HISTORY OF LYSOSOMAL ACID LIPASE
Lysosomal acid lipase (LAL) cleaves cholesteryl esters (CE) and triglycerides (TG) in cell lysosomes. Mutation in the LAL gene results in Wolman disease (WD) of early infantile onset, and cholesteryl ester storage disease (CESD) of late onset. WD was first described by Dr. Wolman[1] in 1956 as severe malnutrition, hepatosplenomegaly, calcified adrenal glands, and death of children within the first few months of life. Affected WD infants display massive accumulations of CE and TG in the lysosomes of hepatocytes and Kupffer cells, as well as in macrophages throughout the viscera, which lead to liver failure, severe hepatosplenomegaly, steatorrhea, pulmonary fibrosis[2,3], and adrenal calcification and insufficiency[4,5]. Lipid engorged macrophages in intestinal villi lead to severe malabsorption and cachexia[2,4]. The average life span of WD is 3.5 mo[6]. CESD was initially described by Fredrickson, Schiff, Langeron, and Infante and their colleagues in 1967[7-10] and named by Partin and Schubert based on phenotype that exhibited hepatomegaly with increased hepatic levels of cholesteryl esters in 1969. CESD can be a more indolent progressive disease, which shows microvesicular steatosis leading to fibrosis and cirrhosis in the liver, increases atherosclerosis and premature demise[11-13]. Wolman disease and CESD result from allelic mutations at the LAL locus on human chromosome 10q23.2-q23.3 and are autosomal recessive traits. The gene spans 45 kb, has 10 exons, and contains no unusual structures, except for a large intron 3. The LIPA mutations found in Wolman disease include deletions and insertions that lead to premature stop codons and the consequent loss of LAL protein and activity[14]. The mutations found in CESD are usually missense mutations, either heteroallelic or homoallelic with another mutant LIPA gene[14].
Recently, some evidence started to emerge, showing altered mononuclear phagocyte differentiation [increased CD14+CD16+ and CD14+CD33+ cells, subsets of human myeloid-derived suppressive cells, or myeloid-derived suppressive cells (MDSCs)] in humans that were heterozygote carriers of LAL mutations[15]. Furthermore, patients with mutations in the LAL gene have been reported to be associated with carcinogenesis[16]. These clinical observations support the extensive characterization in animal models as described below.
LAL PROPERTIES
LAL is a key player in the modulation of cholesterol metabolism in all cells. On the surface membranes of various cells, there are multiple receptors that can deliver LDL-bound cholesteryl esters/triglycerides to lysosomes, but LAL is the only lipase in the lysosomes that hydrolyzes cholesteryl esters and triglycerides. Once cleaved by LAL, the free cholesterol and fatty acids enter the cytosol from lysosome. In LAL deficiency, cholesteryl esters and triglycerides cannot be cleaved; therefore, free cholesterol and fatty acids cannot leave the lysosome[17,18]. Cells sense this as an intracellular (cytosolic) cholesterol deficiency, and the cholesterol biosynthetic pathway is up-regulated to compensate.
Synthesized in the rough endoplasmic reticulum, LAL is a typical soluble lysosomal hydrolase, which is co-translationally glycosylated when it emerges into the endoplasmic reticulum lumen[18,19]. Following the removal of the leader sequence (21 amino acids), LAL is decorated with oligosaccharides that are remodeled during transit through the Golgi apparatus. The N-linked oligosaccharides are remodeled from high mannosyl to complex forms, with a mannose 6-phosphate being added, which serves as the lysosomal sorting targeting signal. The mannose 6-phosphate receptor system is used to deliver the newly synthesized LAL to the lysosome. LAL is not known to require cofactors for optimal hydrolysis, and it functions as a monomer. Unmodified mature protein (378 amino acids) has a predicted molecular weight approximately 42.5 kDa. Different molecular weights have been reported for purified human LAL[20-24]. Occupancy of the LAL N-glycosylation is essential for enzyme stability, i.e., protection from rapid degradation[25].
LAL has significant similarity to other acidic lipases, for example, lingual lipase and gastric lipases that cleave similar substrates in the stomach. However, LAL is distinct from other lipases, including hormone-sensitive lipase, pancreatic lysophospholipid lipase, lecithin cholesterol acyl transferase, lipoprotein lipase, hepatic lipase, and pancreatic lipase[26]. All such lipases share a motif, Gly-X-Ser-X-Gly, that is an essential pentapeptide in the active site[27,28]. This pentapeptide occurs twice in LAL at serine 99 and serine 153, and specific mutation of serine 153 identified this residue as important to catalytic activity[23]. Like other lipases, LAL also has a catalytic triad of Ser153, Asp423 and His353[27].
GENE KNOCK-OUT PHENOTYPES AND MDSCS IN MICE
A Lipa knock-out mouse (lal-/-) has been created to understand the functional roles of LAL in disease pathophysiology, lipid metabolism, and therapeutic approaches[29,30]. The lal-/- phenotype resembles human CESD. It’s histopathologic and biochemical phenotypes are similar to human WD. The lal-/- mice are normal appearing at birth, but develop liver enlargement by 4 wk and have a grossly enlarged abdomen with hepatosplenomegaly, lymph node enlargement, and intestinal villus infiltration by foamy macrophages by 16 wk. Massive accumulation of CE and TG and macrophage storage develops in these and other organs[29,31-34]. Enzyme therapy has been studied in this model using human recombinant LAL (rhLAL) produced in several different eukaryotic systems[24,35,36]. These studies clearly show the potential for correction of the manifestations if enzyme therapy is begun early in the course of the disease[36,37].
Many phenotypes of seemingly unrelated diseases in various organs co-exist in lal-/- mice. Therefore, these diseases must share common cellular and molecular mechanisms that link these pathological processes. Extensive characterization of lal-/- mice shows that elevation of systemic MDSCs is a major manifestation in association with most of the pathogenic conditions (e.g., > 70% in the bone marrow and > 40% in the blood), suggesting that MDSCs play a central role in mediating LAL deficiency-induced pathogenic progression[29,31-34,36,38-41]. MDSCs was originally identified in tumor pathogenesis[42]. Recent studied have linked this cell population to many other chronic inflammatory diseases[43-50]. MDSCs are a mixture of myeloid cells that express CD11b and Gr-1 antigens in mice. In certain disease conditions (cancer), MDSCs are categorized into granulocytic (CD11b+, Ly6G+) and monocytic (CD11b+Ly6C+) MDSC[51]. Interestingly, most gated lal-/- CD11b+ cells show Ly6C+ and Ly6G+ double positive, making them CD11b+Ly6C+ Ly6G+ cells[34]. Normally, healthy immature myeloid lineage cells differentiate into dendritic cells (DCs), macrophages, or granulocytes in response to environmental changes. However, this process is blocked by LAL deficiency, leading to accumulation and expansion of MDSCs with immune suppressive function[51-53]. This is similar to what has been observed in the tumor environment[54]. It is conceivable that through paracrine and autocrine mechanisms, abnormally elevated MDSCs generate and secrete growth factors, chemokines and cytokines to influence cell differentiation, cell proliferation, cell apoptosis and gene expression in residing organ tissues, contributing to the physiological progression of various diseases. Direct cell-cell contact by MDSCs and other cells through the juxtacrine mechanism also contributes to this pathogenic process.
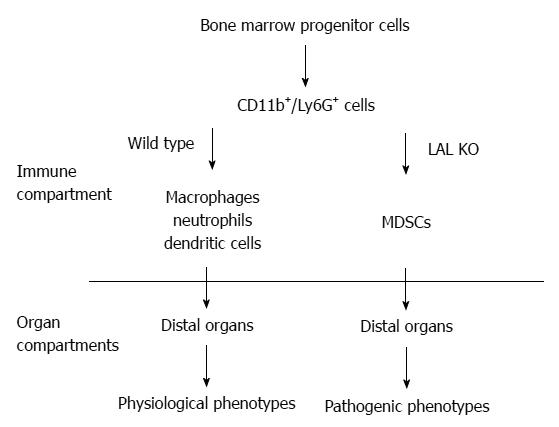
The functional roles of LAL in myeloid cells have been specifically evaluated by creating a myeloid-specific doxycycline-inducible c-fms-rtTA/(tetO)7-CMV-hLAL; lal-/- triple mouse model, in which human LAL is expressed in myeloid cells under the control of the 7.2 kb c-fms promoter/intron2 regulatory sequence in lal-/- mice[32,34,55]. The hLAL expression in myeloid lineage cells in this triple mouse model significantly reduced systemic MDSCs accumulation[34], reversed aberrant gene expression, and ameliorated pathogenic phenotypes[32]. Therefore, the normal biological function of myeloid cells requires normal neutral lipid metabolism (Figure 1).
Figure 1 The functional role of Lysosomal acid lipase in myeloid lineage cells.
In the wild type mice, the CD11b+Ly6G+ cells are myeloid lineage precursors for monocytes/macrophages, neutrophils, and dendritic cells, which participate in the normal physiological functions of the distal organs (e.g., lung, liver, etc.), such as clearance of invading pathogens. The lysosomal acid lipase (LAL) activity is essential for normal myeloid lineage cell development, differentiation and function. LAL deficiency leads to neutral lipid accumulation in myeloid cells and blocks CD11b+Ly6G+ cells from further differentiation into mature myeloid lineage cells. The accumulated CD11b+Ly6G+ cells possess various malfunctions that participate in the pathogenic conditions in the residing organs. MDSCs: Myeloid-derived suppressive cells.
MDSCS DIFFERENTIATION AND DEVELOPMENT
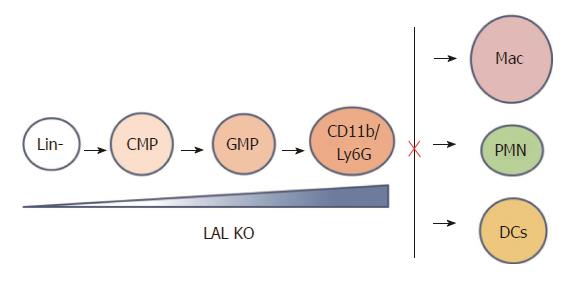
The myeloid linage cells undergo the sequentially differentiated and proliferated from hematopoietic stem cells through an increasingly lineage-restricted intermediate progenitors including common myeloid progenitors (CMPs) and granulocyte-macrophage progenitors (GMPs) in the bone marrow[56,57]. The number and frequency of primitive LSK (Lin-/Sca-1+/c-kit+), CMP, and GMP populations in the bone marrow, systemic myeloid cell distribution are changed in lal-/- mice, leading to an expansion in CD11b+/Gr-1+ MDSCs[41]. Both increased proliferation and decreased apoptosis contribute to the expansion of MDSCs in lal-/- mice. Lal-/- mice also display increased numbers of high proliferative potential colony-forming cells (HPP-CFC), colony-forming unite of granulocyte and macrophage progenitor cells (CFU-GM), colony-forming unite of granulocytes (CFU-G) and colony-forming unite of macrophages (CFU-M) colonies from cultured bone marrow cells. When lal-/- bone marrow cells are transplanted into wild type mice, the donor CD11b+/GR-1+ myeloid cells in the blood, spleen, lung and bone marrow of recipient mice are increased, confirming that the MDSCs increase is primarily due to the intrinsic defect in myeloid lineage progenitor cells. In addition to the intrinsic progenitor problem, the environment in lal-/- mice also contributes to myeloid cell hyperexpansion, since the donor CD11b+/GR-1+ myeloid cell population in lal-/- recipient mice that are transplanted with wild type bone marrow cells is expanded. Therefore, the lal-/- environment does not normally support hematopoiesis. Deregulated bone marrow progenitor cell differentiation is a primary cause for expansion of lal-/- MDSCs, which is attributed to both cell-autonomous and environmental factors. Taken together, LAL expression in myeloid lineage cells is critical to maintain hematopoiesis and myelopoiesis. After MDSCs infiltration into distal organs, at least two mechanisms can explain how the cell-autonomous defect and environmental factors influence each other. Firstly, MDSCs and other regional cells in distal organs influence each other by the paracrine mechanism as both sides secrete cytokines and chemokines. Secondly, MDSCs and other cells can influence each other by direct contact (juxtacrine mechanism). Starting at the GMP stage, hLAL expression in myeloid cells reverses abnormal myeloid development in the bone marrow, and reduces systemic expansion of MDSCs in c-fms-rtTA/(tetO)7-CMV-hLAL; lal-/- triple mice. In addition, differentiation from Lin- progenitor cells to CD11b+GR-1+ cells is abnormally increased in lal-/- mice (Figure 2). This further supports that the cell-autonomous effect of MDSCs expansion in lal-/- mice. Myeloid hLAL expression in c-fms-rtTA/(tetO)7-CMV-hLAL; lal-/- triple mice successfully reverses this abnormality[32]. The environmental effects on MDSCs malformation are further supported by an observation that when the Stat3 pathway is overly activated in lung epithelial cells[58], secretion of Stat3-induced pro-inflammatory cytokines in epithelial cells reversed mature myeloid lineage cells to MDSCs[59].
Figure 2 Lysosomal acid lipase is required for normal myeloid lineage cell development and differentiation.
Lysosomal acid lipase (LAL) deficiency leads to increased myeloid-derived suppressive cells differentiation from Lin- progenitor cells in the bone barrow, and decreased differentiation to mature macrophages, neutrophils, and dendritic cells in other compartments. Lin-: Lineage negative progenitor; CMP: Common myeloid progenitor; GMP: Granulocyte-macrophage progenitor; Mac: Macrophage; PMN: Polymorphonuclear cell, or neutrophil; DC: Dendritic cell.
MDSCS IMMUNOSUPPRESSION
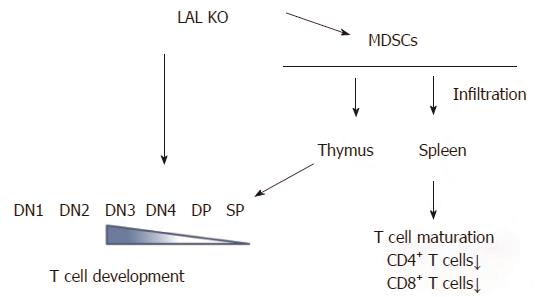
In contrast to myeloid lineage cells, T cells are systemically decreased in lal-/- mice. Lal-/- T cells behave abnormally. In response to stimulation of anti-CD3 plus anti-CD28 antibodies, or phorbol-12-myristate-13-acetate (agonist to activate PKC) and ionomycin (calcium ionophore), there is severely diminished T cell proliferation, decreased CD69 expression, and decreased expression of T cell lymphokines. LAL deficiency does not drive effector T cells into either Th1 or Th2 status[33]. The thymus is the most important organ for T cell development, which is divided into different developmental stages that are marked by CD4-CD8- double negative (DN) 1 to 4 stages, CD4+CD8+ double positive (DP) stage and CD4+ or CD8+ single positive (SP) stage. The earliest stage for thymocyte paucity appears at the DN4 (CD25-CD44-) stage in the lal-/- thymus. After this developmental point, thymocytes are declining at all stages, suggesting that the blockage of T cell development initially occurs at the DN3 to DN4 transition (Figure 3)[33]. Decrease of T cell development and maturation was also observed in lal-/- mice due to the defects in lymphoid progenitors in the bone marrow chimeras study. This notion has been supported by the bone marrow profile analysis, in which common lymphoid progenitor development is blocked in the bone marrow of lal-/- mice[33,41].
Figure 3 Lysosomal acid lipase is required for normal T cell development and differentiation.
Lysosomal acid lipase (LAL) deficiency can cause the intrinsic defect in T cell development, starting at the double negative 3 (DN3) stage. In addition, myeloid-derived suppressive cells infiltrate into the thymus and spleen, resulting in blockage of normal T cell development, differentiation, and maturation. DN: CD4 and CD8 double negative; DP: CD4 and CD8 double positive; SP: CD4 or CD8 single positive.
In addition to the above intrinsic defect, extensive analyses have revealed a second mechanism that contributes to systemic reduction of T cell populations. Strikingly, LAL deficiency dramatically increases MDSCs expansion and infiltration in the thymus and the spleen of lal-/- mice, leading to neutral lipid accumulation and abnormal organization of the thymus and spleen[33]. Infiltration of MDSCs in these important T cell organs affects T cell development, differentiation and maturation. Functional analyses have shown that MDSCs from lal-/- mice strongly inhibit proliferation and function of T cells (Figure 3)[34,40,41].
Direct connection between LAL in MDSCs and T cell abnormalities comes from the c-fms-rtTA/(tetO)7-CMV-hLAL;lal-/- triple mouse study. MDSCs expansion and infiltration into the thymus and spleen are reduced in this mouse model. This leads to restoration of T cell proliferation in the spleen and normal T cell development in the thymus[34]. Stat3 and NFκB p65 signaling play a critical role in lal-/- MDSCs immune suppressive function[34]. The above observations are further proved by an MDSCs depletion study, in which anti-Gr-1 antibody treatment recovers T cell numbers in lal-/- mice[34]. lal-/- MDSCs also inhibits T cell lymphokine production, which is resulted from inactivation of the pZAP-70/Syk intracellular signaling, loss of expression of TCR ξ chain and CD69, a failure to respond to TCR stimulation[33]. These defects can also be reversed by myeloid hLAL expression[34]. Lastly, Treg cells inhibit CD4+ T cell lymphokine production and proliferation[60]. LAL deficiency substantially increases CD4+FoxP3+ Treg cells in lal-/-mice[33].
GENE PROFILES IN LAL DEFICIENCY-INDUCED MDSCS
Since LAL controls homeostasis and development of MDSCs, which have profound pathogenic impact on various disease development, it is essential to identify the intrinsic defects that are involved in the MDSCs homeostasis and function for future targeting. In a comprehensive gene transcriptional profile study by Affymetrix GeneChip microarray analysis, multiple pathways have been revealed in lal-/- bone marrow MDSCs. Below are lists of some major (but not limited) changed pathways in lal-/- MDSCs.
Genes of G-protein superfamily
Expression changes of both large and small GTPases have been detected in lal-/- MDSCs, which have diverse functions in cells[61,62]. They include: (1) Rab GTPases, which control vesicle formation, receptor internalization, and trafficking to the nucleus, lysosome and plasma membrane. Rab GTPases regulate cellular proliferation, apoptosis and migration by integrating signaling pathways; (2) Rho GTPases, which organize actin cytoskeleton, cell adhesion and cell motility[63]; and (3) Ras GTPases mediate cell-cycle entry, cell growth, cell survival, cell growth and cellular metabolism by phosphorylating transcription factors through activation of the Raf/Mek/Erk pathway. Activation of Erk and p38 phosphorylation has been observed in lal-/- MDSCs[41].
Histone cluster genes and cell cycle genes
Cell cycle regulating genes are upregulated in lal-/- MDSCs. They include: (1) Histone-variants cluster genes, which favor the epigenetic microenvironment change to promote MDSCs expansion. Histone-variants exchange also contributes to formation of centromeric and telomeric chromatin during cell cycles. Indeed, G1/M phases of lal-/- MDSCs are increased in a cell cycle analysis[64]; (2) Cell cycle related genes[65], including Cdk1, Cdk2, Cdk5, Cdk9, and all Cdk regulatory cyclins (A, B, D, E-type), suggesting constitutive mitogenic signaling and defective responses to anti-mitogenic signals; and (3) Ubiquitination and proteasome enzymes/protein factors, which direct proteins to proteolysis within proteasome for recycling[66].
Metabolism and bioenergetics
Bioenergetic and metabolic genes are abnormally upregulated in lal-/- MDSCs, which control mitochondrial oxidative phosphorylation and energy (ATP production) for cellular activities. These include: (1) lactate dehydrogenase A and B, which produce large quantities of secreted lactate, suggesting that lal-/- MDSCs use an aerobic glycolysis; (2) nitric oxide/reactive oxygen species (ROS) production genes, glutathione peroxidase/glutathione reductase genes, and glucose 6-phosphate dehydrogenase gene, which are involved in production of ROS. The concentration of ROS is significantly increased in lal-/- MDSCs; (3) enzymes and proteins in glycolysis and citric acid cycles; and (4) respiratory chain proteins (NADH dehydrogenases, cytochrome proteins, ATPases and mitochondrial ribosomal proteins).
The mTOR pathway in LAL deficiency induced MDSCs
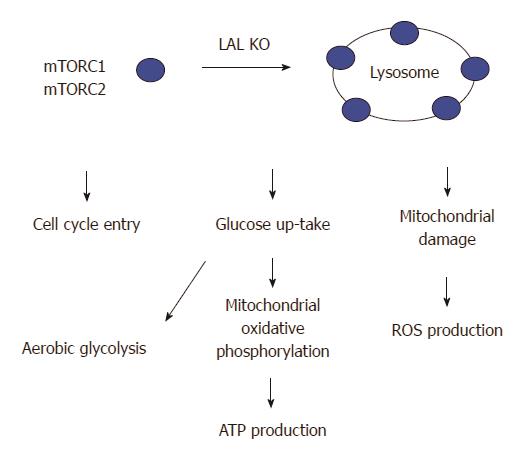
PI3K/thymoma viral proto-oncogene (AKT)/mammalian target of rapamycin (mTOR) is activated in lal-/- MDSCs[64]. mTOR is a lysosomal membrane-bound protein, which controls apoptosis, promotes influx of glucose and amino acids into the cells, stimulates ATP production[67], contributes to cell growth, cell cycle entry, cell survival, and cell motility[68,69]. Lack of the LAL activity changes lipid composition and dynamics on the lysosomal membrane that potentially influence endomembrane trafficking and stimulate the mTOR activity, which in turn coordinates the cellular metabolism[64,69,70]. It has been demonstrated that mTOR plays a critical role in modulating cellular immune functions[71,72], activation of the mTOR pathway contributes to lal-/- MDSCs production and function[40]. mTOR is the catalytic subunit of two distinctive complexes; mTOR complex 1 (mTORC1) and mTOR complex (mTORC2). mTORC1 contains unique regulatory associated proteins of mTOR (RAPTOR) while mTORC2 contains rapamycin-insensitive companion of mTOR (RICTOR)[67,72-75]. Inhibition of mTOR and associated proteins (Raptor, Rictor, and Akt1) corrects lal-/- MDSCs development, increased cell proliferation, decreased cellular apoptosis, and immune suppression in association with decreased ROS production, recovery from impairment of the mitochondrial membrane potential, increased ATP synthesis, and increased cell cycling. Potentially, the mTOR pathway can serve as a target to modulate the emergence of MDSCs in various pathophysiologic states where these cells play an immunosuppressive role (Figure 4).
Figure 4 Lysosomal acid lipase deficiency induces overactivation of the mTOR pathway in myeloid-derived suppressive cells.
Lysosomal acid lipase (LAL) is a lysosome-associated enzyme. LAL deficiency increases mTOR complexes anchoring on lysosomes and stimulates the mTOR1 activity to influence the cellular metabolism and proliferation of lal-/- myeloid-derived suppressive cells (MDSCs). These include an increased influx of glucose through aerobic glycolysis, an increased mitochondrial oxidative phosphorylation and ATP production, an impairment of the mitochondrial membrane potential in association with increased reactive oxygen species (ROS) production, and an increased cell cycle entry in lal-/- MDSCs.
The Stat3 and NFκB pathways
Although upregulation of Signal Transducer and Activator of Transcription (Stat) family members and NFκB family members are not detected by microarray analysis, phosphorylation of Stat3 and NFκB has been detected in expanded lal-/- MDSCs[34,41]. Activation of Stat3 directly leads to MDSCs expansion in vivo[58,59].
STUDY OF LAL DOWNSTREAM GENES
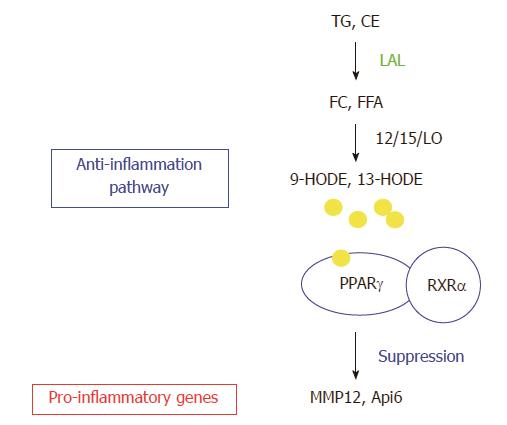
The gene profile study in the lung of lal-/- mice by Affymetrix GeneChip microarray analysis has also been performed. This is because the lung is a lipid rich organ and highly responsive to inflammation. Neutral lipids account for 10% of the composition of pulmonary surfactant that protects alveoli from collapse during respiratory cycles[76]. LAL deficiency results in massive myeloid cell infiltration, hyperplasia and emphysema in the lal-/- lung[32,39]. Comparison between the changed gene lists of bone marrow MDSCs and the whole lung by Affymetrix GeneChip microarray analyses reveals a few overlapping genes. Therefore, LAL performs differential roles in different compartments. LAL exerts its biological effects through its downstream genes. In order to fully understand the LAL functions, it is necessary and essential to characterize its downstream genes. From the whole lung gene list, the two most up-regulated genes matrix metal proteinase 12 (MMP12) and apoptosis inhibitor 6 (Api6) are characterized extensively. The functional role of LAL downstream effector peroxisome proliferator-activated receptor gamma (PPARγ) has also been studied in depth. Figure 5 shows the relationship between LAL and its downstream effectors.
Figure 5 Lysosomal acid lipase and its downstream effector genes.
Lysosomal acid lipase (LAL) cleaves cholesteryl esters (CE) and triglycerides (TG) to produce free cholesterol (FC) and fatty acids (FFA) in lysosomes of cells. The lipid derivatives (9-HODE, 13-HODE) of FFA serve as ligands for PPARγ in coupling with retinoid X receptor α (RXRα, which suppresses gene expression of a variety of pro-inflammatory cytokines. The LAL/PPARγ axis serves as an anti-inflammatory pathway. LAL deficiency blocks this metabolic pathway to provoke up-regulation of pro-inflammatory cytokines (e.g., Api6, MMP12). TGF: Transforming growth factor beta; IL: Interleukin; MCP: Monocyte chemotactic protein; TNF: Tumor necrosis factor; NF: Nuclear factor.
PPARγ
Involvement of the receptor network in the metabolic programming of myeloid lineage cells is essential to the innate immune system[77,78]. PPARγ is of high interest for several reasons. Firstly, the metabolites of LAL hydrolysis, 9-hydroxyoctadecanoic acids (9-HODE) or 13-HODE from linoleic acid, serve as ligands for PPARγ. Upon binding to the ligands, PPAR interacts with the retinoid X receptor (RXR) to form the PPARγ/RXR dimer on target genes. Secondly, PPARγ plays an important role in anti-inflammation of various tissues[77,79,80]. It has been shown that PPARγ agonists suppress gene expression of inflammatory cytokines[79]. In the lal-/- lung, these pro-inflammatory cytokines are up-regulated (Figure 5)[39]. Therefore, LAL deficiency causes inactivation of PPARγ by depleting ligand production. Using the lung as a model system, reintroduction of LAL downstream metabolic derivative 9-HODE (a natural occurring ligand for PPARγ) and a synthetic ligand compound ciglitazone for PPARγ improves the inflammatory status and pathogenesis in the lal-/- lung. Therefore, the ligands/PPARγ axis controls inflammation-triggered elevated gene expression and pathogenesis in the lal-/- mice[31].
To directly evaluate functional role of LAL downstream effector PPARγ in myeloid cells, dominant negative PPARγ (dnPPARγ) is overexpressed in a myeloid-specific c-fms-rtTA/(TetO)7-CMV-dnPPARγ bitransgenic mouse model[81]. In this bitransgenic system, total numbers and frequencies of LK, LSK, CMP and GMP progenitor cells in the bone marrow are abnormally elevated. DnPPARγ overexpression leads to up-regulation of IL-1β, IL-6 and TNFα in the blood plasma. MDSCs from this bitransgenic mouse model inhibit the proliferation and lymphokine production of wild type CD4+ T cells in vitro. Both CD4+ and CD8+ T cell populations are decreased in doxycycline-induced dnPPARγ expressed mice. Bone marrow transplantation reveals that a myeloid autonomous defect is responsible for MDSC expansion, immunosuppression and tumorigenesis in this myeloid-specifically expressed dnPPARγ bitransgenic mice. Multiple forms of carcinoma and sarcoma in various organs (the lung, liver, spleen and lymph nodes) are observed in this mouse model. Therefore, the LAL/hormonal ligands/PPARγ axis is critical to control inflammation and the induction of various tumors. Disruption of this pathway in myeloid cells, either by blocking ligand synthesis (as in lal -/- mice), or inhibition of PPARγ (as in c-fms-rtTA/(TetO)7-CMV-dnPPARγ bitransgenic mice) can initiate up-regulation of inflammatory molecules which cause hematopoietic progenitors skewing towards myeloid lineage expansion to form MDSCs.
Matrix metalloproteinases12
Zinc-dependent MMPs act as modulators for inflammation and innate immunity by activating, deactivating or modifying the activities of signaling cytokines, chemokines and receptors through proteolytic and nonproteolytic functions[82-84]. Among MMPs, MMP12 is a 22-kDa secretory proteinase that is predominantly expressed in macrophages as previously reported[85]. MMP12 degrades extracellular matrix components, such as type IV collagen, fibronectin, laminin, gelatin, vitronectin, entactin, heparin, and chondroitin sulphates, to facilitate tissue remodeling[86]. The expression of MMP12 in macrophages is induced in the lung of cigarette smokers[87]. Inactivation of the MMP12 gene in knock-out mice demonstrates a critical role of MMP12 in smoking-induced chronic obstructive pulmonary disease (COPD)[88], a disease highly related to lung cancer. From clinical studies, MMP12 correlates with early cancer-related deaths in non-small cell lung cancer (NSCLC), especially with those associated with tobacco cigarette smoke exposure[89,90]. In the lal-/- lung, MMP-12 is the highest upregulated gene[31]. In the lal-/- lung, both macrophages and lung epithelial alveolar type II (AT II) cells are responsible for MMP-12 increase[31,91,92]. Both myeloid-specific and lung epithelia-specific MMP12 bitransgenic mouse models have been created to study the functional roles of this LAL/PPARγ downstream molecule.
In the myeloid-specific c-fms-rtTA/(TetO)7-CMV-MMP12 bitransgenic mouse model, induction of MMP12 abnormally elevates numbers and frequencies of CMP and GMP populations in the bone marrow, similar to that observed in lal-/- mice. Addition of activated MMP12 is able to stimulate wild type Lin- progenitor cells to differentiate into the MDSC population, suggesting that MMP12 directly exerts its effect on hematopoietic progenitor cells. The MDSCs are systemically increased in multiple organs of MMP12 bitransgenic mice. MDSCs from MMP12-overexpred bitransgenic mice suppress T cell proliferation and function. MMP12 directly stimulates differentiation of CD11b+Gr-1+ cells from Lin- progenitor cells. In the lung, the concentration of IL-6 is increased, which aberrantly activates oncogenic Stat3 and increases expression of Stat3 downstream genes in epithelial tumor progenitor cells. As a result, spontaneous emphysema and lung adenocarcinoma are sequentially developed in MMP12-overexpressive bitransgenic mice, suggesting a critical role of MMP12 in the transition from emphysema to lung cancer.
In epithelial-specific CCSP-rtTA/(TetO)7-CMV-MMP12 bitransgenic mice, MMP12 overexpression induces regional MDSCs infiltration and increases epithelial growth. Again, spontaneous emphysema and bronchioalveolar adenocarcinoma are developed sequentially. Importantly, MMP12 upregulation is highly associated with COPD and lung cancer in human patients. Together, these studies support that LAL/PPARγ downstream MMP12 plays a critical role in emphysema to lung cancer transition that is facilitated by inflammation. Clinically, it has been reported that there is a pathophysiological connection between emphysema/COPD and lung cancers[93,94].
Apoptosis inhibitor 6
Apoptosis inhibitor 6 (Api6) belongs to the macrophage scavenger receptor cysteine-rich domain superfamily (SRCR-SF)[95,96]. Api6 expression is the second highest induced gene in the lal-/- lung. Api6 is regulated by LAL metabolic derivatives (e.g., 9-HODE) and PPARγ[31]. In a myeloid-specific c-fms-rtTA/(TetO)7-CMV-Api6 bitransgenic mouse model, many phenotypes are similar to those observed in lal-/- mice. Overexpression of Api6 abnormally elevates MDSCs in the bone marrow, blood and lung with increased cell proliferation and decreased apoptotic activities. Api6 overexpression activates Stat3, Erk1/2 and p38 in myeloid lineage cells. Persistent inflammation in myeloid-specific Api6 bitransgenic mice causes lung adenocarcinoma[97].
Pathogenic overexpression of Api6 is also observed in lal-/- AT II cells. In an epithelial-specific CCSP-rtTA/(TetO)7-CMV-Api6 bitransgenic mice, Api6 overexpression in AT II cells increases pro-inflammatory cytokine/chemokine levels in bronchoalveolar lavage fluid and serum, activates oncogenic signaling and inhibits apoptosis, promotes expansion of MDSCs in lung and blood but not in the bone marrow or spleen. Lung MDSCs from this bitransgenic mouse model suppress T cell proliferation and function, which results in occurrence of emphysema and adenocarcinoma.
CONCLUSION
MDSCs play vital roles in various inflammation-induced chronic diseases. Elimination or reduction of MDSCs populations can slow down disease formation and progression. It is important to identify the molecular pathways in order to effectively block MDSCs homeostasis and function. Extensive studies outlined in this review have shown that the role of LAL in controlling neutral lipid metabolism is a key player in MDSCs development, homeostasis and function, therefore, providing a new avenue to develop therapeutic or immunologic approaches for clinical application. Through studies of the LAL function, defective gene expression patterns have been mapped in lal-/- MDSCs. These provide novel targets for controlling MDSCs and associated diseases by designing small molecule inhibitors. Clinically, small molecule inhibitors for c-kit have been tested to target MDSCs[98]. Using the gene profile list from LAL deficiency-induced MDSCs, more small molecule inhibitors can and will be identified to inhibit MDSCs pathogenic functions in various disease conditions.
ACKNOWLEDGMENTS
The authors thank Miss Katlin Walls for proof-reading the manuscript.
P- Reviewer: Gopinath SCB, Schuurman HJ, Saeki K S- Editor: Ji FF L- Editor: A E- Editor: Wang CH