD116 region is implicated in Prader-Willi Syndrome. Eur J Hum Genet. 2015;23:252-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 97. | Langouët M, Glatt-Deeley HR, Chung MS, Dupont-Thibert CM, Mathieux E, Banda EC, Stoddard CE, Crandall L, Lalande M. Zinc finger protein 274 regulates imprinted expression of transcripts in Prader-Willi syndrome neurons. Hum Mol Genet. 2018;27:505-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 98. | Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233-247.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 850] [Article Influence: 94.4] [Reference Citation Analysis (0)] |
| 99. | Cruvinel E, Budinetz T, Germain N, Chamberlain S, Lalande M, Martins-Taylor K. Reactivation of maternal SNORD116 cluster via SETDB1 knockdown in Prader-Willi syndrome iPSCs. Hum Mol Genet. 2014;23:4674-4685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 100. | Reiner D, Seifert L, Deck C, Schüle R, Jung M, Stark H. Epigenetics meets GPCR: inhibition of histone H3 methyltransferase (G9a) and histamine H(3) receptor for Prader-Willi Syndrome. Sci Rep. 2020;10:13558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 101. | Kim Y, Lee HM, Xiong Y, Sciaky N, Hulbert SW, Cao X, Everitt JI, Jin J, Roth BL, Jiang YH. Targeting the histone methyltransferase G9a activates imprinted genes and improves survival of a mouse model of Prader-Willi syndrome. Nat Med. 2017;23:213-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 102] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 102. | Kim Y, Wang SE, Jiang YH. Epigenetic therapy of Prader-Willi syndrome. Transl Res. 2019;208:105-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 103. | Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genet Med. 2010;12:385-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 219] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 104. | Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW Jr, Lee HM, Chen X, Jin J, Bridges AS, Zylka MJ, Roth BL, Philpot BD. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2011;481:185-189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 262] [Cited by in RCA: 281] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 105. | Lee HM, Clark EP, Kuijer MB, Cushman M, Pommier Y, Philpot BD. Characterization and structure-activity relationships of indenoisoquinoline-derived topoisomerase I inhibitors in unsilencing the dormant Ube3a gene associated with Angelman syndrome. Mol Autism. 2018;9:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 106. | Tsagkaris C, Papakosta V, Miranda AV, Zacharopoulou L, Danilchenko V, Matiashova L, Dhar A. Gene Therapy for Angelman Syndrome: Contemporary Approaches and Future Endeavors. Curr Gene Ther. 2020;19:359-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 107. | Fink JJ, Robinson TM, Germain ND, Sirois CL, Bolduc KA, Ward AJ, Rigo F, Chamberlain SJ, Levine ES. Disrupted neuronal maturation in Angelman syndrome-derived induced pluripotent stem cells. Nat Commun. 2017;8:15038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 108. | van den Bos MAJ, Geevasinga N, Higashihara M, Menon P, Vucic S. Pathophysiology and Diagnosis of ALS: Insights from Advances in Neurophysiological Techniques. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 109. | Nowicka N, Juranek J, Juranek JK, Wojtkiewicz J. Risk Factors and Emerging Therapies in Amyotrophic Lateral Sclerosis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 110. | Amado DA, Davidson BL. Gene therapy for ALS: A review. Mol Ther. 2021;29:3345-3358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 111. | Longinetti E, Fang F. Epidemiology of amyotrophic lateral sclerosis: an update of recent literature. Curr Opin Neurol. 2019;32:771-776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 365] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 112. | Mejzini R, Flynn LL, Pitout IL, Fletcher S, Wilton SD, Akkari PA. ALS Genetics, Mechanisms, and Therapeutics: Where Are We Now? Front Neurosci. 2019;13:1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 522] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 113. | Trojsi F, D'Alvano G, Bonavita S, Tedeschi G. Genetics and Sex in the Pathogenesis of Amyotrophic Lateral Sclerosis (ALS): Is There a Link? Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 114. | Wang MD, Little J, Gomes J, Cashman NR, Krewski D. Identification of risk factors associated with onset and progression of amyotrophic lateral sclerosis using systematic review and meta-analysis. Neurotoxicology. 2017;61:101-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 160] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 115. | Kim G, Gautier O, Tassoni-Tsuchida E, Ma XR, Gitler AD. ALS Genetics: Gains, Losses, and Implications for Future Therapies. Neuron. 2020;108:822-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 262] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 116. | Meijboom KE, Brown RH. Approaches to Gene Modulation Therapy for ALS. Neurotherapeutics. 2022;19:1159-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 117. | Ediriweera GR, Chen L, Yerbury JJ, Thurecht KJ, Vine KL. Non-Viral Vector-Mediated Gene Therapy for ALS: Challenges and Future Perspectives. Mol Pharm. 2021;18:2142-2160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 118. | Miller T, Cudkowicz M, Shaw PJ, Andersen PM, Atassi N, Bucelli RC, Genge A, Glass J, Ladha S, Ludolph AL, Maragakis NJ, McDermott CJ, Pestronk A, Ravits J, Salachas F, Trudell R, Van Damme P, Zinman L, Bennett CF, Lane R, Sandrock A, Runz H, Graham D, Houshyar H, McCampbell A, Nestorov I, Chang I, McNeill M, Fanning L, Fradette S, Ferguson TA. Phase 1-2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N Engl J Med. 2020;383:109-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 392] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 119. | Kim A, Lalonde K, Truesdell A, Gomes Welter P, Brocardo PS, Rosenstock TR, Gil-Mohapel J. New Avenues for the Treatment of Huntington's Disease. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 101] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 120. | Fields E, Vaughan E, Tripu D, Lim I, Shrout K, Conway J, Salib N, Lee Y, Dhamsania A, Jacobsen M, Woo A, Xue H, Cao K. Gene targeting techniques for Huntington's disease. Ageing Res Rev. 2021;70:101385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 121. | Tabrizi SJ, Estevez-Fraga C, van Roon-Mom WMC, Flower MD, Scahill RI, Wild EJ, Muñoz-Sanjuan I, Sampaio C, Rosser AE, Leavitt BR. Potential disease-modifying therapies for Huntington's disease: lessons learned and future opportunities. Lancet Neurol. 2022;21:645-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 176] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 122. | Pan L, Feigin A. Huntington's Disease: New Frontiers in Therapeutics. Curr Neurol Neurosci Rep. 2021;21:10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 123. | Feigin A, Evans EE, Fisher TL, Leonard JE, Smith ES, Reader A, Mishra V, Manber R, Walters KA, Kowarski L, Oakes D, Siemers E, Kieburtz KD, Zauderer M; Huntington Study Group SIGNAL investigators. Pepinemab antibody blockade of SEMA4D in early Huntington's disease: a randomized, placebo-controlled, phase 2 trial. Nat Med. 2022;28:2183-2193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 124. | Jurcau A. Molecular Pathophysiological Mechanisms in Huntington's Disease. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 125. | Duan W, Urani E, Mattson MP. The potential of gene editing for Huntington's disease. Trends Neurosci. 2023;46:365-376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 126. | Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, Barker RA, Blair NF, Craufurd D, Priller J, Rickards H, Rosser A, Kordasiewicz HB, Czech C, Swayze EE, Norris DA, Baumann T, Gerlach I, Schobel SA, Paz E, Smith AV, Bennett CF, Lane RM; Phase 1–2a IONIS-HTTRx Study Site Teams. Targeting Huntingtin Expression in Patients with Huntington's Disease. N Engl J Med. 2019;380:2307-2316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 469] [Article Influence: 78.2] [Reference Citation Analysis (0)] |
| 127. | Kumar D, Hasan GM, Islam A, Hassan MI. Therapeutic targeting of Huntington's disease: Molecular and clinical approaches. Biochem Biophys Res Commun. 2023;655:18-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 128. | Adil A, Koritala T, Munakomi S, Singh AK. Neurofibromatosis Type 1. In: StatPearls. Treasure Island (FL): StatPearls Publishing, 2023. Available from: http://www.ncbi.nlm.nih.gov/books/NBK459358/. |
| 129. | Evans DG. Neurofibromatosis type 2 (NF2): a clinical and molecular review. Orphanet J Rare Dis. 2009;4:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 368] [Cited by in RCA: 343] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 130. | Tamura R. Current Understanding of Neurofibromatosis Type 1, 2, and Schwannomatosis. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 132] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 131. | Williams VC, Lucas J, Babcock MA, Gutmann DH, Korf B, Maria BL. Neurofibromatosis type 1 revisited. Pediatrics. 2009;123:124-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 422] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 132. | Kim Z, Lee JH. Neurofibromatosis Symptom-Lacking B-Cell Lineage Acute Lymphoblastic Leukemia with Only an NF1 Gene Pathogenic Variant. Diagnostics (Basel). 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 133. | Parkin B, Ouillette P, Wang Y, Liu Y, Wright W, Roulston D, Purkayastha A, Dressel A, Karp J, Bockenstedt P, Al-Zoubi A, Talpaz M, Kujawski L, Shedden K, Shakhan S, Li C, Erba H, Malek SN. NF1 inactivation in adult acute myelogenous leukemia. Clin Cancer Res. 2010;16:4135-4147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 134. | Yap YS, McPherson JR, Ong CK, Rozen SG, Teh BT, Lee AS, Callen DF. The NF1 gene revisited - from bench to bedside. Oncotarget. 2014;5:5873-5892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 136] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 135. | Ly KI, Blakeley JO. The Diagnosis and Management of Neurofibromatosis Type 1. Med Clin North Am. 2019;103:1035-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 136. | Moutal A, Yang X, Li W, Gilbraith KB, Luo S, Cai S, François-Moutal L, Chew LA, Yeon SK, Bellampalli SS, Qu C, Xie JY, Ibrahim MM, Khanna M, Park KD, Porreca F, Khanna R. CRISPR/Cas9 editing of Nf1 gene identifies CRMP2 as a therapeutic target in neurofibromatosis type 1-related pain that is reversed by (S)-Lacosamide. Pain. 2017;158:2301-2319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 137. | Chi XX, Schmutzler BS, Brittain JM, Wang Y, Hingtgen CM, Nicol GD, Khanna R. Regulation of N-type voltage-gated calcium channels (Cav2.2) and transmitter release by collapsin response mediator protein-2 (CRMP-2) in sensory neurons. J Cell Sci. 2009;122:4351-4362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 123] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 138. | Farrell CJ, Plotkin SR. Genetic causes of brain tumors: neurofibromatosis, tuberous sclerosis, von Hippel-Lindau, and other syndromes. Neurol Clin. 2007;25:925-946, viii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 78] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 139. | Ater JL, Zhou T, Holmes E, Mazewski CM, Booth TN, Freyer DR, Lazarus KH, Packer RJ, Prados M, Sposto R, Vezina G, Wisoff JH, Pollack IF. Randomized study of two chemotherapy regimens for treatment of low-grade glioma in young children: a report from the Children's Oncology Group. J Clin Oncol. 2012;30:2641-2647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 306] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 140. | Ullrich NJ, Prabhu SP, Reddy AT, Fisher MJ, Packer R, Goldman S, Robison NJ, Gutmann DH, Viskochil DH, Allen JC, Korf B, Cantor A, Cutter G, Thomas C, Perentesis JP, Mizuno T, Vinks AA, Manley PE, Chi SN, Kieran MW. A phase II study of continuous oral mTOR inhibitor everolimus for recurrent, radiographic-progressive neurofibromatosis type 1-associated pediatric low-grade glioma: a Neurofibromatosis Clinical Trials Consortium study. Neuro Oncol. 2020;22:1527-1535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 141. | Kawachi Y, Maruyama H, Ishitsuka Y, Fujisawa Y, Furuta J, Nakamura Y, Ichikawa E, Furumura M, Otsuka F. NF1 gene silencing induces upregulation of vascular endothelial growth factor expression in both Schwann and non-Schwann cells. Exp Dermatol. 2013;22:262-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 142. | Randle SC. Tuberous Sclerosis Complex: A Review. Pediatr Ann. 2017;46:e166-e171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 106] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 143. | Zamora EA, Aeddula NR. Tuberous Sclerosis. In: StatPearls. Treasure Island (FL): StatPearls Publishing, 2023. Available from: http://www.ncbi.nlm.nih.gov/books/NBK538492/. |
| 144. | De Waele L, Lagae L, Mekahli D. Tuberous sclerosis complex: the past and the future. Pediatr Nephrol. 2015;30:1771-1780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 145. | Northrup H, Krueger DA; International Tuberous Sclerosis Complex Consensus Group. Tuberous sclerosis complex diagnostic criteria update: recommendations of the 2012 Iinternational Tuberous Sclerosis Complex Consensus Conference. Pediatr Neurol. 2013;49:243-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 1002] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 146. | Miloloza A, Rosner M, Nellist M, Halley D, Bernaschek G, Hengstschläger M. The TSC1 gene product, hamartin, negatively regulates cell proliferation. Hum Mol Genet. 2000;9:1721-1727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 87] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 147. | Cheah PS, Prabhakar S, Yellen D, Beauchamp RL, Zhang X, Kasamatsu S, Bronson RT, Thiele EA, Kwiatkowski DJ, Stemmer-Rachamimov A, György B, Ling KH, Kaneki M, Tannous BA, Ramesh V, Maguire CA, Breakefield XO. Gene therapy for tuberous sclerosis complex type 2 in a mouse model by delivery of AAV9 encoding a condensed form of tuberin. Sci Adv. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 148. | Kohrman MH. Emerging treatments in the management of tuberous sclerosis complex. Pediatr Neurol. 2012;46:267-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 149. | MacKeigan JP, Krueger DA. Differentiating the mTOR inhibitors everolimus and sirolimus in the treatment of tuberous sclerosis complex. Neuro Oncol. 2015;17:1550-1559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 134] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 150. | Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, Schmithorst VJ, Laor T, Brody AS, Bean J, Salisbury S, Franz DN. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358:140-151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1041] [Cited by in RCA: 917] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 151. | Davies DM, de Vries PJ, Johnson SR, McCartney DL, Cox JA, Serra AL, Watson PC, Howe CJ, Doyle T, Pointon K, Cross JJ, Tattersfield AE, Kingswood JC, Sampson JR. Sirolimus therapy for angiomyolipoma in tuberous sclerosis and sporadic lymphangioleiomyomatosis: a phase 2 trial. Clin Cancer Res. 2011;17:4071-4081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 233] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 152. | Verhave J, Boucher A, Dandavino R, Collette S, Senécal L, Hebert MJ, Girardin C, Cardinal H. The incidence, management, and evolution of rapamycin-related side effects in kidney transplant recipients. Clin Transplant. 2014;28:616-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 153. | Fidan K, Kandur Y, Sozen H, Gonul II, Dalgic A, Söylemezoğlu O. How often do we face side effects of sirolimus in pediatric renal transplantation? Transplant Proc. 2013;45:185-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 154. | Śmiałek D, Jóźwiak S, Kotulska K. Safety of Sirolimus in Patients with Tuberous Sclerosis Complex under Two Years of Age-A Bicenter Retrospective Study. J Clin Med. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |