Published online May 9, 2022. doi: 10.5409/wjcp.v11.i3.221
Peer-review started: June 30, 2021
First decision: July 30, 2021
Revised: August 5, 2021
Accepted: April 2, 2022
Article in press: April 2, 2022
Published online: May 9, 2022
Processing time: 310 Days and 12.2 Hours
As a result of the obesity epidemic, non-alcoholic fatty liver disease (NAFLD) represents a global medical concern in childhood with a closely related increased cardiometabolic risk. Knowledge on NAFLD pathophysiology has been largely expanded over the last decades. Besides the well-known key NAFLD genes (including the I148M variant of the PNPLA3 gene, the E167K allele of the TM6SF2, the GCKR gene, the MBOAT7-TMC4 rs641738 variant, and the rs72613567:TA variant in the HSD17B13 gene), an intriguing pathogenic role has also been demonstrated for the gut microbiota. More interestingly, evidence has added new factors involved in the “multiple hits” theory. In particular, omics determinants have been highlighted as potential innovative markers for NAFLD diagnosis and treatment. In fact, different branches of omics including metabolomics, lipidomics (in particular sphingolipids and ceramides), transcriptomics (including micro RNAs), epigenomics (such as DNA methylation), proteomics, and glycomics represent the most attractive pathogenic elements in NAFLD development, by providing insightful perspectives in this field. In this perspective, we aimed to provide a comprehensive overview of NAFLD pathophysiology in children, from the oldest pathogenic elements (including genetics) to the newest intriguing perspectives (such as omics branches).
Core Tip: A large body of evidence supported a complex non-alcoholic fatty liver disease (NAFLD) physiopathology with several factors involved in this tangled puzzle. Considering the cardiometabolic burden of NAFLD even in childhood, a better knowledge of NAFLD physiopathology is fundamental for novel therapeutic strategies.
- Citation: Riccio S, Melone R, Vitulano C, Guida P, Maddaluno I, Guarino S, Marzuillo P, Miraglia del Giudice E, Di Sessa A. Advances in pediatric non-alcoholic fatty liver disease: From genetics to lipidomics. World J Clin Pediatr 2022; 11(3): 221-238
- URL: https://www.wjgnet.com/2219-2808/full/v11/i3/221.htm
- DOI: https://dx.doi.org/10.5409/wjcp.v11.i3.221
Due to the increasing rate in pediatric obesity worldwide, non-alcoholic fatty liver disease (NAFLD) has become the most common chronic liver disease in childhood[1,2]. Current pediatric estimates report a prevalence of 3%-10% in the general population, while a dramatic increase (up to 50%) has been observed in children and adolescents with obesity[2]. Owing to its strong relationship with the metabolic syndrome (MetS) and insulin resistance (IR), both metabolic and cardiovascular risks are increased in children with NAFLD[2-4].
Hepatic fat accumulation represents the hallmark of the disease, that includes a wide spectrum of progressive forms ranging from simple steatosis through non-alcoholic steatohepatitis (NASH) to fibrosis and cirrhosis[5]. Lipolysis of adipose tissue and de novo hepatic lipogenesis are the main biological pathogenic processes contributing to fatty liver and IR[3,6]. Taken together, they result in an increased flux of free fatty acids to the liver and skeletal muscle that might activate lipotoxic pathways responsible for more progressive forms of hepatocellular injury. Interestingly, recent studies have highlighted not only the role of lipotoxicity but also of fatty acid composition as central players in NAFLD[7-9].
Pathophysiological hypotheses of NAFLD have been resumed in the “multiple hits” theory, by assuming the role of genetics, microbial, metabolic, and environmental factors through a complex interplay[1,2,10-12].
Key genetic factors for NAFLD are represented by the I148M variant of the PNPLA3 gene[13], the E167K allele of the TM6SF2[14,15], the MBOAT7-TMC4 rs641738 variant[16], and the rs72613567:TA variant in the HSD17B13 gene[17] (Table 1).
| Gene | Changes | Methods | Ref. |
| FGFR2 | Hypomethylation | Bisulfite pyrosequencing and liver biopsy | Zhang et al[112] |
| MAT1A | Hypomethylation | Bisulfite pyrosequencing and liver biopsy | Zhang et al[112] |
| CASP1 | Hypomethylation | Bisulfite pyrosequencing and liver biopsy | Zhang et al[108] |
| MTND6 | Hypomethylation | Methylation-specific PCR and liver biopsy | Pirola et al[109] |
| PARVB | Hypomethylation | Targeted-bisulfite sequencing and liver biopsy | Kitamoto et al[111] |
| PNPLA3 | Hypomethylation | Targeted-bisulfite sequencing and liver biopsy | Kitamoto et al[111] |
| PPARα | Hypomethylation | Bisulfite pyrosequencing and liver biopsy | Zeybel et al[112] |
| TGFβ1 | Hypomethylation | Bisulfite pyrosequencing and liver biopsy | Zeybel et al[112] |
| Collagen 1A1 | Hypomethylation | Bisulfite pyrosequencing and liver biopsy | Zeybel et al[112] |
| PDGFα | Hypomethylation | Bisulfite pyrosequencing and liver biopsy | Zeybel et al[112] |
| PPARGC1A | Hypomethylation | Methylation-specific PCR and liver biopsy | Sookoian et al[113] |
| cg08309687 (LINC00649) | Hypomethylation | Illumina BeadChip for array analyses | Ma et al[114] |
| NPC1L1 | Methylation | Illumina human methylation 450 Beadchip and liver biopsy | Mwinyi et al[116] |
| STARD | Methylation | Illumina human methylation 450 Beadchip and liver biopsy | Mwinyi et al[116] |
| GRHL | Methylation | Illumina human methylation 450 Beadchip and liver biopsy | Mwinyi et al[116] |
Recently, advances in the understanding of NAFLD pathogenesis have reported the role of specific lipid classes (in particular sphingolipids and ceramides) and their correlation also with IR, by underscoring the strength of the tangled link between NAFLD and IR[9,18-21].
For this reason, we aimed to provide a comprehensive overview from the oldest to the newest pathophysiological evidence on pediatric NAFLD.
One of the most recurrent questions regarding NAFLD concerns the potential progression to more severe forms in certain subjects. This seems to be relevant as hepatic inflammation or fibrosis determine the long-term prognosis of the disease, while simple steatosis does not seem to worsen the outcome[22,23], although some studies would seem to weaken this assumption[24,25].
In an attempt to explain NAFLD pathogenesis, Day et al[26] first proposed the ‘‘two hit’’ model theory, suggesting that after a first hit (i.e., hepatic steatosis), another hit (e.g., gut-derived endotoxin) contributed to NASH development. Later, a more complex model called the “multiple parallel hits model”[23] in which multiple factors (including genetics, obesity, insulin resistance, metabolic and environmental determinants) act together to induce NAFLD development and progression in genetically predisposed or high-risk individuals was proposed. In particular, increased lipid storage, lipogenesis, and adipokine synthesis in adipose and liver tissue, may act as stress signals for the endoplasmic reticulum (ER) with subsequent hepatocellular damage[27]. In addition, certain genes (such as PNPLA3, TM6SF2, GCKR, MBOAT7, and HSD17B13) have been strongly related to NAFLD susceptibility.
PNPLA3: The PNPLA3 gene, discovered by Hobbs and colleagues in 2008, has been largely accepted as the most important genetic determinant in NAFLD development. PNPLA3 is located on chromosome 22 and belongs to the patatin-like phospholipase family. Its expression seems to be influenced by several factors, including diet, obesity, insulin and glucose levels, and gene mutation[28]. PNPLA3 encodes for a protein called adiponutrin, an enzyme found in liver and adipose tissue that appears to confer susceptibility to increased liver fat levels and liver inflammation[29]. The discovery of PNPLA3 has brought new insights into the understanding of fatty liver, specifically lipid remodeling in intracellular droplets has been identified as a common mechanism underlying disease progression independent of environmental triggers. In particular, PNPLA3 is involved in the remodeling of triglycerides, phospholipids, and retinyl ester release, acting as a lipase on lipid droplets[30]. Adiponutrin is an enzyme with retinyl-palmitate lipase function that, in response to insulin, has been shown to be responsible for the release of retinol from lipid droplets in hepatic stellate cells in vitro and ex vivo[31]. It is induced by diet and ΙR[32] and exhibits lipolytic activity on triglycerides[33].
Several studies have investigated the major pathogenic role of the PNPLA3 rs738409 (PNPLA3 I148M) single nucleotide polymorphism (SNP) in NAFLD development. It is a non-synonymous variant in which there is a cytosine to guanosine change leading to an amino acid substitution of isoleucine to methionine at amino acid position 148 of the coding sequence, in the active site of the enzyme (I148M). This amino acid substitution affects the function of the enzyme (loss of-function), leading to intrahepatic triglyceride accumulation and consequent development of microvesicular steatosis. On the other hand, adiponutrin might exhibit a gain of lipogenic function, which could further lead to hepatic fatty acid accumulation[34]. The I148M variant, due to the altered enzymatic activity, determines an altered lipid remodeling, with accumulation of polyunsaturated fatty acids in diacylglycerol and triglycerides, and a parallel depletion in phospholipids[30]. Several studies have reported that the PNPLA3 SNP resulted in decreased retinol metabolism and decreased hepatic protein levels of retinol dehydrogenase 16, which correlate with fibrosis severity[31].
There is strong evidence in the literature for an association between the PNPLA3 148M allele and NAFLD in both adults and children. In 2008, Romeo et al[29] first reported the association between the PNPLA3 gene polymorphism (rs738409C/G) and NAFLD in a multiethnic cohort of Hispanic, African American, and European American adults.
Similarly, a large body of evidence supported the role of this gene in NAFLD development in children. Santoro et al[35], in a multiethnic group of 85 obese youths with magnetic resonance imaging (MRI)-detected steatosis, demonstrated that the prevalence of the G allele was higher in subjects with hepatic steatosis. Another study investigating 1048 obese Italian children, reported that children carrying the 148M allele showed higher aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels, in particular homozygous 148M carriers with a high level of abdominal fat (expressed as Waist/Height ratio greater than 0.62) had a higher odds ratio (OR) for developing pathological ALT. Thus, it was observed for the first time that the extent of PNPLA3 association with liver enzymes was determined by the amount of abdominal fat[36].
Romeo et al[37], in a 2010 study of 475 obese/overweight children and adolescents with steatosis evaluated by liver ultrasound, reported that the I148M variant of the PNPLA3 gene was associated with increased ALT/AST levels in obese children and adolescents, suggesting that it conferred a genetic susceptibility to liver damage at an early age.
In addition, it has been demonstrated that the frequency of the PNPLA3 risk allele rs738409 was lower in African Americans, by suggesting some protection from hepatic steatosis in obese African American youths[38]. In a 2018 study, Hudert et al[39] in a cohort of Berlin adolescents aged 10-17 years with NAFLD observed that the PNPLA3 rs 73844078G variant was significantly associated with the severity of steatosis, with an increased risk of progression to fibrosis.
The association between PNPLA3 gene and the other major genetic variants of NAFLD was also evaluated. Viitasalo et al[40] demonstrated higher serum ALT levels in children carrying the risk alleles for the polymorphisms PNPLA3, MBOAT7 and TM6SF2. Grandone et al[15] reported that homozygous subjects for the PNPLA3 148M allele carrying the rare variant of TM6SF2 showed an OR of 12.2 (confidence interval 3.8-39.6, P = 0.000001) to have hypertransaminasemia compared with the remaining patients. Of interest, an Italian pediatric study also confirmed the combined effect of the 3 major risk variants (PNPLA3, TM6F2 and MBOAT7) on NAFLD risk[16].
Besides, the interaction of the PNPLA3 148M allele with environmental risk factors for NAFLD such as obesity, nutrients (including carbohydrate and polyunsaturated fatty acids), physical activity, and sedentary behaviors have been demonstrated in children with NAFLD[41-45]. Dai et al[28], in a meta-analysis, reported a strong influence of the PNPLA3 rs738409 polymorphism not only on fatty liver but also on histological damage.
More recently, compelling evidence has also supported an intriguing role of this gene in reducing the estimated glomerular filtration rate independently of common renal and metabolic factors both in adults and children[46-49]. This gene seems to promote both fibrogenesis and glomerulosclerosis through the activation of renal pericytes in which the 148M allele is highly expressed[47,48].
Considering its detrimental effect on renal function in childhood[46-48], these findings demonstrated that the PNPLA3 gene acts not only as one of the major genetic player in NAFLD development but also as a harmful factor beyond the liver[46-48].
GCKR: Several studies reported that variations at the GCKR gene locus are associated with NAFLD and appear to influence hepatic fat accumulation. The GCKR protein has an inhibitory action on the activity of the enzyme glucokinase that regulates the hepatic storage and disposal of glucose. In particular, GCKR forms an inactive complex with the enzyme glucokinase and transports it from the cytoplasm to the nucleus, thus controlling both activity and intracellular localization of this key enzyme of glucose metabolism[49].
Fructose-6-phosphate (F6P) enhances GCKR-mediated inhibition. By controlling glucose influx into hepatocytes, GCKR regulates de novo lipogenesis. The mechanism responsible for liver injury is probably due to the lack of inhibition of glucokinase enzymatic activity by F6P and consequently uncontrolled lipogenesis[50].
GCKR gene polymorphisms (rs780094 and rs1260326) have been identified that appear to be important in the pathogenesis of NAFLD. In particular, Beer et al[51] and Valenti et al[52] reported that in the association with NAFLD and consequently in the accumulation of hepatic fat, the common missense loss-of-function GCKR mutation (rs1260326 C>T) encoding for the P446L protein variant plays an important pathogenic role. The P446L variant blocks the inhibitory activity of GCKR on the enzyme glucokinase, resulting in a steady increase in hepatic glucokinase and glucose uptake by the liver. Hepatic glycolysis associated with the minor allele P446L results in lower levels of both glucose and insulin, but leads to increased levels of malonyl-CoA which in turn blocks fatty acid oxidation through inhibition of carnitine-palmytoyltransferase-1 and acts as a substrate for lipogenesis, thus promoting hepatic fat accumulation[53]. The GCKR rs780094 C>T variant has been found to be associated with increased intrahepatic fat accumulation and progressive forms of NAFLD[54,55].
A pediatric study involving 70 obese adolescents demonstrated that the GCKR rs780094 C>T variant was associated with NAFLD and decreased levels of GCKR protein, while the GCKR rs780094C>T and rs1260326C>T variants were associated with fibrosis and decreased levels of GCKR protein[39]. Lin et al[56], in a study examining 797 obese Taiwanese children, reported that the GCKR rs780094T variant was associated with an increased risk of NAFLD, by further demonstrating that the GCKR and PNPLA3 variants were common NAFLD risk genetic factors in obese individuals. In fact, several studies have also reported a combined effect of the PNPLA3 and GCKR SNPs as NAFLD risk polymorphisms. In particular, Santoro et al[57] in a study of 455 obese children and adolescents reported that the GCKR rs1260326 variant was associated with hepatic fat accumulation along with large levels of very-low-density lipoprotein (VLDL) and triglycerides, further demonstrating that GCKR and PNPLA3 synergistically act to convey susceptibility to fatty liver in obese youths.
More recent studies confirmed the strong association of the three major genetic variants such as TM6SF2 rs58542926, PNPLA3rs738409, and GCKR rs1260326 with NAFLD in obese children and adolescents[58].
TM6SF2: TM6SF2 is responsible for the regulation of lipid metabolism in the liver[59]. In particular, TM6SF2 gene contributes to the secretion of VLDL from the liver[60]. As suggested by recent evidence[61], TM6SF2 is a polytopic membrane protein acting as a lipid transporter. It is predominantly expressed in the liver, small intestine, and kidney. TM6SF2 encodes a 351 amino acid protein with 7-10 predicted transmembrane domains[60]. Sliz et al[62] reported an association of the TM6SF2 rs58542926-T allele with lower-risk lipoprotein lipid profile and lower levels of glycerol and glycoprotein acetylation. Specifically, the authors reported that the TM6SF2 variant was associated with lower concentrations of all lipoprotein particle subclasses [including VLDL and low-density lipoprotein (LDL)]. In addition, there was an inverse association between this variant and total serum triglycerides and triglycerides in all lipoprotein subclasses, including high-density lipoprotein (HDL) subclasses. Finally, the TM6SF2 rs58542926-T allele did not appear to affect apolipoprotein A-I concentration, whereas it was associated with lower apolipoprotein B concentration. Furthermore, it was also found to impair the secretory pathway leading to hepatic lipid accumulation and reduced levels of circulating lipids and lipoproteins.
In the last few years, a single nucleotide rs58542926 C>T polymorphism giving rise to the E167K TM6SF2 variant was noted in the complex puzzle of NAFLD pathophysiology[34]. It was associated with increased liver fat content, NASH, advanced liver fibrosis, and cirrhosis[63]. This variant is characterized by an adenine-guanine substitution in nucleotide 499 that replaces glutamate at residue 167 with lysine (c.499A > G; p.Glu167Lys) leading to a loss of function in hepatic secretion of VLDL[61].
Another study on two large histologically characterized adult cohorts (including steatosis, steatohepatitis, fibrosis and cirrhosis) reported an association of the TM6SF2 gene with advanced liver fibrosis, regardless of the PNPLA3 genotype presence[64]. This association was also independently validated in another large European cohort[65].
Thus, TM6SF2 might be considered as a regulator of liver fat metabolism with the opposite effects on triglyceride-rich lipoprotein secretion and hepatic lipid droplet content[34].
Chen et al[59] in a recent meta-analysis, on associations of TM6SF2 polymorphisms with chronic liver disease, suggested that rs58542926 polymorphism may be significantly associated with chronic liver disease in both Asians and Caucasians. In addition, Holmen et al[66] showed in a longitudinal adult Norwegian study an association of the E167K TM6SF2 variant with lower total cholesterol levels resulting in a reduced risk of myocardial infarction. Accordingly, Dongiovanni et al[65] showed an effect of this polymorphism on reducing the risk of carotid atherosclerosis in adults.
The effect of this polymorphism on ALT and cholesterol levels has also been confirmed in children and adolescents. Grandone et al[15] demonstrated in a cohort of 1010 obese Caucasian children and adolescents that the TM6SF2 167K allele in carriers was associated with hepatic steatosis, higher ALT levels and lower total cholesterol, LDL-cholesterol, triglycerides and non-high density lipoproteins. In addition, subjects homozygous for the PNPLA3 148M allele carrying the rare variant of TM6SF2 showed an OR of 12.2 for presenting hypertransaminasemia compared with the remaining patients. Thus, the effect of PNPLA3 and TM6SF2 alleles appeared to be additive in determining pediatric NAFLD. As previously demonstrated in adults, the authors found that the TMS6SF2 E167K variant predisposed to NAFLD in obese children, with a relevant beneficial effect on cardiovascular risk[15].
It is noteworthy that recent data also showed a protective effect of the TM6SF2 gene on renal function both in adults and children through the reduction of lipotoxicity[47,67].
In conclusion, the discovery of the E167K variant adds another piece not only in the complex pathophysiology of NAFLD but also in the larger context of NAFLD-related cardiometabolic risk.
MBOAT7: The pathogenic role of this gene in NAFLD susceptibility has been largely studied both in adults and children. Findings demonstrated its effect in increasing not only the risk (and the severity) of NAFLD but also of other chronic liver diseases (e.g. hepatitis B and C virus-related). MBOAT7 encodes lysophosphatidylinositol acyltransferase, involved in the inflammation cascade through the regulation of arachidonic acid levels and leukotriene synthesis in neutrophils. A combined effect of this gene with the major NAFLD risk polymorphisms (such as PNPLA3 and TM6SF2) has also been highlighted in adult and pediatric studies[16]. Similar to renal effects observed for PNPLA3 and TM6SF2, a role for this gene in kidney dysfunction has also been demonstrated[47].
HSD17B13: The 17β-hydroxysteroid dehydrogenases (HSD17Bs) encompasses a large family of 15 members involved in various metabolic processes such as the metabolism of steroid hormones, cholesterol, fatty acids, and bile acids[68]. In 2008, Horiguchi identified HSD17B13 as a novel lipid droplet (LD) associated protein. The human HSD17B13 gene is located on chromosome 4 (4q22.1) and its expression is highly restricted to the liver, particularly in hepatocytes[69]. The human HSD17B13 gene encodes a 300 amino acid protein, hydroxyl-steroid 17-beta dehydrogenase 13, a liver-specific LD-associated protein which is localized to lipid droplets[70].
To date, the physiological function of HSD17B13 remains largely unclear. HSD17B13 appears to have a role in estradiol metabolism and enzymatic activity against bioactive lipid mediators, such as leukotriene B4, that are involved in lipid-mediated inflammation[71].
In a 2019 study, Ma et al[72] reported that HSD17B13 exerts retinol dehydrogenase activity in vitro, which is closely linked to lipid droplets. Indeed, it was observed that HSD17B13 catalyzes the oxidation of retinol to retinaldehyde, the rate-limiting step in all-trans retinoic acid biosynthesis.
The fact that HSD17B13 is highly abundant in the liver and selectively expressed on the lipid droplet surface suggests a potential critical effect in lipid droplet function, as supported by growing data demonstrating the key role of the HSD17B13 gene in hepatic lipid homeostasis and NAFLD pathogenesis[73].
In contrast, inactivating variants in the HSD17B13 gene have recently been linked with a reduced risk of chronic liver disease in several studies[63]. In 2018, Abul-Husn et al[71] reported that a loss-of-function variation in the HSD17B13 (rs72613567:TA) gene resulting in a truncated protein confers protection against chronic liver damage and attenuates the progression of NAFLD and alcoholic liver disease (ALD) in European Americans through reduced enzymatic activity against several proinflammatory lipid species. Sookoian et al[74] in an exome-wide association study, confirmed that the HSD17B13 rs72613567 variant had an influence on the susceptibility and histological severity of NAFLD. Furthermore, Pirola et al[75] observed a lower risk of progressive NASH in subjects carrying the rs72613567:TA variant compared to non-carriers. However, the exact role of HSD17B13 in the NAFLD pathophysiology remains largely uncharacterized. Recently, interesting studies on the inactivation of HSD17B13 in mice and the identification of an enzymatic active site that metabolizes retinol have been reported[76,77], but pathophysiological evidence on human models is still limited[74,78]. The rs72613567: TA HSD17B13 variant seems to affect liver by modulating hepatic retinol metabolism and by reducing stellate cell activity[78]. Another study, examining a large adult population, reported a protective role of this variant against various liver diseases such as cirrhosis, and hepatocellular carcinoma (HCC). In particular, HSD17B13 rs72613567 was associated with reduced inflammation and fibrosis, and milder disease severity of NAFLD. Thus, HSD17B13 rs72613567 represents an important protective factor in distinct liver diseases (including ALD, cirrhosis, and HCC) and seems to be associated with milder histological progression of NAFLD[79,80]. In 2019, Yang et al[81] in a multicenter European study of a total of 3315 patients with or without HCC but with chronic liver disease, reported that the HSD17B13 loss-of-function variant rs72613567 is protective of HCC development in patients with ALD. Taken together, these findings suggested the potential therapeutic role of the HSD17B13 inhibition[79] in patients at high risk for liver diseases. The rs72613567 variant also appears to interact with PNPLA3 I148M through the additional HSD17B13 TA alleles that reduce the effect of the additional PNPLA3 I148M alleles on serum ALT levels. It also mitigated liver damage in individuals genetically predisposed to hepatic steatosis by PNPLA3 I148M[71]. The protective effect of the rs72613567:TA HSD17B13 variant in reducing liver damage has also been observed in children[17]. By analyzing a large cohort of Italian obese children, carriers of the HSD17B13 variant showed lower NAFLD risk than noncarriers. It is noteworthy that this variant was found to protect against liver damage even among patients stratified on the basis of the number of the steatogenic alleles of the three major NAFLD risk polymorphisms (such as PNPLA3, TM6SF2, and MBOAT7genes). More interestingly, recent pediatric evidence[47,48,82] showed a similar protective effect of this gene also on renal function, by supposing its role in retinol metabolism through modulation of both inflammation and fibrogenesis. Another variant (rs143404524) in the HSD17B13 gene, resulting in a truncated protein has also been associated with a reduced risk of chronic liver disease in the adult population[83]. Finally, it has also been demonstrated that the rs62305723 variant of the HSD17B13 gene, a missense variant that confers loss of enzyme activity was associated with decreased steatohepatitis[72]. In conclusion, the HSD17B13 gene represents a well-known genetic factor with a protective role against liver damage both in adults and children[68] that might be considered an important pharmacological target for NAFLD treatment[17,84].
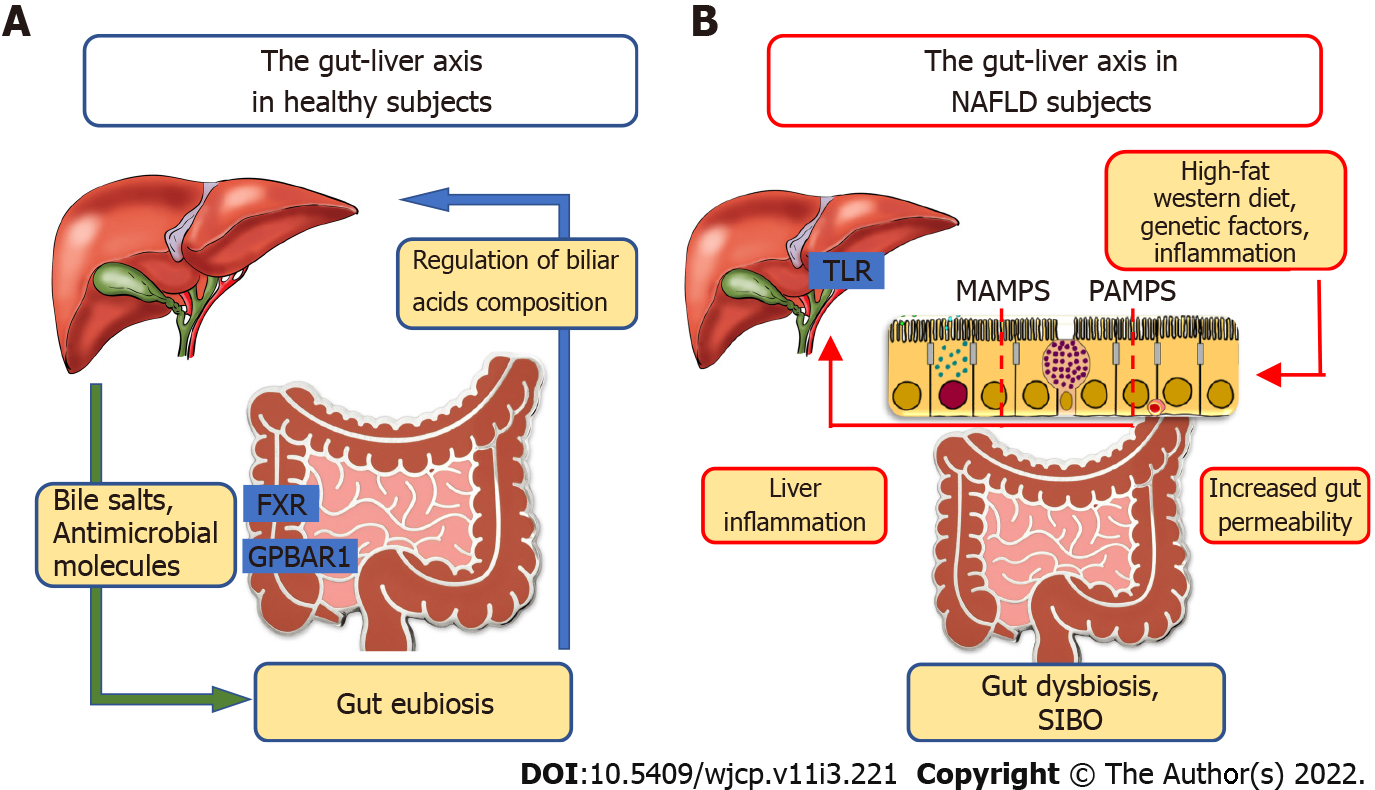
Recently, compelling evidence has supported the close and interdependent relationship between the liver and gut axis in the pathogenesis of numerous chronic liver diseases such as chronic hepatitis B and C, ALD, NAFLD, NASH, development of liver cirrhosis, and HCC (Figure 1).
Bäckhed et al[85] for the first time described the role of gut microbiota in the context of NAFLD and obesity, taking part in the processes of absorption and storage of energy but also in the production of triglycerides, responsible for the infiltration of hepatocytes.
Crosstalk between the liver and gut occurs by means of the biliary tract, portal vein and systemic mediators[86]. The liver contributes to the maintenance of gut eubiosis through the transport of bile salts and antimicrobial molecules to the intestinal lumen. Conversely, the gut regulates bile acids (BAs) composition. In addition, BAs using farnesoid X receptor (FXR) in the enterocytes and G protein-coupled bile acid receptor 1 (also known as TGR5) are involved in the regulation of glucose and lipid metabolism, anti-inflammatory immune responses and host energy expenditure[87-91]. Furthermore, the gut through secretion of the incretin hormones glucagon-like peptide-1 (GLP-1) and glucose-dependent insulinotropic peptide influences the pancreas in regulating both insulin and glucagon secretion[92]. Moreover, GLP-1 interaction with its receptor (also located on the hepatocytes) results in reduced hepatic fat deposition and IR. Finally, it promotes energy expenditure and peripheral utilization of triglycerides for energy production[93].
BAs synthesis is regulated by two hepatic methods: the enterohepatic circulation (with a subsequent negative feedback loop on the expression of CYP7A1) and FGF19, (derived from the activation of FXR by BAs in the ileum and has an inhibitory effect on CYP7A1 gene[94]).
Impaired FXR-FGF19 signaling and elevated circulating BA levels were described both in children and adults with NAFLD. However, experimental therapeutic interventions targeting BA signaling with FXR agonists (obeticholic acid) have produced contradictory results[95].
Some differences were reported in the composition of gut microbiota (i.e. dysbiosis) in healthy controls than in subjects with simple fatty liver disease (FLD) and NASH[96]. In fact, many pediatric studies have reported a decreased gut microbiota alpha diversity, measured with the Shannon index[45,97-99].
In 2006, Turnbaugh et al[100] found that the ratio of Firmicutes to Bacteroidetes increased in obese mice, suggesting a putative role for Firmicutes as a group of obesity-related microbiomes.
Loomba et al[101] in an elegant study showed that NAFLD patients exhibited more Gram-negative and fewer Gram-positive bacteria compared to healthy subjects, with an increase in Proteobacteria and a decrease in Firmicutes in more progressive NAFLD forms.
Michail et al[102] noted that children with NAFLD had more abundant Gammaproteobacteria and Prevotella compared to obese children without NAFLD and healthy controls. In addition, no difference in Firmicutes and Bacteroidetes or their ratio was observed between the groups.
Del Chierico et al[97] in a complex study with an integrated meta-omics-based approach found a significant increment of Actinobacteria and a decrease of Bacteroidetes in NAFLD patients compared to healthy controls.
Stanislawski et al[102] examined 107 adolescents with MRI-detected hepatic steatosis and found that Bilophila was positively correlated with hepatic fat fraction (HFF), while Oscillospira and Bacteroides showed different patterns in relation to HFF.
Schwimmer et al[99] in a prospective, observational, cross-sectional study of 87 children with biopsy-proven NAFLD and 37 obese children without NAFLD noted that a high abundance of Prevotella copri was associated with more severe fibrosis.
In a metagenomic study of gut microbiota by Zhao et al[103] conducted in 58 children and adolescents with NAFLD diagnosed by magnetic resonance spectroscopy, the authors found no significant differences in terms of alpha diversity among the study groups (NAFLD children, obese children without NAFLD and healthy controls). However, Proteobacteria were found to be more represented in NAFLD children than in the control group, while Bacteroidates (Alistipes) were significantly reduced.
Finally, Kravetz et al[45] in a cross-sectional study including 73 obese children and adolescents with and without NAFLD, in which HFF was determined by MRI, the NAFLD group showed a higher Firmicutes to Bacteroidetes ratio and lower levels of Bacteroidetes, Prevotella, Gemmiger and Oscillospira.
Altered gut microbial composition and increased intestinal permeability are linked to several factors (e.g. high-fat Western diet, chronic alcohol consumption, and genetic factors) and promote the influx of microbial-associated molecular patterns or pathogen-associated molecular patterns into the portal system reaching the liver. These molecular patterns are responsible for inflammatory responses mediated by the activation of pattern recognition receptors, like Toll-like receptor, in Kupffer cells and hepatic stellate cells, leading to liver injury and fibrosis[86,104-106].
Potential gut-microbiome-targeted therapies in hepatic diseases are represented by probiotics, prebiotics, antibiotics, fecal microbial transplantation and bacteriophages, but larger validation studies are needed[107].
Several authors have studied the role of epigenetic modifications in the natural history of NAFLD. The main epigenomic modification studied in NAFLD is DNA methylation.
A recent systematic review[108] included twelve studies on DNA methylation and FLD of which two assessed global DNA methylation, five assessed DNA methylation for specific candidate genes and the remaining four used the EWAS approach. The review suggested no consistent associations with FLD in the studies of global DNA methylation evaluated in hepatic tissue samples by quantifying the methylcytosine (5-mC) present in the genome. One of the two studies assessing global DNA methylation found mitochondrial encoded NADH dehydrogenase 6 hypermethylation in the liver of NASH patients compared to those with simple steatosis, and this methylation was significantly associated with NAFLD activity score[109]. On the other hand, another study reported that global liver methylation based on genome-wide methylation arrays was not associated with NAFLD or NASH, but NASH was associated with long-interspersed nuclear element hypomethylation compared to simple steatosis or normal liver[110]. More, studies using a candidate gene approach found that NAFLD was associated with hypomethylation at FGFR2, MAT1A, CASP1 and PARVB genes and hypermethylation at PNPLA3[111], PPARα, TGFβ1, Collagen 1A1 and PDGFα genes[112]. Furthermore, PPARGC1A methylation status was significantly associated with NAFLD[113]. The epigenome-wide DNA methylation studies reported different associations of distinct methylation compounds with NAFLD[114,115]. Finally, a single study reported the role of methylation in NAFLD in the expression of three genes (NPC1L1, STARD and GRHL) involved in lipoprotein particle composition[116].
A recent and interesting prospective cohort study analyzed epigenome-wide DNA methylation data of 785 newborns and 344 10-year-old children in relation to liver fat fraction (measured by MRI) at 10 years. No differential DNA methylation at age 10 years in newborns or 10-year-old children were found[117].
Despite some causative evidence, little is still known about the relationship between these changes in hepatic epigenome and their repercussion in the bloodstream. As a result, the contribution of epigenomics in the non-invasive diagnosis of NAFLD is still very limited but promising.
A growing body of data is derived from micro RNAs (miRNAs), highly conserved noncoding small RNAs, involved in gene expression modulation at the post-transcriptional level (Table 2). MiRNAs are resistant to degradation as well as to several freeze–thaw cycles, suggesting their potential role as ideal biomarkers for use in clinical practice.
| Ref. | Study design | Population (n) | Main findings |
| Yamada et al[118] | Cross-sectional study | 403 male subjects (median age 68.2 ± 10.3 yr); 48 NAFLD subjects (median age 66.2 ± 9.1 yr); 221 female patients (median age 65.5 ± 9.6 yr); 44 women with NAFLD (median age 65.0 ± 8.93 yr). Hepatic steatosis was assessed by ultrasound | Increased serum levels of miR-21, miR-34a, miR-122, and miR-451 were found in NAFLD patients |
| Cheung et al[119] | Cross-sectional study | 50 patients with NASH (median age 52.5 yr) and 25 normal controls (median age 40.3 yr). NAFLD was suspected if abnormal liver enzymes or radiological evidence of a fatty liver and negative study for other common causes of liver disease and absence of clinically significant alcohol consumption | miR-34a and miR-146b were overexpressed in the liver of NASH patients, while miR-122 was underexpressed; miR-451 was not significantly different among the two groups |
| Pirola et al[120] | Case-control study | 48 control patients (median age 47.8 ± 6.81 yr); 16 patients with simple steatosis (median age 51.5 ± 6.81 yr); 16 patients with NASH (median age 49.1 ± 8.6 yr). NAFLD was proven by biopsy | Increased levels of miR-122, miR-19a, miR-192, miR-19b, miR-125b, and miR-375 in serum either in SS or NASH patients were found. Reduced miR-122 levels in the liver of NASH patients were detected |
| Prats-Puig et al[122] | Cross-sectional study | 10 lean children (median age 9.9 ± 1 yr), 5 obese children (median age 8.8 ± 1.8 yr) | Increased miR-486-5p, miR-486-3p, miR-142-3p, miR-130b, miR-423-5p, miR-532-5p, miR140-5p, miR-16-1, miR-222, miR-363, and miR-122; decreased miR-221, miR-28–3p, miR-125b, and miR-328 in obese children |
| Can et al[123] | Case-control study | 86 non obese children (median age 14.44 ± 1.62 yr); 45 obese children (median age 14.71 ± 1.76 yr) | Reduced miR-335, miR-143, miR-758 and increased miR-27, miR-378, and miR-370 in the serum of obese children were detected |
| Cui et al[124] | Cross-sectional study | 535 obese patients (median age 61.0 ± 10.4 yr); 106 OW patients (median age 59.6 ± 11.0 yr); 101 patients with T2D (median age 57.5 ± 12.2 yr); 82 with NGT (median age 49.3 ± 7.73 yr); 146 normal controls (median age 60.4 ± 11.1 yr) | miR-486, miR-146b and miR-15b were increased in the serum of obese children and T2D patients |
| Iacomino et al[125] | Cross-sectional study | 189 children (median age 12.0 ± 1.6 yr) and 94 OW/Ob children (median age 12.3 ± 1.8 yr) | Increased miR-551a and miR-501-5p and reduced miR-10b-5p, miR-191-3p, miR-215-5p, and miR-874-3p levels in the serum of OW/Ob children were found |
Several studies highlighted the association between miR-122 and the severity of steatosis[118]. A reduced hepatic expression of miR-122 was described[119,120], whereas miR-122 levels were upregulated in serum[120].
A systematic review reported 34 miRNAs associated with FLD. Among these, miR-122, miR-34a, miR-192, miR-21 and miR-99a were associated with FLD in two or more independent studies[108].
Specifically, circulating miR-122 and miR-192 not only reflected both histological and molecular processes occurring in the liver, but have also been considered to be able to differentiate simple steatosis from NASH[121].
A cross-sectional validation study disclosed that 15 specific circulating miRNAs were significantly deregulated in prepubertal obesity, including the decreased miR-221 and miR-28 -3p, and increased concentrations in plasma of miR-486-5p, miR-486-3p, miR-142-3p, miR-130b, and miR-423-5p[122].
Can et al[123] showed a significant association between circulating miR-370, miR-33, miR-378, miR-27, miR-335, miR-143 and miR-758 values, and childhood obesity. Low levels of miR-335, miR-143 and miR-758, and high levels of miR-27, miR-378, miR-33 and miR-370 may have been responsible for elevated triglycerides and LDL-C levels, and a low level of HDL-C in obese subjects.
An interesting work by Cui et al[124] highlighted the specific role of three miRNAs, miR-486, miR-146b and miR-15b, by demonstrating their increased circulating expression in obese children and adult patients with type 2 diabetes mellitus (T2DM). In particular, miR-486 was implicated in accelerating preadipocyte proliferation and myotube glucose intolerance, miR-146b and miR-15b were engaged in the suppression of high concentration glucose-induced pancreatic insulin secretion, and they all contributed to the pathological processes of obesity and T2DM.
Iacomino et al[125] in a pilot study (FAMILY Study) conducted in 149 overweight/obese and 159 normal weight children and adolescents demonstrated a panel of miRNAs differentially expressed in these two groups (miR-551a and miR-501-5p were upregulated; miR-10b-5p, miR-191-3p, miR-215-5p, and miR-874-3p were downregulated).
In a transcriptomic study by Sheldon et al[126] a new candidate marker for distinguishing steatosis from NASH was proposed, the soluble factor FCER2, produced from NOCTH2 activation in B cells, whose expression was increased in NASH patients.
Finally, in a recent study interleukin-32 was found as the most significantly upregulated transcript in advanced NAFLD and NASH, being linked to lipid accumulation and disease severity[127].
Although many studies have been investigating the role of miRNAs in the pathogenesis of NAFLD in view of their potential use as non-invasive biomarkers, results are still controversial and scarce. However, the innovative role of transcriptomics in the non-invasive diagnosis of NAFLD contributes to the new “omics” path of NAFLD.
To date, few studies on proteomic analysis in NAFLD have been performed, probably due to technical limitations in the correct detection and identification of proteins and to the changing quantification of blood proteins[128].
Among these proteins, the caspase-generated cytokeratin-18 (CK-18) fragments have been proposed as a noninvasive alternative biomarker of NASH. CK-18 showed a relatively good specificity for NAFLD, NASH and fibrosis but limited overall sensitivity[129].
Another protein being studied is the soluble intercellular adhesion molecule-1, with promising results also in NASH detection[130].
The mitochondrial enzyme carbamoyl-phosphate synthase 1 and the heat shock protein family A member 5 have been indicated as potential tools to stratify the different phenotypes associated with liver disease severity[131-133].
In a recent study by Malecki et al[134], a proteome analysis in a group of 30 children (16 with a previous NAFLD diagnosis by ultrasound) identified a total of 297 proteins. Thirty-seven distinct proteins (responsible for inflammation, stress response, and regulation of these processes) were identified. Up-regulated proteins included afamin, retinol-binding protein-4, complement components, and hemopexin, while serum protease inhibitors, clusterin, immunoglobulin chains, and vitamin D binding protein were found in the down-regulated group[134].
Bălănescu et al[135] confirmed the role of the heat shock protein-90 (Hsp90) isoforms as biomarkers for NAFLD in obese and overweight children. While the Hsp90β isoform was higher, the Hsp90α isoform was lower in overweight and obese NAFLD patients.
Hence, proteomics represents one of the most challenging fields that might contribute to the development of new noninvasive targeted tools for NAFLD diagnosis and treatment. See Table 3.
| Ref. | Study design | Population (n) | Main findings |
| Cusi et al[129] | Case-control study | 300 subjects with NAFLD (median age 52 ± 1 yr) and 124 without NAFLD (median age 51 ± 1 yr). NAFLD was proven by MRS, biopsy, or US | Increased plasma CK-18 in steatosis, inflammation, and fibrosis |
| Sookoian et al[130] | Cross- sectional study | 101 subjects with simple steatosis (median age 52.3 yr) and 60 NASH patients (median age 54.6 yr). NAFLD was proven by biopsy | sICAM-1 is able to differentiate between patients with simple steatosis and NASH |
| Rodriguez-Suarez et al[131] | Cross- sectional study | 18 controls, 6 obese patients with NAFLD, 6 obese patients with early stage of NASH. Liver disease diagnosis was by biopsy | CPS1 could stratify different phenotypes associated with liver disease severity |
| Małecki et al[134] | Cross- sectional study | 30 children (mean age 10.62 yr), 16 children with NAFLD (mean age 11.06 yr). NAFLD was proven by US | Afamin, retinol-binding protein-4, complement components, and hemopexin were upregulated; serum protease inhibitors, clusterin, immunoglobulin chains, vitamin D binding protein were down-regulated |
| Bălănescu et al[135] | Cross- sectional study | 68 overweight and obese children (mean age 10 yr) and 10 healthy controls. NAFLD was proven by US or elevated alanine transaminase levels | HSP-90 isoforms could be used as NAFLD biomarkers in obese and overweight patients |
Most of the glycomics studies in NAFLD have tried to identify glycans or glycoproteins that can serve as blood biomarkers for differentiating between NAFLD and NASH or for detection of the presence of liver fibrosis and its stage.
Changes in glycosylation represent a potential good marker of liver damage due to the hepatic production of several serum glycoproteins[136].
The findings of these studies demonstrated that higher concentrations of fucosylated, sialylated and agalactosylated glycans were observed in NAFLD and its progressive forms. Circulating sialic acid levels were also positively associated with metabolic syndrome and with NAFLD[128].
Furthermore, changes in fucosylation were observed in other inflammatory conditions, such as in chronic pancreatitis, Crohn's disease, rheumatoid arthritis and sickle cell disease[137].
Finally, hypogalactosylation (especially of IgG) was also associated with some autoimmune diseases and inflammatory pathways[138].
The first glycomic analysis in a pediatric NAFLD population was conducted by Blomme et al[136]. In agreement with adult findings, B cells were found to play a dominant role in the N-glycan alterations of pediatric NASH patients. Serum protein N-glycosylation patterns of 51 pediatric NAFLD patients were assessed with deoxyribonucleic acid sequencer-assisted fluorophore-assisted capillary electrophoresis and compared with histology. Analysis of the N-glycans on IgG confirmed the under-galactosylation status typical of chronic inflammatory conditions.
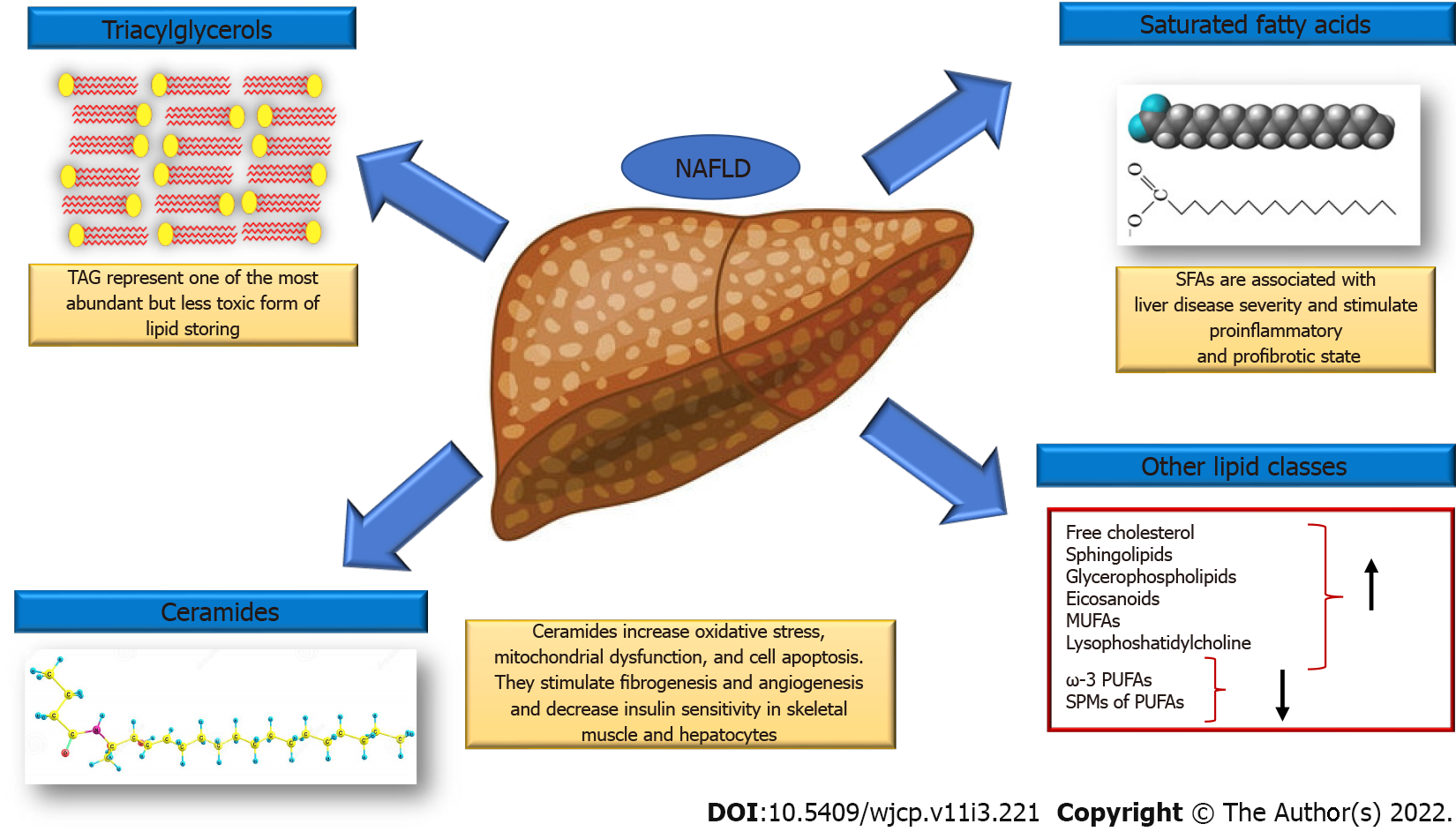
To date, both metabolomics and lipidomics represent the most investigated omics branches in NAFLD with promising results for the development of new targeted strategies (Figure 2). Of interest, robust and extensive changes were observed both in the hepatic as well as in the circulating lipidome, which have led to the development of numerous diagnostic models for NAFLD and the identification of novel therapeutic targets. Many studies have reported several diagnostic models based on metabolomics, lipidomics alone or combined with other biochemical and clinical parameters for the diagnosis and staging of NAFLD.
Lipidomic studies have described specific changes in hepatic lipidome in patients with NAFLD. The hepatic concentrations of triacylglycerols, saturated fatty acids (SFAs and specifically of palmitic acid, C16:0 and stearate acid, C18:0), free cholesterol, sphingolipids, glycerophospholipids and eicosanoids increase, whereas ω-3 polyunsaturated fatty acids (PUFAs) and specialized proresolving mediators of PUFAs decrease. Monounsaturated fatty acids, lysophosphatidylcholine (LPC) and ceramide are also increased[21].
SFAs accumulation is associated with liver disease severity. They work in two different ways: on the hepatocytes stimulating proinflammatory cytokine secretion, enhancing oxidative stress, inducing apoptosis and on nonparenchymal liver cells stimulating secretion of proinflammatory and profibrotic cytokines (Kupffer cells) and induce proinflammatory M1 polarization of macrophages. Finally, SFAs stimulate the secretion of chemokines from hepatic stellate cells that recruit more macrophages in the liver[128].
LPC also stimulates ER stress, causes mitochondrial dysfunction and increases apoptosis[139]. Increased activity of the enzyme phospholipase A2 that catalyzes the formation of LPC from PC, leads to the rapid depletion of PC which affects hepatocyte membrane integrity and results in hepatocyte apoptosis, high release of lipotoxic lipids and increased inflammation. Additionally, PC deficiency reduces VLDL secretion resulting in higher intrahepatic lipid degradation and the formation of toxic intermediates[140].
Ceramides correlate positively with hepatic disease severity[141]. These lipids have been found to decrease insulin sensitivity in skeletal muscle and hepatocytes[142] and are involved in increased oxidative stress, mitochondrial dysfunction, and cell apoptosis[142,143]. Finally, ceramide stimulates fibrogenesis and angiogenesis by increasing extracellular matrix deposition and the secretion of pro-angiogenic factors by hepatic stellate cells[144].
The attractive omics field might greatly contribute to improving not only knowledge on NAFLD pathophysiology but also its management.
Given the global relentless spread of childhood obesity, NAFLD and its cardiometabolic burden (including MetS, IR, cardiovascular disease, prediabetes, and type 2 diabetes) in childhood represent a major health challenge for clinicians[145]. Moreover, the close relationship of NAFLD with the metabolic milieu has recently been highlighted in the new definition of NAFLD as metabolic associated fatty liver disease[146,147].
To date, diet and lifestyle interventions remain the cornerstone of NAFLD treatment. Over the last few years, promising approaches have been proposed, but larger validation studies are required. In particular, omics represents the most intriguing strategy in this field, due to its potential effectiveness in preventing NAFLD as a noninvasive diagnostic and therapeutic tool.
Further novel therapeutic insights for this insidious disease might be provided only by advances in the knowledge of NAFLD pathophysiology.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Pediatrics
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhou J, Singapore S-Editor: Zhang H L-Editor: Webster JR P-Editor: Zhang H
| 1. | Goldner D, Lavine JE. Nonalcoholic Fatty Liver Disease in Children: Unique Considerations and Challenges. Gastroenterology. 2020;158:1967-1983.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 2. | Shaunak M, Byrne CD, Davis N, Afolabi P, Faust SN, Davies JH. Non-alcoholic fatty liver disease and childhood obesity. Arch Dis Child. 2021;106:3-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 73] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 3. | Morandi A, Di Sessa A, Zusi C, Umano GR, El Mazloum D, Fornari E, Miraglia Del Giudice E, Targher G, Maffeis C. Nonalcoholic Fatty Liver Disease and Estimated Insulin Resistance in Obese Youth: A Mendelian Randomization Analysis. J Clin Endocrinol Metab. 2020;105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 4. | Di Bonito P, Valerio G, Licenziati MR, Miraglia Del Giudice E, Baroni MG, Morandi A, Maffeis C, Campana G, Spreghini MR, Di Sessa A, Morino G, Crinò A, Chiesa C, Pacifico L, Manco M. High uric acid, reduced glomerular filtration rate and non-alcoholic fatty liver in young people with obesity. J Endocrinol Invest. 2020;43:461-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 5. | Barshop NJ, Francis CS, Schwimmer JB, Lavine JE. Nonalcoholic fatty liver disease as a comorbidity of childhood obesity. Ped Health. 2009;3:271-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Flisiak-Jackiewicz M, Lebensztejn DM. Update on pathogenesis, diagnostics and therapy of nonalcoholic fatty liver disease in children. Clin Exp Hepatol. 2019;5:11-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 7. | Kim HY. Recent advances in nonalcoholic fatty liver disease metabolomics. Clin Mol Hepatol. 2021;27:553-559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 8. | Kartsoli S, Kostara CE, Tsimihodimos V, Bairaktari ET, Christodoulou DK. Lipidomics in non-alcoholic fatty liver disease. World J Hepatol. 2020;12:436-450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (5)] |
| 9. | Pei K, Gui T, Kan D, Feng H, Jin Y, Yang Y, Zhang Q, Du Z, Gai Z, Wu J, Li Y. An Overview of Lipid Metabolism and Nonalcoholic Fatty Liver Disease. Biomed Res Int. 2020;2020:4020249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 10. | Bonsembiante L, Targher G, Maffeis C. Non-alcoholic fatty liver disease in obese children and adolescents: a role for nutrition? Eur J Clin Nutr. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 11. | Peng L, Wu S, Zhou N, Zhu S, Liu Q, Li X. Clinical characteristics and risk factors of nonalcoholic fatty liver disease in children with obesity. BMC Pediatr. 2021;21:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 12. | Goyal NP, Schwimmer JB. The Genetics of Pediatric Nonalcoholic Fatty Liver Disease. Clin Liver Dis. 2018;22:59-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Tang S, Zhang J, Mei TT, Guo HQ, Wei XH, Zhang WY, Liu YL, Liang S, Fan ZP, Ma LX, Lin W, Liu YR, Qiu LX, Yu HB. Association of PNPLA3 rs738409 G/C gene polymorphism with nonalcoholic fatty liver disease in children: a meta-analysis. BMC Med Genet. 2020;21:163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Goffredo M, Caprio S, Feldstein AE, D'Adamo E, Shaw MM, Pierpont B, Savoye M, Zhao H, Bale AE, Santoro N. Role of TM6SF2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: A multiethnic study. Hepatology. 2016;63:117-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 103] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 15. | Grandone A, Cozzolino D, Marzuillo P, Cirillo G, Di Sessa A, Ruggiero L, Di Palma MR, Perrone L, Miraglia Del Giudice E. TM6SF2 Glu167Lys polymorphism is associated with low levels of LDL-cholesterol and increased liver injury in obese children. Pediatr Obes. 2016;11:115-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 16. | Di Sessa A, Umano GR, Cirillo G, Del Prete A, Iacomino R, Marzuillo P, Del Giudice EM. The Membrane-bound O-Acyltransferase7 rs641738 Variant in Pediatric Nonalcoholic Fatty Liver Disease. J Pediatr Gastroenterol Nutr. 2018;67:69-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 17. | Di Sessa A, Umano GR, Cirillo G, Marzuillo P, Arienzo MR, Pedullà M, Miraglia Del Giudice E. The rs72613567: TA Variant in the Hydroxysteroid 17-beta Dehydrogenase 13 Gene Reduces Liver Damage in Obese Children. J Pediatr Gastroenterol Nutr. 2020;70:371-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 18. | Yki-Järvinen H. Ceramides: A Cause of Insulin Resistance in Nonalcoholic Fatty Liver Disease in Both Murine Models and Humans. Hepatology. 2020;71:1499-1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Samuel VT, Shulman GI. Nonalcoholic Fatty Liver Disease, Insulin Resistance, and Ceramides. N Engl J Med. 2019;381:1866-1869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 20. | Apostolopoulou M, Gordillo R, Gancheva S, Strassburger K, Herder C, Esposito I, Schlensak M, Scherer PE, Roden M. Role of ceramide-to-dihydroceramide ratios for insulin resistance and non-alcoholic fatty liver disease in humans. BMJ Open Diabetes Res Care. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Di Sessa A, Riccio S, Pirozzi E, Verde M, Passaro AP, Umano GR, Guarino S, Miraglia Del Giudice E, Marzuillo P. Advances in paediatric nonalcoholic fatty liver disease: Role of lipidomics. World J Gastroenterol. 2021;27:3815-3824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol. 2010;5:145-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 586] [Cited by in RCA: 646] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 23. | Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836-1846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1543] [Cited by in RCA: 1819] [Article Influence: 121.3] [Reference Citation Analysis (0)] |
| 24. | McPherson S, Hardy T, Henderson E, Burt AD, Day CP, Anstee QM. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: implications for prognosis and clinical management. J Hepatol. 2015;62:1148-1155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 687] [Cited by in RCA: 793] [Article Influence: 79.3] [Reference Citation Analysis (0)] |
| 25. | Pais R, Charlotte F, Fedchuk L, Bedossa P, Lebray P, Poynard T, Ratziu V; LIDO Study Group. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. J Hepatol. 2013;59:550-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 384] [Article Influence: 32.0] [Reference Citation Analysis (1)] |
| 26. | Day CP, James OF. Steatohepatitis: a tale of two "hits"? Gastroenterology. 1998;114:842-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2953] [Cited by in RCA: 3129] [Article Influence: 115.9] [Reference Citation Analysis (36)] |
| 27. | Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1376] [Cited by in RCA: 2914] [Article Influence: 416.3] [Reference Citation Analysis (1)] |
| 28. | Dai G, Liu P, Li X, Zhou X, He S. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: A meta-analysis. Medicine (Baltimore). 2019;98:e14324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 29. | Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461-1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2701] [Cited by in RCA: 2596] [Article Influence: 152.7] [Reference Citation Analysis (0)] |
| 30. | Trépo E, Valenti L. Update on NAFLD genetics: From new variants to the clinic. J Hepatol. 2020;72:1196-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 274] [Article Influence: 54.8] [Reference Citation Analysis (1)] |
| 31. | Woodside M. Research on children of alcoholics: past and future. Br J Addict. 1988;83:785-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 32. | Marzuillo P, Miraglia del Giudice E, Santoro N. Pediatric fatty liver disease: role of ethnicity and genetics. World J Gastroenterol. 2014;20:7347-7355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 59] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 33. | Dongiovanni P, Donati B, Fares R, Lombardi R, Mancina RM, Romeo S, Valenti L. PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol. 2013;19:6969-6978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 170] [Cited by in RCA: 203] [Article Influence: 16.9] [Reference Citation Analysis (1)] |
| 34. | Marzuillo P, Grandone A, Perrone L, Miraglia Del Giudice E. Understanding the pathophysiological mechanisms in the pediatric non-alcoholic fatty liver disease: The role of genetics. World J Hepatol. 2015;7:1439-1443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 35. | Santoro N, Kursawe R, D'Adamo E, Dykas DJ, Zhang CK, Bale AE, Calí AM, Narayan D, Shaw MM, Pierpont B, Savoye M, Lartaud D, Eldrich S, Cushman SW, Zhao H, Shulman GI, Caprio S. A common variant in the patatin-like phospholipase 3 gene (PNPLA3) is associated with fatty liver disease in obese children and adolescents. Hepatology. 2010;52:1281-1290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 183] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 36. | Giudice EM, Grandone A, Cirillo G, Santoro N, Amato A, Brienza C, Savarese P, Marzuillo P, Perrone L. The association of PNPLA3 variants with liver enzymes in childhood obesity is driven by the interaction with abdominal fat. PLoS One. 2011;6:e27933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Romeo S, Sentinelli F, Cambuli VM, Incani M, Congiu T, Matta V, Pilia S, Huang-Doran I, Cossu E, Loche S, Baroni MG. The 148M allele of the PNPLA3 gene is associated with indices of liver damage early in life. J Hepatol. 2010;53:335-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 38. | Tricò D, Caprio S, Rosaria Umano G, Pierpont B, Nouws J, Galderisi A, Kim G, Mata MM, Santoro N. Metabolic Features of Nonalcoholic Fatty Liver (NAFL) in Obese Adolescents: Findings From a Multiethnic Cohort. Hepatology. 2018;68:1376-1390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 39. | Hudert CA, Selinski S, Rudolph B, Bläker H, Loddenkemper C, Thielhorn R, Berndt N, Golka K, Cadenas C, Reinders J, Henning S, Bufler P, Jansen PLM, Holzhütter HG, Meierhofer D, Hengstler JG, Wiegand S. Genetic determinants of steatosis and fibrosis progression in paediatric non-alcoholic fatty liver disease. Liver Int. 2019;39:540-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 40. | Viitasalo A, Eloranta AM, Atalay M, Romeo S, Pihlajamäki J, Lakka TA. Association of MBOAT7 gene variant with plasma ALT levels in children: the PANIC study. Pediatr Res. 2016;80:651-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 41. | Marzuillo P, Grandone A, Perrone L, del Giudice EM. Weight loss allows the dissection of the interaction between abdominal fat and PNPLA3 (adiponutrin) in the liver damage of obese children. J Hepatol. 2013;59:1143-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 42. | Wang S, Song J, Shang X, Chawla N, Yang Y, Meng X, Wang H, Ma J. Physical activity and sedentary behavior can modulate the effect of the PNPLA3 variant on childhood NAFLD: a case-control study in a Chinese population. BMC Med Genet. 2016;17:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 43. | Davis JN, Lê KA, Walker RW, Vikman S, Spruijt-Metz D, Weigensberg MJ, Allayee H, Goran MI. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am J Clin Nutr. 2010;92:1522-1527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 44. | Santoro N, Savoye M, Kim G, Marotto K, Shaw MM, Pierpont B, Caprio S. Hepatic fat accumulation is modulated by the interaction between the rs738409 variant in the PNPLA3 gene and the dietary omega6/omega3 PUFA intake. PLoS One. 2012;7:e37827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 45. | Monga Kravetz A, Testerman T, Galuppo B, Graf J, Pierpont B, Siebel S, Feinn R, Santoro N. Effect of Gut Microbiota and PNPLA3 rs738409 Variant on Nonalcoholic Fatty Liver Disease (NAFLD) in Obese Youth. J Clin Endocrinol Metab. 2020;105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 46. | Mantovani A, Zusi C, Sani E, Colecchia A, Lippi G, Zaza GL, Valenti L, Byrne CD, Maffeis C, Bonora E, Targher G. Association between PNPLA3rs738409 polymorphism decreased kidney function in postmenopausal type 2 diabetic women with or without non-alcoholic fatty liver disease. Diabetes Metab. 2019;45:480-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 47. | Di Sessa A, Guarino S, Passaro AP, Liguori L, Umano GR, Cirillo G, Miraglia Del Giudice E, Marzuillo P. NAFLD and renal function in children: is there a genetic link? Expert Rev Gastroenterol Hepatol. 2021;15:975-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 48. | Marzuillo P, Di Sessa A, Guarino S, Capalbo D, Umano GR, Pedullà M, La Manna A, Cirillo G, Miraglia Del Giudice E. Nonalcoholic fatty liver disease and eGFR levels could be linked by the PNPLA3 I148M polymorphism in children with obesity. Pediatr Obes. 2019;14:e12539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 49. | Fang YL, Chen H, Wang CL, Liang L. Pathogenesis of non-alcoholic fatty liver disease in children and adolescence: From "two hit theory" to "multiple hit model". World J Gastroenterol. 2018;24:2974-2983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 191] [Cited by in RCA: 267] [Article Influence: 38.1] [Reference Citation Analysis (5)] |
| 50. | Nobili V, Alisi A, Valenti L, Miele L, Feldstein AE, Alkhouri N. NAFLD in children: new genes, new diagnostic modalities and new drugs. Nat Rev Gastroenterol Hepatol. 2019;16:517-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 218] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 51. | Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PR, Orho-Melander M, Gloyn AL. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081-4088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 327] [Cited by in RCA: 315] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 52. | Valenti L, Alisi A, Nobili V. Unraveling the genetics of fatty liver in obese children: additive effect of P446L GCKR and I148M PNPLA3 polymorphisms. Hepatology. 2012;55:661-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 53. | Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J Hepatol. 2018;68:268-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 732] [Cited by in RCA: 690] [Article Influence: 98.6] [Reference Citation Analysis (0)] |
| 54. | Speliotes EK, Butler JL, Palmer CD, Voight BF; GIANT Consortium; MIGen Consortium; NASH CRN, Hirschhorn JN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904-912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 302] [Cited by in RCA: 293] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 55. | Petta S, Miele L, Bugianesi E, Cammà C, Rosso C, Boccia S, Cabibi D, Di Marco V, Grimaudo S, Grieco A, Pipitone RM, Marchesini G, Craxì A. Glucokinase regulatory protein gene polymorphism affects liver fibrosis in non-alcoholic fatty liver disease. PLoS One. 2014;9:e87523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 56. | Lin YC, Chang PF, Chang MH, Ni YH. Genetic variants in GCKR and PNPLA3 confer susceptibility to nonalcoholic fatty liver disease in obese individuals. Am J Clin Nutr. 2014;99:869-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 57. | Santoro N, Zhang CK, Zhao H, Pakstis AJ, Kim G, Kursawe R, Dykas DJ, Bale AE, Giannini C, Pierpont B, Shaw MM, Groop L, Caprio S. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology. 2012;55:781-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 189] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 58. | Zusi C, Mantovani A, Olivieri F, Morandi A, Corradi M, Miraglia Del Giudice E, Dauriz M, Valenti L, Byrne CD, Targher G, Maffeis C. Contribution of a genetic risk score to clinical prediction of hepatic steatosis in obese children and adolescents. Dig Liver Dis. 2019;51:1586-1592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 59. | Chen X, Zhou P, De L, Li B, Su S. The roles of transmembrane 6 superfamily member 2 rs58542926 polymorphism in chronic liver disease: A meta-analysis of 24,147 subjects. Mol Genet Genomic Med. 2019;7:e824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 60. | Mahdessian H, Taxiarchis A, Popov S, Silveira A, Franco-Cereceda A, Hamsten A, Eriksson P, van't Hooft F. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111:8913-8918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 286] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 61. | Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 724] [Cited by in RCA: 930] [Article Influence: 84.5] [Reference Citation Analysis (0)] |
| 62. | Sliz E, Sebert S, Würtz P, Kangas AJ, Soininen P, Lehtimäki T, Kähönen M, Viikari J, Männikkö M, Ala-Korpela M, Raitakari OT, Kettunen J. NAFLD risk alleles in PNPLA3, TM6SF2, GCKR and LYPLAL1 show divergent metabolic effects. Hum Mol Genet. 2018;27:2214-2223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 63. | Carlsson B, Lindén D, Brolén G, Liljeblad M, Bjursell M, Romeo S, Loomba R. Review article: the emerging role of genetics in precision medicine for patients with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2020;51:1305-1320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 64. | Liu YL, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JB, Allison ME, Alexander GJ, Piguet AC, Anty R, Donaldson P, Aithal GP, Francque S, Van Gaal L, Clement K, Ratziu V, Dufour JF, Day CP, Daly AK, Anstee QM. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 362] [Cited by in RCA: 460] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 65. | Dongiovanni P, Petta S, Maglio C, Fracanzani AL, Pipitone R, Mozzi E, Motta BM, Kaminska D, Rametta R, Grimaudo S, Pelusi S, Montalcini T, Alisi A, Maggioni M, Kärjä V, Borén J, Käkelä P, Di Marco V, Xing C, Nobili V, Dallapiccola B, Craxi A, Pihlajamäki J, Fargion S, Sjöström L, Carlsson LM, Romeo S, Valenti L. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 418] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 66. | Holmen OL, Zhang H, Fan Y, Hovelson DH, Schmidt EM, Zhou W, Guo Y, Zhang J, Langhammer A, Løchen ML, Ganesh SK, Vatten L, Skorpen F, Dalen H, Pennathur S, Chen J, Platou C, Mathiesen EB, Wilsgaard T, Njølstad I, Boehnke M, Chen YE, Abecasis GR, Hveem K, Willer CJ. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46:345-351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 243] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 67. | Marzuillo P, Di Sessa A, Cirillo G, Umano GR, Pedullà M, La Manna A, Guarino S, Miraglia Del Giudice E. Transmembrane 6 superfamily member 2 167K allele improves renal function in children with obesity. Pediatr Res. 2020;88:300-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 68. | Su W, Mao Z, Liu Y, Zhang X, Zhang W, Gustafsson JA, Guan Y. Role of HSD17B13 in the liver physiology and pathophysiology. Mol Cell Endocrinol. 2019;489:119-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 69. | Dong XC. A closer look at the mysterious HSD17B13. J Lipid Res. 2020;61:1361-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 70. | Su W, Wang Y, Jia X, Wu W, Li L, Tian X, Li S, Wang C, Xu H, Cao J, Han Q, Xu S, Chen Y, Zhong Y, Zhang X, Liu P, Gustafsson JÅ, Guan Y. Comparative proteomic study reveals 17β-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc Natl Acad Sci U S A. 2014;111:11437-11442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 180] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 71. | Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, Liu Y, Kozlitina J, Stender S, Wood GC, Stepanchick AN, Still MD, McCarthy S, O'Dushlaine C, Packer JS, Balasubramanian S, Gosalia N, Esopi D, Kim SY, Mukherjee S, Lopez AE, Fuller ED, Penn J, Chu X, Luo JZ, Mirshahi UL, Carey DJ, Still CD, Feldman MD, Small A, Damrauer SM, Rader DJ, Zambrowicz B, Olson W, Murphy AJ, Borecki IB, Shuldiner AR, Reid JG, Overton JD, Yancopoulos GD, Hobbs HH, Cohen JC, Gottesman O, Teslovich TM, Baras A, Mirshahi T, Gromada J, Dewey FE. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med. 2018;378:1096-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 610] [Article Influence: 87.1] [Reference Citation Analysis (0)] |
| 72. | Ma Y, Belyaeva OV, Brown PM, Fujita K, Valles K, Karki S, de Boer YS, Koh C, Chen Y, Du X, Handelman SK, Chen V, Speliotes EK, Nestlerode C, Thomas E, Kleiner DE, Zmuda JM, Sanyal AJ; (for the Nonalcoholic Steatohepatitis Clinical Research Network), Kedishvili NY, Liang TJ, Rotman Y. 17-Beta Hydroxysteroid Dehydrogenase 13 Is a Hepatic Retinol Dehydrogenase Associated With Histological Features of Nonalcoholic Fatty Liver Disease. Hepatology. 2019;69:1504-1519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 224] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 73. | Lin YC, Wu CC, Ni YH. New Perspectives on Genetic Prediction for Pediatric Metabolic Associated Fatty Liver Disease. Front Pediatr. 2020;8:603654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 74. | Sookoian S, Arrese M, Pirola CJ. Genetics Meets Therapy? Hepatology. 2019;69:907-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 75. | Pirola CJ, Garaycoechea M, Flichman D, Arrese M, San Martino J, Gazzi C, Castaño GO, Sookoian S. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res. 2019;60:176-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 76. | Ma Y, Karki S, Brown PM, Lin DD, Podszun MC, Zhou W, Belyaeva OV, Kedishvili NY, Rotman Y. Characterization of essential domains in HSD17B13 for cellular localization and enzymatic activity. J Lipid Res. 2020;61:1400-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 77. | Ma Y, Brown PM, Lin DD, Ma J, Feng D, Belyaeva OV, Podszun MC, Roszik J, Allen JN, Umarova R, Kleiner DE, Kedishvili NY, Gavrilova O, Gao B, Rotman Y. 17-Beta Hydroxysteroid Dehydrogenase 13 Deficiency Does Not Protect Mice From Obesogenic Diet Injury. Hepatology. 2021;73:1701-1716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 78. | Gellert-Kristensen H, Nordestgaard BG, Tybjaerg-Hansen A, Stender S. High Risk of Fatty Liver Disease Amplifies the Alanine Transaminase-Lowering Effect of a HSD17B13 Variant. Hepatology. 2020;71:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 79. | Wang P, Wu CX, Li Y, Shen N. HSD17B13 rs72613567 protects against liver diseases and histological progression of nonalcoholic fatty liver disease: a systematic review and meta-analysis. Eur Rev Med Pharmacol Sci. 2020;24:8997-9007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 80. | Stickel F, Lutz P, Buch S, Nischalke HD, Silva I, Rausch V, Fischer J, Weiss KH, Gotthardt D, Rosendahl J, Marot A, Elamly M, Krawczyk M, Casper M, Lammert F, Buckley TWM, McQuillin A, Spengler U, Eyer F, Vogel A, Marhenke S, von Felden J, Wege H, Sharma R, Atkinson S, Franke A, Nehring S, Moser V, Schafmayer C, Spahr L, Lackner C, Stauber RE, Canbay A, Link A, Valenti L, Grove JI, Aithal GP, Marquardt JU, Fateen W, Zopf S, Dufour JF, Trebicka J, Datz C, Deltenre P, Mueller S, Berg T, Hampe J, Morgan MY. Genetic Variation in HSD17B13 Reduces the Risk of Developing Cirrhosis and Hepatocellular Carcinoma in Alcohol Misusers. Hepatology. 2020;72:88-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 81. | Yang J, Trépo E, Nahon P, Cao Q, Moreno C, Letouzé E, Imbeaud S, Bayard Q, Gustot T, Deviere J, Bioulac-Sage P, Calderaro J, Ganne-Carrié N, Laurent A, Blanc JF, Guyot E, Sutton A, Ziol M, Zucman-Rossi J, Nault JC. A 17-Beta-Hydroxysteroid Dehydrogenase 13 Variant Protects From Hepatocellular Carcinoma Development in Alcoholic Liver Disease. Hepatology. 2019;70:231-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 82. | Di Sessa A, Umano GR, Cirillo G, Passaro AP, Verde V, Cozzolino D, Guarino S, Marzuillo P, Miraglia Del Giudice E. Pediatric non-alcoholic fatty liver disease and kidney function: Effect of HSD17B13 variant. World J Gastroenterol. 2020;26:5474-5483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 83. | Kozlitina J, Stender S, Hobbs HH, Cohen JC. HSD17B13 and Chronic Liver Disease in Blacks and Hispanics. N Engl J Med. 2018;379:1876-1877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 84. | Stender S, Romeo S. HSD17B13 as a promising therapeutic target against chronic liver disease. Liver Int. 2020;40:756-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 85. | Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718-15723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4530] [Cited by in RCA: 4410] [Article Influence: 210.0] [Reference Citation Analysis (4)] |
| 86. | Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 872] [Cited by in RCA: 970] [Article Influence: 138.6] [Reference Citation Analysis (0)] |
| 87. | Copple BL, Li T. Pharmacology of bile acid receptors: Evolution of bile acids from simple detergents to complex signaling molecules. Pharmacol Res. 2016;104:9-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 179] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 88. | Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1341] [Cited by in RCA: 1399] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 89. | Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol. 2011;54:1263-1272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 327] [Cited by in RCA: 321] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 90. | Broeders EP, Nascimento EB, Havekes B, Brans B, Roumans KH, Tailleux A, Schaart G, Kouach M, Charton J, Deprez B, Bouvy ND, Mottaghy F, Staels B, van Marken Lichtenbelt WD, Schrauwen P. The Bile Acid Chenodeoxycholic Acid Increases Human Brown Adipose Tissue Activity. Cell Metab. 2015;22:418-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 338] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 91. | Slijepcevic D, van de Graaf SF. Bile Acid Uptake Transporters as Targets for Therapy. Dig Dis. 2017;35:251-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 92. | Svegliati-Baroni G, Patrício B, Lioci G, Macedo MP, Gastaldelli A. Gut-Pancreas-Liver Axis as a Target for Treatment of NAFLD/NASH. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 93. | Jiang X, Zheng J, Zhang S, Wang B, Wu C, Guo X. Advances in the Involvement of Gut Microbiota in Pathophysiology of NAFLD. Front Med (Lausanne). 2020;7:361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 94. | Arab JP, Karpen SJ, Dawson PA, Arrese M, Trauner M. Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology. 2017;65:350-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 457] [Article Influence: 57.1] [Reference Citation Analysis (1)] |
| 95. | Hernandez GV, Smith VA, Melnyk M, Burd MA, Sprayberry KA, Edwards MS, Peterson DG, Bennet DC, Fanter RK, Columbus DA, Steibel JP, Glanz H, Immoos C, Rice MS, Santiago-Rodriguez TM, Blank J, VanderKelen JJ, Kitts CL, Piccolo BD, La Frano MR, Burrin DG, Maj M, Manjarin R. Dysregulated FXR-FGF19 signaling and choline metabolism are associated with gut dysbiosis and hyperplasia in a novel pig model of pediatric NASH. Am J Physiol Gastrointest Liver Physiol. 2020;318:G582-G609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 96. | Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 555] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 97. | Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D, Furlanello C, Zandonà A, Paci P, Capuani G, Dallapiccola B, Miccheli A, Alisi A, Putignani L. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65:451-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 527] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 98. | Schwimmer JB, Johnson JS, Angeles JE, Behling C, Belt PH, Borecki I, Bross C, Durelle J, Goyal NP, Hamilton G, Holtz ML, Lavine JE, Mitreva M, Newton KP, Pan A, Simpson PM, Sirlin CB, Sodergren E, Tyagi R, Yates KP, Weinstock GM, Salzman NH. Microbiome Signatures Associated With Steatohepatitis and Moderate to Severe Fibrosis in Children With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2019;157:1109-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 99. | Stanislawski MA, Lozupone CA, Wagner BD, Eggesbø M, Sontag MK, Nusbacher NM, Martinez M, Dabelea D. Gut microbiota in adolescents and the association with fatty liver: the EPOCH study. Pediatr Res. 2018;84:219-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 100. | Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9796] [Cited by in RCA: 8752] [Article Influence: 460.6] [Reference Citation Analysis (1)] |
| 101. | Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, Jones MB, Sirlin CB, Schnabl B, Brinkac L, Schork N, Chen CH, Brenner DA, Biggs W, Yooseph S, Venter JC, Nelson KE. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2019;30:607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 102. | Michail S, Lin M, Frey MR, Fanter R, Paliy O, Hilbush B, Reo NV. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol. 2015;91:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 236] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 103. | Zhao Y, Zhou J, Liu J, Wang Z, Chen M, Zhou S. Metagenome of Gut Microbiota of Children With Nonalcoholic Fatty Liver Disease. Front Pediatr. 2019;7:518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 104. | Compare D, Coccoli P, Rocco A, Nardone OM, De Maria S, Cartenì M, Nardone G. Gut--liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2012;22:471-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 323] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 105. | He X, Ji G, Jia W, Li H. Gut Microbiota and Nonalcoholic Fatty Liver Disease: Insights on Mechanism and Application of Metabolomics. Int J Mol Sci. 2016;17:300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 106. | Seki E, Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol. 2012;590:447-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 339] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 107. | Iruzubieta P, Medina JM, Fernández-López R, Crespo J, de la Cruz F. A Role for Gut Microbiome Fermentative Pathways in Fatty Liver Disease Progression. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 108. | Zhang X, Asllanaj E, Amiri M, Portilla-Fernandez E, Bramer WM, Nano J, Voortman T, Pan Q, Ghanbari M. Deciphering the role of epigenetic modifications in fatty liver disease: A systematic review. Eur J Clin Invest. 2021;51:e13479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 109. | Pirola CJ, Gianotti TF, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castaño GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 280] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 110. | de Mello VD, Matte A, Perfilyev A, Männistö V, Rönn T, Nilsson E, Käkelä P, Ling C, Pihlajamäki J. Human liver epigenetic alterations in non-alcoholic steatohepatitis are related to insulin action. Epigenetics. 2017;12:287-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 111. | Kitamoto T, Kitamoto A, Ogawa Y, Honda Y, Imajo K, Saito S, Yoneda M, Nakamura T, Nakajima A, Hotta K. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes encoding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J Hepatol. 2015;63:494-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 112. | Zeybel M, Hardy T, Robinson SM, Fox C, Anstee QM, Ness T, Masson S, Mathers JC, French J, White S, Mann J. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin Epigenetics. 2015;7:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (1)] |
| 113. | Sookoian S, Rosselli MS, Gemma C, Burgueño AL, Fernández Gianotti T, Castaño GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology. 2010;52:1992-2000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 114. | Ma J, Nano J, Ding J, Zheng Y, Hennein R, Liu C, Speliotes EK, Huan T, Song C, Mendelson MM, Joehanes R, Long MT, Liang L, Smith JA, Reynolds LM, Ghanbari M, Muka T, van Meurs JBJ, Alferink LJM, Franco OH, Dehghan A, Ratliff S, Zhao W, Bielak L, Kardia SLR, Peyser PA, Ning H, VanWagner LB, Lloyd-Jones DM, Carr JJ, Greenland P, Lichtenstein AH, Hu FB, Liu Y, Hou L, Darwish Murad S, Levy D. A Peripheral Blood DNA Methylation Signature of Hepatic Fat Reveals a Potential Causal Pathway for Nonalcoholic Fatty Liver Disease. Diabetes. 2019;68:1073-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 115. | Nano J, Ghanbari M, Wang W, de Vries PS, Dhana K, Muka T, Uitterlinden AG, van Meurs JBJ, Hofman A; BIOS consortium, Franco OH, Pan Q, Murad SD, Dehghan A. Epigenome-Wide Association Study Identifies Methylation Sites Associated With Liver Enzymes and Hepatic Steatosis. Gastroenterology. 2017;153:1096-1106.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 116. | Mwinyi J, Boström AE, Pisanu C, Murphy SK, Erhart W, Schafmayer C, Hampe J, Moylan C, Schiöth HB. NAFLD is associated with methylation shifts with relevance for the expression of genes involved in lipoprotein particle composition. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:314-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 117. | Geurtsen ML, Jaddoe VWV, Salas LA, Santos S, Felix JF. Newborn and childhood differential DNA methylation and liver fat in school-age children. Clin Epigenetics. 2019;12:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 118. | Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, Hamajima N, Hashimoto S. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. 2013;424:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 257] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 119. | Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810-1820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 583] [Cited by in RCA: 551] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 120. | Pirola CJ, Fernández Gianotti T, Castaño GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, Mirshahi F, Sanyal AJ, Sookoian S. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 444] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 121. | Pirola CJ, Sookoian S. Multiomics biomarkers for the prediction of nonalcoholic fatty liver disease severity. World J Gastroenterol. 2018;24:1601-1615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 56] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 122. | Prats-Puig A, Ortega FJ, Mercader JM, Moreno-Navarrete JM, Moreno M, Bonet N, Ricart W, López-Bermejo A, Fernández-Real JM. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab. 2013;98:E1655-E1660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 165] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 123. | Can U, Buyukinan M, Yerlikaya FH. The investigation of circulating microRNAs associated with lipid metabolism in childhood obesity. Pediatr Obes. 2016;11:228-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 124. | Cui X, You L, Zhu L, Wang X, Zhou Y, Li Y, Wen J, Xia Y, Ji C, Guo X. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism. 2018;78:95-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 125. | Iacomino G, Russo P, Marena P, Lauria F, Venezia A, Ahrens W, De Henauw S, De Luca P, Foraita R, Günther K, Lissner L, Molnár D, Moreno LA, Tornaritis M, Veidebaum T, Siani A. Circulating microRNAs are associated with early childhood obesity: results of the I.Family Study. Genes Nutr. 2019;14:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 126. | Sheldon RD, Kanosky KM, Wells KD, Miles L, Perfield JW 2nd, Xanthakos S, Inge TH, Rector RS. Transcriptomic differences in intra-abdominal adipose tissue in extremely obese adolescents with different stages of NAFLD. Physiol Genomics. 2016;48:897-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 127. | Baselli GA, Dongiovanni P, Rametta R, Meroni M, Pelusi S, Maggioni M, Badiali S, Pingitore P, Maurotti S, Montalcini T, Taliento AE, Prati D, Rossi G, Fracanzani AL, Mancina RM, Romeo S, Valenti L. Liver transcriptomics highlights interleukin-32 as novel NAFLD-related cytokine and candidate biomarker. Gut. 2020;69:1855-1866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 128. | Perakakis N, Stefanakis K, Mantzoros CS. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metabolism. 2020;111S:154320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (1)] |
| 129. | Cusi K, Chang Z, Harrison S, Lomonaco R, Bril F, Orsak B, Ortiz-Lopez C, Hecht J, Feldstein AE, Webb A, Louden C, Goros M, Tio F. Limited value of plasma cytokeratin-18 as a biomarker for NASH and fibrosis in patients with non-alcoholic fatty liver disease. J Hepatol. 2014;60:167-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 184] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 130. | Sookoian S, Castaño G, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A diagnostic model to differentiate simple steatosis from nonalcoholic steatohepatitis based on the likelihood ratio form of Bayes theorem. Clin Biochem. 2009;42:624-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 131. | Rodriguez-Suarez E, Mato JM, Elortza F. Proteomics analysis of human nonalcoholic fatty liver. Methods Mol Biol. 2012;909:241-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 132. | Younossi ZM, Baranova A, Ziegler K, Del Giacco L, Schlauch K, Born TL, Elariny H, Gorreta F, VanMeter A, Younoszai A, Ong JP, Goodman Z, Chandhoke V. A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology. 2005;42:665-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 165] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 133. | Ulukaya E, Yilmaz Y, Moshkovskii S, Karpova M, Pyatnitskiy M, Atug O, Dolar E. Proteomic analysis of serum in patients with non-alcoholic steatohepatitis using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Scand J Gastroenterol. 2009;44:1471-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 134. | Małecki P, Tracz J, Łuczak M, Figlerowicz M, Mazur-Melewska K, Służewski W, Mania A. Serum proteome assessment in nonalcoholic fatty liver disease in children: a preliminary study. Expert Rev Proteomics. 2020;17:623-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 135. | Bălănescu A, Stan I, Codreanu I, Comănici V, Bălănescu E, Bălănescu P. Circulating Hsp90 Isoform Levels in Overweight and Obese Children and the Relation to Nonalcoholic Fatty Liver Disease: Results from a Cross-Sectional Study. Dis Markers. 2019;2019:9560247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 136. | Blomme B, Fitzpatrick E, Quaglia A, De Bruyne R, Dhawan A, Van Vlierberghe H. Serum protein N-glycosylation in paediatric non-alcoholic fatty liver disease. Pediatr Obes. 2012;7:165-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 137. | Li J, Hsu HC, Mountz JD, Allen JG. Unmasking Fucosylation: from Cell Adhesion to Immune System Regulation and Diseases. Cell Chem Biol. 2018;25:499-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 160] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 138. | Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol. 2019;15:346-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1362] [Article Influence: 227.0] [Reference Citation Analysis (0)] |
| 139. | Hollie NI, Cash JG, Matlib MA, Wortman M, Basford JE, Abplanalp W, Hui DY. Micromolar changes in lysophosphatidylcholine concentration cause minor effects on mitochondrial permeability but major alterations in function. Biochim Biophys Acta. 2014;1841:888-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 140. | Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3:321-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 516] [Cited by in RCA: 569] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 141. | Zhou Y, Orešič M, Leivonen M, Gopalacharyulu P, Hyysalo J, Arola J, Verrijken A, Francque S, Van Gaal L, Hyötyläinen T, Yki-Järvinen H. Noninvasive Detection of Nonalcoholic Steatohepatitis Using Clinical Markers and Circulating Levels of Lipids and Metabolites. Clin Gastroenterol Hepatol. 2016;14:1463-1472.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 120] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 142. | Holland WL, Summers SA. Sphingolipids, insulin resistance, and metabolic disease: new insights from in vivo manipulation of sphingolipid metabolism. Endocr Rev. 2008;29:381-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 456] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 143. | Marí M, Colell A, Morales A, Caballero F, Moles A, Fernández A, Terrones O, Basañez G, Antonsson B, García-Ruiz C, Fernández-Checa JC. Mechanism of mitochondrial glutathione-dependent hepatocellular susceptibility to TNF despite NF-kappaB activation. Gastroenterology. 2008;134:1507-1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 144. | Moles A, Tarrats N, Morales A, Domínguez M, Bataller R, Caballería J, García-Ruiz C, Fernández-Checa JC, Marí M. Acidic sphingomyelinase controls hepatic stellate cell activation and in vivo liver fibrogenesis. Am J Pathol. 2010;177:1214-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 145. | Marzuillo P, Grandone A, Perrone L, Miraglia Del Giudice E. Controversy in the diagnosis of pediatric non-alcoholic fatty liver disease. World J Gastroenterol. 2015;21:6444-6450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 146. | Kang SH, Cho Y, Jeong SW, Kim SU, Lee JW; Korean NAFLD Study Group. From nonalcoholic fatty liver disease to metabolic-associated fatty liver disease: Big wave or ripple? Clin Mol Hepatol. 2021;27:257-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 147. | Di Sessa A, Guarino S, Umano GR, Arenella M, Alfiero S, Quaranta G, Miraglia Del Giudice E, Marzuillo P. MAFLD in Obese Children: A Challenging Definition. Children (Basel). 2021;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |