Published online Nov 19, 2019. doi: 10.5492/wjccm.v8.i7.120
Peer-review started: May 8, 2019
First decision: August 2, 2019
Revised: August 21, 2019
Accepted: October 27, 2019
Article in press: October 27, 2019
Published online: November 19, 2019
Processing time: 212 Days and 21.9 Hours
With the recent change in the definition (Sepsis-3 Definition) of sepsis and septic shock, an electronic search algorithm was required to identify the cases for data automation. This supervised machine learning method would help screen a large amount of electronic medical records (EMR) for efficient research purposes.
To develop and validate a computable phenotype via supervised machine learning method for retrospectively identifying sepsis and septic shock in critical care patients.
A supervised machine learning method was developed based on culture orders, Sequential Organ Failure Assessment (SOFA) scores, serum lactate levels and vasopressor use in the intensive care units (ICUs). The computable phenotype was derived from a retrospective analysis of a random cohort of 100 patients admitted to the medical ICU. This was then validated in an independent cohort of 100 patients. We compared the results from computable phenotype to a gold standard by manual review of EMR by 2 blinded reviewers. Disagreement was resolved by a critical care clinician. A SOFA score ≥ 2 during the ICU stay with a culture 72 h before or after the time of admission was identified. Sepsis versions as V1 was defined as blood cultures with SOFA ≥ 2 and Sepsis V2 was defined as any culture with SOFA score ≥ 2. A serum lactate level ≥ 2 mmol/L from 24 h before admission till their stay in the ICU and vasopressor use with Sepsis-1 and-2 were identified as Septic Shock-V1 and-V2 respectively.
In the derivation subset of 100 random patients, the final machine learning strategy achieved a sensitivity-specificity of 100% and 84% for Sepsis-1, 100% and 95% for Sepsis-2, 78% and 80% for Septic Shock-1, and 80% and 90% for Septic Shock-2. An overall percent of agreement between two blinded reviewers had a k = 0.86 and 0.90 for Sepsis 2 and Septic shock 2 respectively. In validation of the algorithm through a separate 100 random patient subset, the reported sensitivity and specificity for all 4 diagnoses were 100%-100% each.
Supervised machine learning for identification of sepsis and septic shock is reliable and an efficient alternative to manual chart review.
Core tip: This study presents and validates a supervised machine learning model for the identification of sepsis and septic shock cases using electronic medical records as an alternative to manual chart review. This method showed to be an efficient, fast and reliable option for retrospective data abstraction, with the potential to be applied to other clinical conditions.
- Citation: Dhungana P, Serafim LP, Ruiz AL, Bruns D, Weister TJ, Smischney NJ, Kashyap R. Machine learning in data abstraction: A computable phenotype for sepsis and septic shock diagnosis in the intensive care unit. World J Crit Care Med 2019; 8(7): 120-126
- URL: https://www.wjgnet.com/2220-3141/full/v8/i7/120.htm
- DOI: https://dx.doi.org/10.5492/wjccm.v8.i7.120
Septicemia was identified as one of the most costly in-hospital conditions in the United States[1]. The incidence and burden of Systematic Inflammatory Response Syndrome in the acutely ill is around 35% and almost half of all patients hospitalized develop this condition at least once during their stay[2]. It also contributes to as many as half of all hospital deaths[3]. These statistics outline the overall burden of sepsis as a leading cause of critical illness associated with significant mortality and morbidity[4,5]. The Third International Consensus Definition for Sepsis and Septic Shock defined sepsis as life-threatening organ dysfunction caused by a dysregulated host response to infection[6]. The identification of cases of sepsis in the intensive care unit (ICU) is important to further gain knowledge on the subject and improve outcomes.
The use of electronic medical records (EMR’s) as a tool to reduce cost and improve safety has been increasing over the years in both clinical practice and health care research[7]. Despite increasing use, there is a lack of structured search strategies and data capturing to identify cases of sepsis or septic shock. With the change in definition of sepsis and septic shock, a search phenotype is useful to identify cases and help additional studies related to sepsis. Machine learning methods to identify comorbidities, post-operative complications, and extubation failure have been developed and validated[8-10]. These algorithms are valuable in research to identify conditions of interest with a sensitivity and specificity approaching and improving on manual review.
The objective of the study was to develop and validate a computable phenotype via supervised machine learning method for retrospectively identifying sepsis and septic shock in the ICU based on the Sepsis-3 criteria using information available from the EMR. Identification of these cases is necessary for improving research related to this condition, and also as a stepping stone to design machine learning models for real-time sepsis detection. Our secondary aim was to validate the results obtained from the computable phenotype by comparing it with a gold standard (i.e., manual review) performed by two independent blinded reviewers.
The study was approved by the Mayo Clinic Institutional Review Board for the use of existing medical records of patients who gave prior research authorization. The guidelines of the STROBE statement have been adopted.
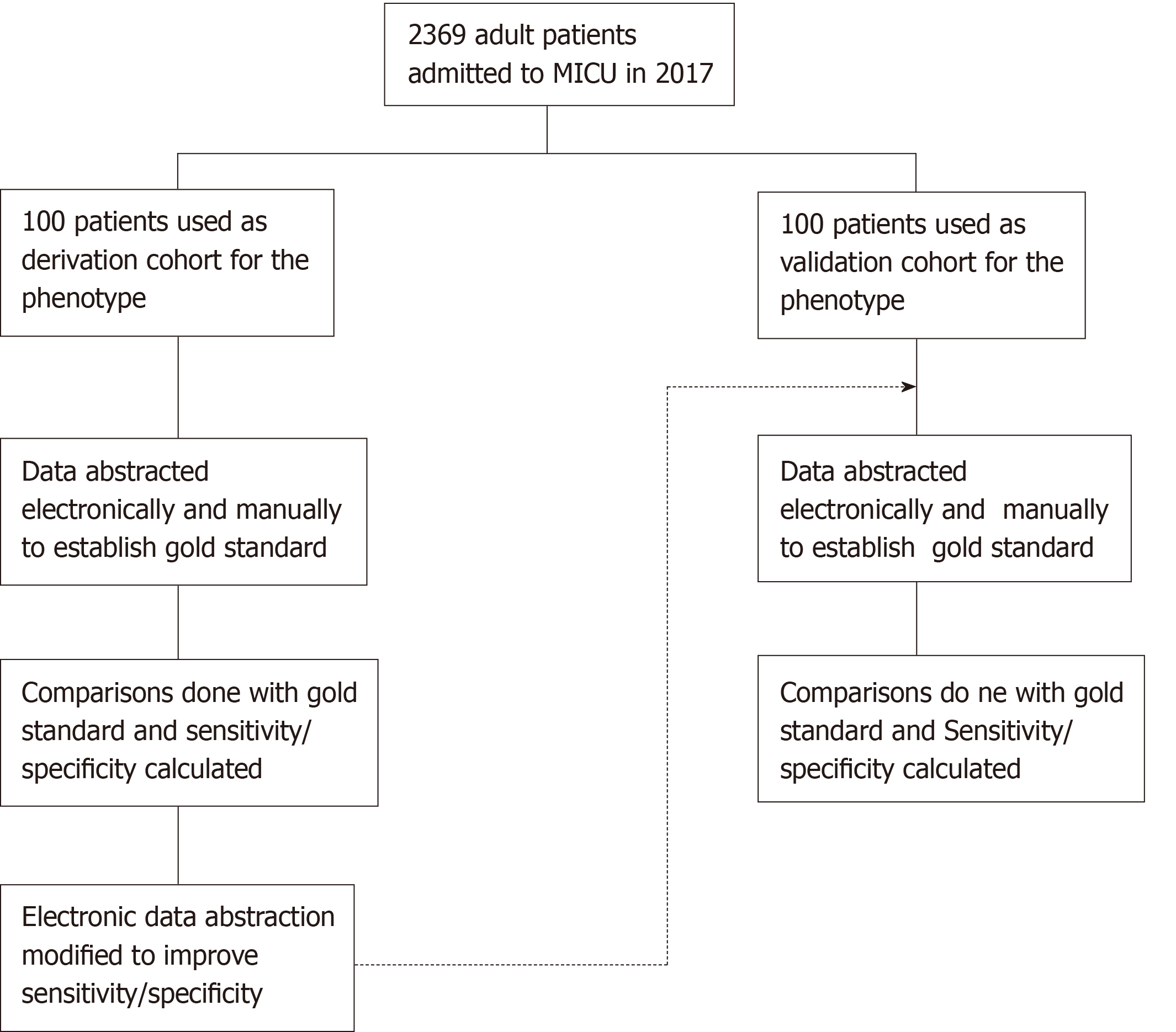
The study population consisted of patients above 18 years of age admitted to the medical ICU at Mayo Clinic, Rochester, with prior research authorization. A random subset of 100 patients each was used for derivation and validation of the computable phenotype (Figure 1). The selection of 200 total patients for the study population was to be able to have a comparable sample size between the two cohorts while keeping the time and effort for manual review reasonable.
The medical records of the derivation and validation cohort were manually reviewed by two independent blinded reviewers. The data obtained from the two reviewers were compared to each other and disagreements were reviewed by a third reviewer. The final set obtained from this process was taken as the gold standard for the study (Figure 1). The reviewers collected data for cultures, Sequential Organ Failure Assessment (SOFA) scores, serum lactate levels, and vasopressor use for the cohorts during their ICU admission. Serum lactate and Cultures were abstracted within 24 h and 72 h before or after ICU admission date respectively, while the other parameters were abstracted during the entire period of their ICU stay.
Data for the retrospective study was used from Mayo Clinic ICU DataMart and Unified Data Platform, which are extensive data warehouses containing a near real-time normalized replica of Mayo Clinic’s EMR. These databases contain patient information along with their laboratory test results, clinical and pathological information from sources within the institution and have been previously validated. A web-based software tool set (Advanced Cohort Explorer) was used for data access.
The data for cultures, SOFA scores, serum lactate levels and vasopressor use was abstracted and cases were identified as having or not having sepsis and septic shock. The computable phenotype was refined continuously in several iterations to improve the sensitivity and specificity of the derivation subset to more than 90%. The phenotype algorithm was validated using sensitivity and specificity calculated by comparing the results to the gold standard obtained by manual review (Figure 1). The machine learning model for the cohort was done under supervision of an independent critical care researcher.
Sepsis 1 and 2 was defined as blood culture and any culture drawn within 72 h of ICU admission and SOFA score ≥ 2 on any ICU admission days 1-7 respectively. Septic Shock 1 and 2 was defined as Sepsis 1 or 2 criteria plus a serum lactate ≥ 2 mmol/L and at least one vasopressor infusion during ICU admission respectively.
Sensitivity and specificity of the computable phenotype was calculated by comparing the results to the gold standard obtained by manual review of the charts using JMP statistical software, SAS 13.0, Cary, North Carolina.
In the initial derivation cohort, the supervised machine learning model achieved a sensitivity of 100% for sepsis (Table 1) and 80% for septic shock (Table 2). There were 4 disagreements between the manual review and the computable phenotype in the dataset. The disagreements were reviewed and it was identified that 2 of the disagreements were due to the results being obtained at a time before the patient was admitted to the ICU. The 2 other disagreements were due to the sample culture sent. The reviewers identified the cases as not being septic even though a culture was sent for the cases and fulfilling sepsis-3 criteria. The supervised machine-learning phenotype algorithm was refined and the results compared to obtain a final derivation sensitivity-specificity of 100%-100% for sepsis (Table 1) and 90%-100% for septic shock (Table 2). Correction was then made for the timing of the lactate data and data was abstracted for the validation cohort by both manual review and the computable phenotype. The sensitivity-specificity for the validation cohort was found to be 100%-100% for both sepsis and septic shock (Tables 1 and 2).
The results of the study demonstrate that the development of an automated phenotype algorithm (computable phenotype) based on supervised machine learning is an effective and reliable method when compared to manual review. It is also faster than manual review and the sensitivity-specificity achieved in this study provides a highly effective method for reliable retrospective data extraction. Computable phenotypes via machine learning methods have been used as alternatives to manual chart review[8-13]; however, to date, it has never been employed for sepsis and the new sepsis-3 criteria, representing a novel application of this technology.
With widespread use of EMRs, the emphasis on “big data” has increased. The accumulation of vast amounts of data opens doors for opportunities to improve the processes of care and treatment by conducting studies on the available data. Time might be a constraining factor when trying to identify the correct study population through manual review as it takes significant time. Traditional billing searches for conditions may not be completely accurate[9] and changes in coding guidelines make them even less reliable.
The study has several strengths. It allows for a quick and reliable way to retrospectively identify cases of sepsis and septic shock based on the Sepsis-3 guidelines. This provides strong support to the educational and research activity at our institution and demonstrates a simple, yet effective, method that can be applied to other clinical conditions and institutions. It also demonstrates the capability of an algorithm to identify cases based on data stream and hence is an important step towards algorithm/model for real time detection of sepsis.
The limitation of our study is that we used ICU DataMart and Advanced Cohort Explorer systems for electronic data abstraction which are specific to Mayo Clinic. This reduces the generalizability of supervised machine learning model for sepsis and septic shock. The computable phenotype however used laboratory, culture and medication data which are well recorded through various EMR systems and therefore should be easy to recreate with modification based on the institutions’ database. Our machine learning model also offers an opportunity for other researchers and institutions to build additional phenotype algorithms to identify conditions/cases objectively to further the improvement in care and knowledge of the medical community.
In conclusion, computable phenotypes based on machine learning are able to correctly identify sepsis and septic shock with high sensitivity and specificity in a cohort of retrospective data. This method can help expedite clinical research by reducing cost and time required for cohort identification. It will also allow the use of larger cohorts thereby enabling the researcher to perform larger studies to ultimately improve clinical outcomes. Finally, the supervised machine learning model can be incorporated into a near real time identification tool to pick up cases of sepsis and septic shock and aid clinical practice as part of a sniffer system.
With the recent change in the definition (Sepsis-3 Definition) of sepsis and septic shock, an electronic search algorithm was required to identify the cases for data automation.
This supervised machine learning method would help screen a large amount of electronic medical records (EMR) for efficient research purposes.
The main objective was to develop and validate a computable phenotype via supervised machine learning method for retrospectively identifying sepsis and septic shock in critical care patients.
A supervised machine learning method was developed based on culture orders, Sequential Organ Failure Assessment (SOFA) scores, serum lactate levels and vasopressor use in the intensive care units (ICUs). The computable phenotype was derived from a retrospective analysis of a random cohort of 100 patients admitted to the medical ICU. This was then validated in an independent cohort of 100 patients. We compared the results from computable phenotype to a gold standard by manual review of EMR by 2 blinded reviewers. Disagreement was resolved by a critical care clinician. A SOFA score ≥ 2 during the ICU stay with a culture 72 h before or after the time of admission was identified. Sepsis versions as V1 was defined as blood cultures with SOFA ≥ 2 and Sepsis V2 was defined as any culture with SOFA score ≥ 2. A serum lactate level ≥ 2 mmol/L from 24 h before admission till their stay in the ICU and vasopressor use with Sepsis-1 and-2 were identified as Septic Shock-V1 and-V2 respectively.
In the derivation subset of 100 random patients, the final machine learning strategy achieved a sensitivity-specificity of 100% and 84% for Sepsis-1, 100% and 95% for Sepsis-2, 78% and 80% for Septic Shock-1, and 80% and 90% for Septic Shock-2. An overall percent of agreement between two blinded reviewers had a k = 0.86 and 0.90 for Sepsis 2 and Septic shock 2 respectively. In validation of the algorithm through a separate 100 random patient subset, the reported sensitivity and specificity for all 4 diagnoses were 100%-100% each.
Supervised machine learning for identification of sepsis and septic shock is reliable and an efficient alternative to manual chart review.
This study presents and validates a supervised machine learning model for the identification of sepsis and septic shock cases using EMRs as an alternative to manual chart review. This method showed to be an efficient, fast and reliable option for retrospective data abstraction, with the potential to be applied to other clinical conditions.
Manuscript source: Invited manuscript
Specialty type: Critical care medicine
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhang ZH S-Editor: Yan JP L-Editor: A E-Editor: Liu MY
| 1. | Torio CM, Andrews RM. National Inpatient Hospital Costs: The Most Expensive Conditions by Payer, 2011: Statistical Brief #160. Healthcare Cost and Utilization Project (HCUP) Statistical Briefs. Rockville, MD, United States. 2006;. [PubMed] |
| 2. | Churpek MM, Zadravecz FJ, Winslow C, Howell MD, Edelson DP. Incidence and Prognostic Value of the Systemic Inflammatory Response Syndrome and Organ Dysfunctions in Ward Patients. Am J Respir Crit Care Med. 2015;192:958-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 217] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 3. | Liu V, Escobar GJ, Greene JD, Soule J, Whippy A, Angus DC, Iwashyna TJ. Hospital deaths in patients with sepsis from 2 independent cohorts. JAMA. 2014;312:90-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 685] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 4. | Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K; International Forum of Acute Care Trialists. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am J Respir Crit Care Med. 2016;193:259-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1639] [Cited by in RCA: 2313] [Article Influence: 257.0] [Reference Citation Analysis (0)] |
| 5. | Vincent JL, Marshall JC, Namendys-Silva SA, François B, Martin-Loeches I, Lipman J, Reinhart K, Antonelli M, Pickkers P, Njimi H, Jimenez E, Sakr Y; ICON investigators. Assessment of the worldwide burden of critical illness: the intensive care over nations (ICON) audit. Lancet Respir Med. 2014;2:380-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 606] [Cited by in RCA: 838] [Article Influence: 76.2] [Reference Citation Analysis (0)] |
| 6. | Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315:801-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15803] [Cited by in RCA: 17185] [Article Influence: 1909.4] [Reference Citation Analysis (2)] |
| 7. | Zlabek JA, Wickus JW, Mathiason MA. Early cost and safety benefits of an inpatient electronic health record. J Am Med Inform Assoc. 2011;18:169-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 8. | Rishi MA, Kashyap R, Wilson G, Hocker S. Retrospective derivation and validation of a search algorithm to identify extubation failure in the intensive care unit. BMC Anesthesiol. 2014;14:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Singh B, Singh A, Ahmed A, Wilson GA, Pickering BW, Herasevich V, Gajic O, Li G. Derivation and validation of automated electronic search strategies to extract Charlson comorbidities from electronic medical records. Mayo Clin Proc. 2012;87:817-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 179] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 10. | Tien M, Kashyap R, Wilson GA, Hernandez-Torres V, Jacob AK, Schroeder DR, Mantilla CB. Retrospective Derivation and Validation of an Automated Electronic Search Algorithm to Identify Post Operative Cardiovascular and Thromboembolic Complications. Appl Clin Inform. 2015;6:565-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Amra S, O'Horo JC, Singh TD, Wilson GA, Kashyap R, Petersen R, Roberts RO, Fryer JD, Rabinstein AA, Gajic O. Derivation and validation of the automated search algorithms to identify cognitive impairment and dementia in electronic health records. J Crit Care. 2017;37:202-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 12. | Guru PK, Singh TD, Passe M, Kashani KB, Schears GJ, Kashyap R. Derivation and Validation of a Search Algorithm to Retrospectively Identify CRRT Initiation in the ECMO Patients. Appl Clin Inform. 2016;7:596-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Smischney NJ, Velagapudi VM, Onigkeit JA, Pickering BW, Herasevich V, Kashyap R. Derivation and validation of a search algorithm to retrospectively identify mechanical ventilation initiation in the intensive care unit. BMC Med Inform Decis Mak. 2014;14:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |