Published online Oct 10, 2014. doi: 10.5306/wjco.v5.i4.730
Revised: June 7, 2014
Accepted: June 18, 2014
Published online: October 10, 2014
Processing time: 208 Days and 4.4 Hours
Cancer initiation and development engage extremely complicated pathological processes which involve alterations of a large number of cell signaling cascades and functional networks in temporal and spatial orders. During last decades, microRNAs (miRNAs), a class of non-coding RNAs, have emerged as critical players in cancer pathogenesis and progression by modulating many pathological aspects related to tumor development, growth, metastasis, and drug resistance. The major function of miRNAs is to post-transcriptionally regulate gene expression depending on recognition of complementary sequence residing in target mRNAs. Commonly, a particular miRNA recognition sequence could be found in a number of genes, which allows a single miRNA to regulate multiple functionally connected genes simultaneously and/or chronologically. Furthermore, a single gene can be targeted and regulated by multiple miRNAs. However, previous studies have demonstrated that miRNA functions are highly context-dependent, which leads to distinct pathological outcomes in different types of cancer as well as at different stages by alteration of the same miRNA. Here we summarize recent progress in studies on miRNA function in cancer initiation, metastasis and therapeutic response, focusing on breast cancer. The varying functions of miRNAs and potential application of using miRNAs as biomarkers as well as therapeutic approaches are further discussed in the context of different cancers.
Core tip: MicroRNAs (miRNAs) have been shown to play critical roles in cancer pathogenesis and progression by modulating tumor initiation, growth, metastasis, and therapeutic resistance. In this review, we discuss the recent progress in understanding miRNA function in cancer development and therapeutic response, especially in breast cancer, as well as the potential application of using miRNA signatures as biomarkers for predicting therapeutic response and for personalizing anti-cancer regimens.
- Citation: Xue J, Niu J, Wu J, Wu ZH. MicroRNAs in cancer therapeutic response: Friend and foe. World J Clin Oncol 2014; 5(4): 730-743
- URL: https://www.wjgnet.com/2218-4333/full/v5/i4/730.htm
- DOI: https://dx.doi.org/10.5306/wjco.v5.i4.730
In the human genome, protein coding genes (around 20000) only represent approximately 1.5% of entire DNA sequences[1]. Since little discernible function was known about the majority of non-coding sequences, they were called “junk DNA”. However, recent data provided by ENCODE project and other research progress have revealed that a large portion of “junk DNA” may pertain to important biological functions, such as host regulatory DNA sequences, long interspersed elements, short interspersed elements and non-coding RNA genes. Non-coding RNA (ncRNA) comprises several classes of functional RNA transcripts, including ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small neucleolar RNA (snoRNA), long non-coding RNA (lincRNA), small interfering RNA (siRNA) and microRNA (miRNA), which have been shown to exert critical roles such as regulating transcription, stability and translation of protein-coding genes[2,3]. The most studied ncRNAs to date are miRNAs, owing to their crucial functions in control of varying biological and pathological processes, such as development, cell proliferation, differentiation, programmed cell death and stress response. Most miRNAs are highly conserved across different species, while exhibiting high specificity to tissue/cell types and developmental stages. The involvement of miRNAs in cancer initiation and progression was first reported in chronic lymphocytic leukemia[4]. Later studies further revealed that miRNAs may serve as either tumor suppressors or oncogenes (also known as oncomiRs) in a variety of cancers, depending on which genes or pathways were regulated/dysregulated by particular miRNA in a specific cancer type[5,6]. Meanwhile, an individual gene could be regulated by multiple miRNAs, which further underscored that the potential pathological impact of specific miRNA or miRNA cluster may vary in different types of cancer[5,7,8]. High throughput analyses using expression microarrays or next-generation sequencing have revealed that miRNAs are dysregulated in most types of human cancer[9,10]. Moreover, the aberrant expression signatures of miRNAs have been suggested to have diagnostic, prognostic, predictive and therapeutic values[11-14].
As many outstanding reviews have comprehensively described the current understanding of the pathophysiological functions of miRNAs and their involvement in cancer initiation, progression and metastasis[3,5,6,14,15] , here we only summarize the recent progress in these research areas and focus on miRNA function in cancer therapeutic response, especially in breast cancer. The potential application of using miRNA signatures as predictive biomarkers for cancer therapeutic response and how these insights can be used for the development of novel anti-cancer regimens targeting miRNAs are further discussed.
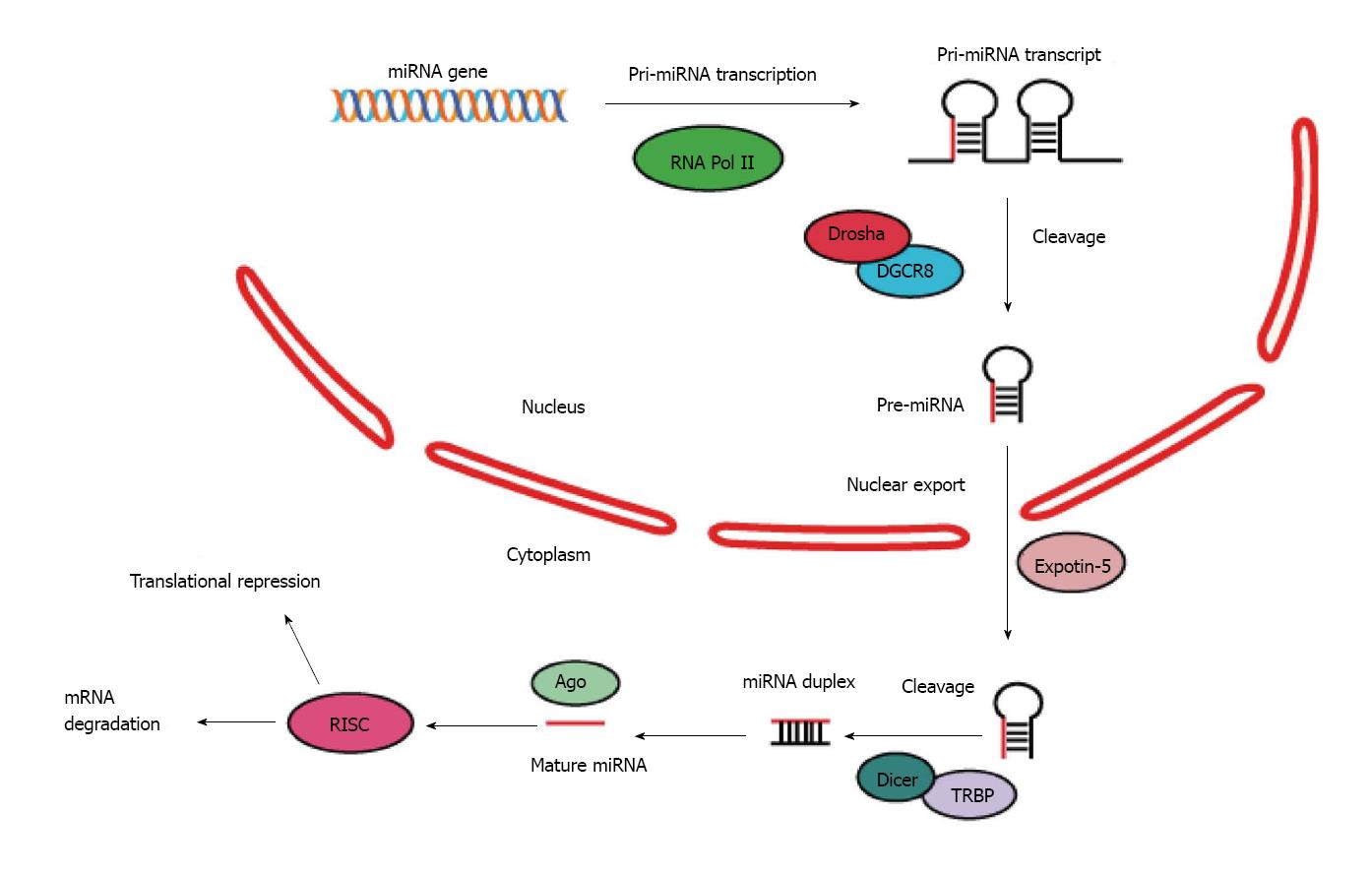
MiRNAs are a family of single-strand RNAs ranging from 19 to 24 nucleotides, which are predominantly transcribed from the genome as primary miRNAs by RNA polymerase II[16,17]. The primary transcripts then undergo a multi-step process to generate precursor and mature form of miRNAs. The initial transcription of miRNA genes yields long, capped and polyadenylated primary miRNA transcripts, varying from several hundreds to several thousands of nucleotides. These primary transcripts are subsequently processed by a nuclear microprocessor complex consisting of ribonuclease (RNase) III Drosha and DGCR8 (DiGeorge syndrome critical region 8, also known as Pasha), yielding precursor miRNAs (Pre-miRs)[18]. Besides the core components of the Drosha microprocessor, several additional proteins, including the DEAD-box helicase proteins p68/DDX17, NF90 and NF45, have been shown to interact with Drosha and facilitate the processing of nearly one-third of pri-miRNAs. Alternatively, pre-miRNAs can also be generated by RNA splicing when the miRNA sequence resides in intron of a protein-coding gene and is co-transcribed with the mRNA of the host gene. Precursor miRNAs form a complex with exportin 5 (XPO5) and Ran-GTP, which is then transported into the cytoplasm. The hairpin structure of pre-miRNAs can be recognized by cytoplasmic type III RNase Dicer which further cleaves pre-miRNAs into double-stranded miRNAs. Then, the miRNA double strands are separated, and the mature miRNA can be loaded into the RNA-induced silencing complex (RISC) along with its target mRNAs, leading to posttranscriptional gene silencing[19-21] (Figure 1).
In general, miRNAs post-transcriptionally decrease gene expression by inhibiting ribosome-dependent translation and/or destabilizing mRNAs of target genes. Typically, mature miRNAs recognize their target genes through sequence-complementarity within the 3ʹ-untranslated region (3′-UTR) of mRNA to the miRNA seed region (nt. 2-7). However, several lines of evidence indicated that miRNAs can also bind to 5′-UTRs or open reading frames (ORFs) of a target mRNA and repress its expression[22,23]. By comparison, ORF targeting appears less frequent and less effective than 3′-UTR targeting but still much more frequent than 5′-UTR targeting based on the computational and experimental genome-wide analyses.
The overall function of miRNAs is believed to negatively regulate gene expression. However, recent evidence suggested that certain miRNAs, such as miR-369, can enhance the translation via binding to the AU-rich elements of the target gene mRNA[24]. Moreover, miR-328 was found to indirectly increase the protein output by promoting release of translation-inhibiting hnRNP E2 from the target gene mRNA, thereby de-repressing translation[25]. In addition, miR-373 was shown to induce expression of genes with complementary promoter sequences, while miR-10a can bind to the 5′-UTR of ribosomal protein gene transcripts and augments their translation[26]. Nevertheless, in contrast to the well-studied consensus mechanism of miRNA-dependent gene silencing, much remains to be learned about how miRNA up-regulate gene expression.
In addition to intracellular functions, abundant miRNAs are detected in the serum and other biological fluids. Circulating miRNAs are released from source cells into extracellular milieu and can be taken up by recipient cells via endocytosis. This process may be mediated by as yet unidentified miRNA-binding membrane receptors on the recipient cells[27]. The circulating miRNAs can act on neighboring cells as well as at remote sites in a paracrine and endocrine fashion, suggesting their roles in mediating both short- and long-range intercellular communication[28,29]. The uptaken miRNA could regulate target gene expression via RISC in recipient cells, or induce pro-metastatic inflammatory response by binding to intracellular Toll-like receptors (TLRs)[28].
MiRNAs have been shown to play pivotal roles in cancer initiation and progression. It was found that miRNA genes were frequently located within cancer-associated genomic regions, either amplified or deleted, and miRNAs can act either as oncogenes or tumor suppressors, depending on regulated genes and cancer types[30]. Furthermore, a particular miRNA can exploit both tumor-suppressive and oncogenic functions depending on the cellular context of its target genes in different cancers.
The first evidence that miRNAs may be involved in human cancer pathogenesis was derived from a study aiming to identify tumor suppressors within the 13q14 region which is frequently deleted in chronic lymphocytic leukemia (CLL)[4]. It was found that, in 69% of CLL patients analyzed, polycistronic miR-15a and miR-16-1 were deleted or downregulated by epigenetic inhibition. Since downregulation of miR-15a/16 is observed in the majority of indolent CLLs, loss of miR-15a and miR-16-1 is likely among the initiating events in the pathogenesis of this disease[4]. Subsequent studies revealed additional tumor suppressor miRNAs including the let-7 family and miR-34 family[15]. These tumor suppressor miRNAs were shown to target varying protein-coding oncogenes for translation inhibition or mRNA decay[5]. For example, miR-15a and miR-16-1 inhibit the expression of anti-apoptotic gene BCL2 (B cell lymphoma 2) and MCL1 (myeloid cell leukemia 1), both of which are well-established oncogenes in hematopoietic cancers.
The tumor suppressive effects of let-7 have been attributed to inhibition of a list of oncogenes, such as KRAS, CCND1, CDK6, HOXA9, TLR4 and MYC[31-33]. Let-7 was also found to negatively regulate self-renewal and tumorigenicity of breast cancer initiating cells through repressing cell proliferation and growth as well as inducing cell cycle exit and terminal differentiation. It has been proposed that cancer stem-like cells or tumor initiating cells may play important roles in breast cancer incidence. Consistently, ectopic expression of miRNAs from the let-7 family was shown to suppress mammary tumor development in mouse models[33]. Furthermore, by comparing miRNAs in normal stem cells from the mammary gland and cancer stem-like cells from DCIS tumors, miR-140 was found to be significantly decreased in cancer stem-like cells but not in normal stem cells, indicating that miR-140 may play a critical role in preventing malignant transformation of mammary stem cells. The downregulation of miR-140 may be associated with DNA hypermethylation in breast cancer cells. Further investigation found that expression of stem cell marker SOX9 and ALDH1, which are significantly upregulated in DCIS stem-like cells, are repressed by miR-140. Accordingly, overexpression of miR-140 decreased expression of SOX9 and ALDH1 in ERα-/basal-like DCIS mammospheres, and reduced tumor growth in vivo[34]. These studies indicated that tumor suppressive miRNAs may antagonize tumor formation by reducing cancer initiating cell population.
Tumor suppressive miRNAs may also work downstream of coding tumor suppressor genes, which may directly regulate miRNA expression. For instance, tumor suppressive miR-205 can be directly transactivated by the tumor suppressor p53[35]. Moreover, a study evaluating miR-205 expression in a panel of highly aggressive triple-negative breast cancer cell lines demonstrated that miR-205 is substantially downregulated compared with normal cells. Accordingly, reconstitution of miR-205 expression strongly reduced cancer cell proliferation and tumorigenic potential in cell culture models and inhibited tumor growth in animal models. The miR-205-dependent tumor suppression is attributed, at least in part, to repressing E2F1 and LAMC1, resulting in blockade of cell cycle progression as well as reduced cell adhesion and migration[35].
Similarly, oncogenes may also promote tumor growth by inhibiting tumor suppressor miRNAs. It was found that estrogen downregulated miR-34b in ERα-positive/p53 wild-type breast cancer cells, as well as in ovarian and endometrial cells, but not in ERα-negative or p53 mutant breast cancer cells[36]. The negative association between ERα and miR-34b expression levels has also been validated in ERα-positive breast cancer patients. miR-34b was found to inhibit expression of cyclin D1 and Jagged-1 (JAG1) in breast cancer cells. It may also mediate tumor suppressor p53 signaling by repressing oncogenes such as cyclin-dependent kinase 4 (CDK4), MYC and MET[37]. In addition, overexpression of miR-34b could inhibit tumor growth in an orthotopic xenograft mouse model transplanted with ERα-positive breast cancer cells, but not with ERα-negative or p53 mutant breast cancer cells[36]. These results suggest that downregulation of miR-34b may play an important role in ERα-driven breast cancer tumorigenesis.
In contrast to the anti-tumorigenic functions described above, oncogenic miRNAs, such as miR-21, miR-17-92 cluster and miR-155, exhibit strong cancer-promoting activity in various human malignancies. As a well-characterized oncomiR, miR-21 was found to be upregulated in almost all cancer types[38]. In vivo studies revealed that cancer cells showed strong addiction to miR-21, whereas inactivation of miR-21 induced complete tumor regression in a pre-B-cell lymphoma model[39]. In breast cancer, repression of the tumor suppressors such as PTEN, PDCD4 and TPM1 by miR-21 was demonstrated to mediate cancer cell survival and proliferation. A strong correlation between the high expression level of miR-21 and advanced clinical stage, lymph node metastasis and poor prognosis has also been found in breast cancer patients[40-42]. It was found that the increased ratio between miR-21 and PDCD4 level in bone marrow of breast cancer patients showed a significant correlation with shortened disease-free survival (DFS) and overall survival (OS)[43]. Meanwhile, the expression of miR-17-92 cluster, another well-studied oncomiR located within 13q22, was found to be frequently increased in a variety of cancers such as breast cancer, lung cancer, colon cancer and lymphoma, through gene amplification or transcriptional activation[38]. Interestingly, this miRNA cluster was found to be a direct downstream target of the MYC oncogene, and the increased expression of miR-17-92 could attenuate apoptosis induced by MYC amplification, thereby promoting B cell lymphoma development in a mouse model[44,45]. These findings suggest that upregulating miR-17-92 may contribute significantly to the oncogenicity of MYC. In contrast, blockade of miR-17 decreased the breast cancer cell invasion/ migration in vitro and metastasis in vivo[46]. Moreover, a series of studies have shown that miR-17-92 cluster acts as an oncogenic gene to promote breast cancer cell invasion and migration through diverse signaling cascades such as Wnt/β-catenin pathway and miR-17-92/ZBTB4/Sp axis[46-51]. Likewise, miR-27a was also found to play oncogenic roles though ZBTB/Sp cascade in breast cancer cells. Upregulation of miR-27a has been shown to repress ZBTB10 and Myt-1 expression in breast cancer cells, which promoted cancer cell proliferation[52]. Inhibiting miR-27a by miR-27a antagmir or anti-cancer drugs, such as betulinic acid, resulted in increased ZBTB10 and decreased Sp family of transcription factors expression in breast cancer cells, thereby leading to consequent growth suppression[52-54].
Recent studies also demonstrated that differentially expressed miRNAs correlated to specific pathological features in breast cancer, such as tumor grade, disease stage, proliferation index and vascular invasion[55]. Moreover, a panel of differentially expressed miRNAs have been identified between ER+ and ER- breast cancer patients. In this panel, miR-191 and miR-26 are the most significantly upregulated miRNAs, while miR-206 was at the opposite end of spectrum[54]. In another study, miR-7, miR-128a, miR-210, and miR-516-3p were identified to be significantly associated with ER+ luminal signature and breast cancer aggressiveness[56]. All these studies support the potential application of miRNA profiling in cancer diagnosis and subtype characterization.
It has been demonstrated that miRNAs also play critical roles in tumor metastasis by regulating the migration and invasion of cancer cells[57,58]. A pair of basal-like subtype breast cancer-specific miRNAs, miR-221 and miR-222, could promote epithelial-to-mesenchymal transition (EMT), a phenotype strongly associated with cancer invasion and metastasis, by repressing epithelial genes while enhancing mesenchymal gene expression[59-61]. The transcription of miR-221/222 can be directly upregulated by FOSL1 (also known as Fra-1), a RAS-activated FOS family transcription factor, and the level of miR-221/222 decreased when MEK (mitogen-activated or extracellular signal-regulated protein kinase) was inhibited, which suggest that miR-221/222 cluster is a transcription target of the oncogenic RAS-RAF-MEK signaling pathway. The miR-221/222-mediated decrease of the mesenchymal signature gene E-cadherin is dependent on repression of trichorhinophalangeal syndrome type 1 (TRPS1), which is a miR-221/222 target and a transcriptional repressor belonging to GATA family. Decrease of TRPS1 could enhance the transcription of zinc finger E-box-binding homeobox 2 (ZEB2), thereby inhibiting E-Cadherin expression while up-regulating Vimentin[60]. Besides, miR-9 was found to directly repress E-Cadherin expression in breast cancer cells, resulting EMT transition as well as activation of β-Catenin signaling[62]. The activated β-Catenin promotes VEGF (vascular endothelial growth factor) transcription, which in turn enhances angiogenesis during the secondary tumor formation. Therefore, these data strongly support the hypothesis that miRNAs may contribute to the aggressiveness of breast cancer by promoting EMT.
In contrast, miR-126 decreases the ability of breast cancer cells to remodel the metastatic niche by recruiting endothelial cells from the tumor microenvironment, leading to reduced metastatic colonization[63]. Meanwhile, miR-335 was found to target the stemness transcription factor SOX4 and extracellular matrix protein tenascin C, resulting in suppression of breast cancer cell metastasis and migration. Consistently, decrease of miR-335 was found in the majority of primary tumors from breast cancer patients who eventually relapsed, which was also associated with poor DMFS[64]. Interestingly, it was found that treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and selective AHR modulator 6-methyl-1,3,-trichlorodibenzofuran (MCDF) could induce miR-335 expression and SOX4 downregulation in several breast cancer cell lines, which inhibited cancer cell growth and lung metastasis in vivo[65,66]. In addition, it was found that significant decrease of miR-34a/c was associated with breast cancer metastasis and lymph node invasion. Overexpression of miR-34a/c leads to decreased breast cancer cell motility and invasiveness as well as reduced distal lung metastasis in animal models. Additional studies found that Fra-1 mRNA and protein levels were decreased by miR-34a/c upregulation, indicating that FOSL1 is a downstream target of miR-34a/c. Furthermore, significant decrease of miR-34a in breast cancer metastases inversely correlates with Fra-1 expression, confirming the critical role of miR-34a-Fra-1 axis in regulating metastasis[67]. As we discussed above, FOSL1/Fra-1-dependent upregulation of miR-221/222 has been linked with aggressiveness of breast cancer[60], and miR-221/222 may serve as indirect downstream target of miR-34a/c for its anti-metastatic activity. The similar crosstalk between miRNAs and their critical roles in cancer progression have been further supported by other studies. A recent study showed that miR-22 may suppress expression of anti-metastatic miR-200, which increased the population of breast cancer stem-like cells and promote metastasis in mice. It was found that miR-22 directly targets the TET (Ten eleven translocation) family of methylcytosine dioxygenases, which promotes DNA demethylation of the miR-200 promoter and its transcriptional activation. The negative correlation between miR-22 and TET-miR-200 axis was also confirmed in metastatic breast cancer patients, supporting the clinical relevance of miRNA crosstalk in breast cancer progression[68].
Some miRNAs can promote oncogenesis in one cancer type while suppressing tumor development in the other. For instance, although the miR-221 has critical roles in antagonizing breast cancer metastasis and aggressive behaviors, overexpression of miR-221 in liver cancer promoted tumor initiation by inhibiting the expression of the tumor suppressor phosphatase and tensin homolog (PTEN)[69]. On the contrary, miR-221 acts again as a tumor suppressor in erythroblastic leukemia by reducing the expression of the KIT oncogene[70]. The complex interactions between miRNAs and protein-coding genes in various cancers indicate that therapies targeting both miRNA and protein-coding genes may be required for effectively controlling cancer progression. However, when miRNAs are considered as emerging therapeutic targets, the selection of miRNA may be highly cancer type- and even stage-specific.
Therapeutic resistance is the major obstacle of effective cancer treatment, and plays paramount roles in cancer relapse and cancer-related deaths. Previous studies have demonstrated that drug resistance of cancer cells can be mediated by a variety of mechanisms including the removal or detoxification of the drug, upregulation of anti-apoptotic processes, or alteration of drug transporters such that the therapeutic agent cannot gain entry into the target cells or is immediately exported. Recently, dysregulation of miRNAs was found to not only affect cellular processes involved in carcinogenesis, but also have a direct impact on the cancer therapeutic response. A number of oncogenic miRNAs (e.g., miR-9,-155, -21) have been shown to induce chemoresistance in vitro by modulating expression of key resistance-associated genes[5,71-73]. In breast cancer, miRNAs have been implicated to affect the response to different types of treatments, including anti-endocrine therapies, targeted therapies, chemotherapy, and radiotherapy, via treatment-specific or shared mechanisms.
The importance of ATP-binding cassette (ABC) transporter proteins, including multidrug resistance protein 1 (MDR1/P-glycoprotein/ABCB1), breast cancer resistance protein (BRCP/ABCG2) and multidrug resistance-associated proteins 1/2 (MRP1/ABCC1, MRP2/ABCC2), in the development of chemoresistance has been well-established[74]. Recently, miRNAs were reported as key regulators of drug transporter gene expression in cancer cells. For example, miR-328 was shown to negatively regulate BRCP by directly targeting 3′-UTR of ABCG2 and consequently influence drug disposition in breast cancer cells. miR-328-directed downregulation of ABCG2 expression significantly improved response to the chemotherapeutic agent mitoxantrone in a cell culture model[75]. Besides, several other miRNAs have been found to target MDR1, including miR-451[76], miR-7, miR-345[77], and miR-326[78], all of which increased cancer cell sensitivity to numerous chemotherapeutic agents. In addition, miR-128-depedent repression of BMI1 and ABCC5 (MRP5) has been implicated in the improved response to doxorubicin treatment, both in vitro and in breast cancer patients[79]. Reduction of miR-128 leads to upregulation of Bmi-1 and ABCC5 in breast tumor initiating cells, which contributes to chemotherapeutic resistance in breast cancer. Ectopic expression of miR-128 sensitizes breast tumor initiating cells to the pro-apoptotic and DNA-damaging effects of doxorubicin, indicating the potential therapeutic value of miR-128 in reducing drug resistance. Moreover, miRNAs may indirectly repress drug resistance gene expression by repressing their upstream activators. It was reported that miR-137 was significantly decreased in cancer cells resistant to doxorubicin. Further investigation revealed that miR-137 can directly repress the expression of constitutive androstane receptor, a nuclear receptor promoting MDR1 gene transcription, thereby downregulating MDR1 expression[80]. In contrast, miR-21 may upregulate MDR1 expression through repressing PDCD4, a miR-21 target gene, leading to reduced apoptosis and chemotherapeutic resistance[81]. The increase of MDR1 may be associated with decreased interaction between PDCD4 and eIF4A, resulting in enhanced protein translation.
It has been recognized that one miRNA could simultaneously target multiple genes sharing the same miRNA-recognition sequence, and these genes could be involved in a specific signaling network and play additive or synergistic roles in regulating cellular processes, such as apoptosis, proliferation or cell survival processes. For instance, chemotherapeutic drugs could upregulate miR-21 expression in breast cancer cells, which in turn inhibits chemodrug-induced apoptosis and promotes therapeutic resistance. Both PTEN and PDCD4 were identified as miR-21 target genes in this experimental setting, whose downregulation could promote cancer cell survival and proliferation[82]. Moreover, miR-34a was found to regulate human breast cancer cell response to docetaxel through targeting BCL-2 and CCND1[83]. Furthermore, miR-125b was found to inhibit pro-apoptotic BCL-2 antagonist killer 1(BAK1) expression. Upregulation of miR-125b in breast cancer cells could markedly inhibit taxol-induced cytotoxicity and apoptosis, and consequently increase the resistance to taxol[84]. In parallel, miR-125b directly targets the E2F3 gene, which regulates cell proliferation and apoptosis[85]. Interestingly, circulating miR-125b in breast cancer patient serum may also serve as a biomarker predicting chemoresistance. Tumor samples from breast cancer patients with high levels of circulating miR-125b showed increased cancer cell proliferation and decreased apoptosis. Accordingly, ectopic miR-125b expression correlated with increased resistance to anticancer drugs, whereas suppressing miR-125b level improved breast cancer cell sensitivity to chemotherapy[85]. Additional studies also revealed that inhibition of miR-21 and miR-200b enhanced sensitivity to doxorubicin in breast cancer cells, which may be mediated by repression of PDCD4 and PTEN, as well as upregulation of pro-apoptotic genes due to ZEB1 inhibition, respectively[81,86,87] .
All these findings indicate that miRNAs may regulate cancer cell response to chemotherapeutic drugs by targeting multiple genes, simultaneously or in parallel, which lead to complex changes in varying cellular processes, such as proliferation and apoptosis. A list of miRNAs involved in sensitivity to chemotherapy in breast cancer can be found in Table 1.
| miRNA | Drug | Target(s) | Ref. |
| miR-328 | Mitoxantrone | BRCP/ABCG2 | [75] |
| miR-21 | Doxorubicin | MDR1, PDCD4 | [81] |
| miR-451 | Doxorubicin | MDR1 | [76] |
| miR-130/107 | Cisplatin | RAD51D | [125] |
| miR-345 | Cisplatin | MDR1 | [77] |
| miR-326 | VP16/Doxorubicin | MRP1 | [78] |
| miR-128 | Doxorubicin | BMI1, ABCC5 | [79] |
| miR-34a | Docetaxel | BCL-2, CCND1 | [83] |
| miR-125b | Doxorubicin | BAK1 | [84] |
| Paclitaxel | E2F3 | [85] | |
| miR-200b/c | Doxorubicin | ZEB1, PTEN | [86,87] |
Anti-estrogen therapy is prescribed as standard treatment for ER-positive breast cancer and tamoxifen has been shown to reduce mortality in these patients by 31%[88]. However, many patients subsequently display resistance and clinical progression, and the involved mechanisms have not been fully understood. Multiple miRNAs have been implicated in mediating anti-estrogen therapy resistance by modulating ERα expression, receptor tyrosine kinase signaling, cell survival signaling, and apoptosis[89,90]. MiR-221/222 were shown to be key regulators of anti-endocrine resistance in vitro, and the resistance could be achieved by downregulation of cell-cycle inhibitor p27/Kip1 and ERα[91,92]. Overexpression of miR-221/222 promoted resistance to tamoxifen in MCF-7 cells. Moreover, knockdown of miR-221/222 sensitized ER+ breast cancer cells to tamoxifen-induced cell growth arrest and apoptosis. The protein level of p27/Kip1, which has been demonstrated as a direct target of miR-221/222, decreased along with miR-221/222 overexpression in MCF-7 cells. Additionally, ectopic p27/Kip1 increased cell death in the endocrine resistant cells exposed to tamoxifen. In parallel, overexpression of miR-221/222 in MCF-7 and T47D cells decreased ERα protein, but not mRNA, level, while miR-221/222 depletion partially rescued ERα downregulation in these cells[92]. Besides, the miR-221/222-mediated resistance to fulvestrant, a drug inducing ERα degradation, was also attributed, at least in part, to the activation of β-catenin and estrogen-independent growth of breast cancer cells[93]. In an in vivo study using miRNAs as therapeutic targets, it was found that when anti-miR222 and -181b were directly delivered into mammary tumor xenografts, the growth of tamoxifen-resistant xenografts was suppressed[94]. The authors subsequently found that miR-221, -222, and -181b all target TIMP3 directly, and knockdown of TIMP3 in MCF7 cells enabling tumors to grow in the presence of tamoxifen. A significant association was found between the expression level of miR-221 and hormone receptor (HR) status in breast cancer patients. High plasma miR-221 level was associated with HR-negative genotype as well as poorer overall response rate to neoadjuvant chemotherapy and shorter overall survival. Additional studies also found that plasma miR-221 may have predictive value in assessing chemoresistance in patients with breast cancer who received neoadjuvant chemotherapy[95].
In contrast, miR-30c was shown to be associated with a better response to endocrine therapy in advanced ER+ breast cancer. Higher expression level of miR-30c was found in patients with longer progression-free survival (PFS) after tamoxifen treatment, which may be attributed to inhibited HER and RAC1 signaling pathways[96]. It was also found that miR-301 was upregulated in mammary tumor vs normal tissue. Moreover, miR-301 expression increased again in patients that relapsed following tamoxifen treatment, but not in those who remained relapse-free[97]. Consistently, downregulation of miR-301 in tamoxifen-resistant cells restored the sensitivity to the drug. A summary of miRNAs involved in sensitivity to antiendocrine therapy in breast cancer can be seen in Table 2.
| miRNA | Drug | Target(s) | Ref. |
| miR-221/222 | Tamoxifen | P27Kip1 | [91] |
| ERα | |||
| Fulvestrant | TGF-β | [93] | |
| miR-181b | Tamoxifen | TIMP3 | [94] |
| miR-30c | Tamoxifen | [96,97] | |
| miR-342 | Tamoxifen | cyclin B1 | [126] |
| miR-451 | Tamoxifen | 14-3-3ζ | [127] |
| Let-7 | Tamoxifen | ER-α36 | [128] |
| miR-128a | Letrozole | TGFαR1 | [129] |
Targeted therapy has brought great success in treating selected subtypes of breast cancer, but the subsequent drug resistance is not rare. Compared with chemotherapy and endocrine therapy, much less was known about the mechanisms involved in resistance to targeted therapies. It has been shown that hyperactivation of the PI3K-Akt pathway played a critical role in HER2+ breast cancer resistance to trastuzumab (a therapeutic antibody targeting HER2/ERBB2)[98], and miR-21-mediated PTEN repression may contribute to the increased Akt activation in trastuzumab-resistant breast cancer cells[99]. Resistant cells can be re-sensitized to the trastuzumab treatment by blocking miR-21 with antisense oligonucleotides, which leads to proliferation inhibition and G1-S cell cycle arrest. Consistent resensitizing effect of miR-21 inhibitor can be also observed in breast cancer xenograft models, while miR-21 mimics promoted resistance to trastuzumab. Importantly, upregulation of miR-21 was found in tumor biopsies obtained from patients receiving trastuzumab treatment, which associated with poor therapeutic response, indicating a potential role of miR-21 level in predicting trastuzumab response. On the contrary, restoration of miR-205 expression, which is downregulated in breast tumors, was found to sensitize breast cancer cells to gefitinib and lapatinib, two tyrosine kinase inhibitors targeting downstream signaling of HER2[100]. The authors further identified HER3 as a direct target of miR-205 in breast cancer cells. HER3 is a critical partner of HER2 to activate downstream tumorigenic signaling pathways, and HER3 upregulation has been implicated as a potential mechanism in breast cancer resistance to HER2-tageting therapies[101-103]. Therefore, reintroduction of miR-205 likely could improve the response to HER2-targeted therapy by silencing HER3[100]. Moreover, plasma miR-210 has been found to correlate with sensitivity to trastuzumab, tumor presence, and lymph node metastases in breast cancer patients. High baseline circulating miR-210 level in patients before receiving neoadjuvant chemotherapy combined with trastuzumab significantly correlated with pathologic complete response, suggesting that miR-210 may serve as a predictive marker for better response to regimens combining trastuzumab[104].
PARP1 [poly (ADP-ribose) polymerase] inhibitors hold high promise as a novel targeted therapy for treating cancers harboring BRCA1/2 mutation, such as breast and ovarian cancers[105]. Recent studies have demonstrated that inhibiting PARP1 results in synthetic lethality in cancers with defect in DNA repair due to BRCA1/2 mutation, which highly overlap with basal-like triple negative breast cancer (TNBC)[106]. Interestingly, miR-182 has been shown to sensitize breast cancer cells to both radiotherapy and to PARP1 inhibitors by repressing BRCA1 expression and inhibiting DNA repair[107]. Antagonizing miR-182 in breast cancer cells increases BRCA1 protein level, leading to reduction of IR-induced cell death and resistance to PARP1 inhibitors. These results suggested that inhibiting miR-182 may further enhance the therapeutic efficiency of PARP1 inhibitors in combination with genotoxic therapies.
Besides targeted therapy and chemotherapy, miRNAs may also modulate cancer cell response to radiotherapy. For example, inhibition of miR-155 was shown to sensitize breast cancer cells to ionizing radiation[108]. Likewise, anti-miR-21 treatment decreased breast cancer cell survival after irradiation, which was associated with failed G2/M checkpoint upon DNA damage[109]. Cell cycle arrest at the G2/M checkpoint is essential for DNA repair machinery after DNA damage, whose failure likely will increase apoptosis upon genotoxic stress. It was found that miR-21 expression was transiently increased after irradiation in T47D cells and MDA-MB-361 cells, which was required for proper cell cycle arrest in these cells after irradiation. These observations indicated that miR-21 may contribute to radiation resistance in breast cancer cells through enhancing cell cycle checkpoint and subsequent DNA repair[109]. Similarly, increased miR-34a has been linked to poor response to radiotherapy in breast cancer cells[110]. It appeared that miR-34 promotes apoptosis while antagonizes non-apoptotic cell death in nematodes upon radiation, which was also confirmed in human breast cancer cell lines. Moreover, it has been shown that miR-34 is a target gene of p53 and can be upregulated in response to radiation[111]. Thus, inhibiting miR-34 with antagonist may be of potential therapeutic utility for sensitizing p53-mutant breast cancer to radiotherapy.
miRNA therapeutics can be devised to downregulate or block the function of pathogenic miRNAs as well as to upregulate the expression of disease-defensive miRNAs. For instance, miravirsen is a locked nucleic acid-modified DNA phosphorothioate antisense oligonucleotide that forms a highly stable heteroduplex with mature miR-122, resulting in its inactivation. It is one of the earliest miRNA-targeting therapeutics approved for clinical trials and has recently shown positive results in treating hepatitis C infection in a phase II trial[112]. In chronic HCV infection patients, miravirsen treatment reduced HCV RNA level in blood samples in a dose-dependent manner without obvious viral resistance.
Since the pathogenesis of cancer involves a number of gene mutations, amplifications and deletions, usually an effective cancer therapeutic regimen needs to target multiple genes involved in same and/or parallel functional networks as well as several signaling cascades. A significant advantage of using miRNAs as therapeutic targets lies in that one miRNA generally has multiple coding genes or non-coding RNAs as targets, which may be involved in a single pathway or in parallel pathways regulating cancer progression[6]. Compared with the strategy using siRNAs, which are usually aiming to repress one specific target gene, miRNA therapeutics appear to be superior to a mixture of siRNAs. For example, several genes associated with the epidermal growth factor receptor (EGFR) signaling pathway, such as p38 mitogen-activated protein kinase (p38), signal transducer and activator of transcription 3 (STAT3) and AKT2, are miR-124 targets[113]. As aberrant activation of EGFR signaling has been found in a number of human malignancies, such as lung and breast cancers[114], it is plausible that enhancing miR-124 expression may concurrently repress the expression of p38, STAT3 and AKT2, thereby effectively inactivating the EGFR signaling pathway.
Antagomirs are synthetic RNAs with a 2'-O-methyl linkage and phosphorothioate modification and conjugated to cholesterol. They can be used as miRNA antagonizers by complementarily binding to the targeted miRNA, thereby deblocking other endogenous miRNA target gene expression. It was found that miR-10b antagomirs remarkably reduced the lung metastasis formation in a breast cancer xenograft model. Further investigation demonstrated that silencing of miR-10b with antagomirs significantly decreased miR-10b levels and enhanced expression of Hoxd10, a miR-10b target gene playing critical roles in breast cancer metastasis[115]. Another type of miRNA inhibitors, miRNA sponges, are RNA traps that are constructed with tandem binding sites complementary to the seed sequence of the miRNA of interest. It was shown that a single sponge inhibitor can block an entire miRNA family sharing the identical seed sequence. In a study using miRNA sponges targeting miR-9, it was found that sponge-dependent miR-9 inhibition dramatically reduced mouse breast cancer cell lung metastasis, potentially by alleviating miR-9-modulated E-Cadherin downregulation and inhibiting EMT[62].
Although the preclinical studies implied high promise of using miRNA therapeutics in cancer treatment, there are also significant challenges remaining to be overcome before practical clinical applications of miRNA-targeted therapy[6]. First of all, miRNA activity highly depends on the cellular context and cancer types. Different target genes may be repressed by the same miRNA in different cell types and consequently opposite biological effects may occur within the same organism. Consequently, targeting a specific miRNA may be beneficial in one cell type but harmful in another. Thus, selective delivery of miRNA therapeutics to target tissues is a major obstacle for effective miRNA-based therapy and minimal side effects. A specific and efficient delivery system that targets only cancer cells has yet to be developed. Moreover, double-stranded RNAs (≥ 21 base pairs) can elicit a sequence-independent interferon response[116]. Since macrophages and monocytes were found to remove complexed RNAs from extracellular spaces, systemically delivered miRNAs or RNA-based miRNA inhibitors might trigger and be eliminated by the host immune response[117]. Nevertheless, the recent advance of using antibody-conjugated nanoparticles to encapsulate miRNA agents may represent a promising future of cancer cell-selective delivery approach with minimal adverse immune response in patients[118].
Beyond being directly delivered as pharmacological intervention, miRNAs may also serve as mediators for carrying out tumoricidal effect of other anti-cancer agents. For instance, Enoxacin, an antibacterial compound, was found to inhibit cancer growth through upregulating tumor suppressive miRNA production[119]. Similarly, curcumin analogue CDF was shown to inhibit pancreatic tumor growth in vitro and in xenograft models, which involved CDF-induced EZH2 downregulation and consequent activation of tumor suppressor miRNAs, such as let-7, miR-26a, and miR-101[120]. It is worth noting that induction of miRNAs may lead to complicated pathological responses in cancer cells, which can be exemplified by the observation that retinoid (ATRA)-induced miR-21 upregulation in MCF-7 cells reduced cell motility but promoted cell proliferation[121]. Meanwhile, general inhibition of miRNA biogenesis and function by small molecules was shown to reverse tumorigenesis[122]. Therefore, the pharmacological effects of conventional drug-induced miRNAs in cancer treatment are highly drug- and cancer type-specific.
As combination therapies have been proved to be more effective than single agents in various cancer regimens, it is not surprising that combinations of varying miRNA agents as well as along with conventional cytotoxic drugs or molecularly targeted agents have been studied in treating cancers. It was shown that overexpression of miR-30c, which is a favorable prognostic marker in breast cancer, sensitized TNBC cells to doxorubicin treatment in an animal model. The chemo-sensitizing effect of miR-30c was attributed to the downregulation of miR-30c target gene, twinfilin 1, which promotes EMT and induces drug resistance by upregulating IL11[123]. Similarly, adenoviral delivery of miR-145 along with 5-fluorouracil achieved greater tumor reduction than using 5-fluorouracil alone in breast cancer models[124]. These preclinical studies suggest that selective miRNA therapeutics may be applicable as sensitizing agents in clinic when used in combination with conventional therapies.
Dysregulated miRNAs in breast cancer play critical roles in the cancer initiation and progression. However, the signatures of miRNA alteration may also help to stratify patients into different risk groups as well as to predict therapeutic response. Accumulating evidence support the notion that miRNAs may sever as both therapeutic targets and tools in anticancer therapy. We envision that novel miRNA therapeutic approaches will be developed and approved for clinical application in the near future, which are combined with chemotherapy, anti-endocrine therapy or radiotherapy, based on small RNAs regulated pathway. To this end, better understanding of the regulatory mechanisms involved in miRNA biogenesis and function is pivotal for developing successful application of miRNA therapeutics. Although there are still big challenges in developing miRNA-based therapy, such as the effective delivery with high selectivity and safety evaluation, miRNAs have emerged with great potential as diagnostic/prognostic biomarkers as well as promising therapeutic targets in cancer treatment.
P- Reviewer: Ke YQ, Safe S S- Editor: Wen LL L- Editor: Wang TQ E- Editor: Lu YJ
| 1. | International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3224] [Cited by in RCA: 2878] [Article Influence: 137.0] [Reference Citation Analysis (0)] |
| 2. | Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 537] [Cited by in RCA: 567] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 3. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16056] [Article Influence: 1003.5] [Reference Citation Analysis (2)] |
| 4. | Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524-15529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3675] [Cited by in RCA: 3787] [Article Influence: 164.7] [Reference Citation Analysis (0)] |
| 5. | Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 730] [Cited by in RCA: 793] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 6. | Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847-865. [RCA] [DOI] [Full Text] [Cited by in Crossref: 965] [Cited by in RCA: 1156] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 7. | Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646-3653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 380] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 8. | Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 216] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 9. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 10. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4471] [Cited by in RCA: 4518] [Article Influence: 237.8] [Reference Citation Analysis (0)] |
| 11. | Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3151] [Cited by in RCA: 3674] [Article Influence: 262.4] [Reference Citation Analysis (0)] |
| 12. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [PubMed] |
| 13. | Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 946] [Cited by in RCA: 980] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 14. | Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2468] [Cited by in RCA: 2461] [Article Influence: 153.8] [Reference Citation Analysis (0)] |
| 15. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [PubMed] |
| 16. | Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-4060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2902] [Cited by in RCA: 2993] [Article Influence: 142.5] [Reference Citation Analysis (0)] |
| 17. | Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957-1966. [PubMed] |
| 18. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3628] [Article Influence: 164.9] [Reference Citation Analysis (0)] |
| 19. | Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631-640. [PubMed] |
| 20. | Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 499] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 21. | MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci USA. 2008;105:512-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 297] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 22. | Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1052] [Cited by in RCA: 1064] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 23. | Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci USA. 2007;104:9667-9672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 900] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 24. | Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931-1934. [PubMed] |
| 25. | Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 441] [Cited by in RCA: 394] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 26. | Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5‘UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 1019] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 27. | Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1009] [Cited by in RCA: 1128] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 28. | Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110-E2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1064] [Cited by in RCA: 1256] [Article Influence: 96.6] [Reference Citation Analysis (0)] |
| 29. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6300] [Article Influence: 370.6] [Reference Citation Analysis (0)] |
| 30. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 31. | Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635-647. [PubMed] |
| 32. | Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762-9770. [PubMed] |
| 33. | Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13:1215-1222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 195] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 34. | Li Q, Yao Y, Eades G, Liu Z, Zhang Y, Zhou Q. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene. 2014;33:2589-2600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 35. | Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G, Tortoreto M, Sasso M, Plantamura I, Triulzi T. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol. 2012;6:458-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 36. | Lee YM, Lee JY, Ho CC, Hong QS, Yu SL, Tzeng CR, Yang PC, Chen HW. miRNA-34b as a tumor suppressor in estrogen-dependent growth of breast cancer cells. Breast Cancer Res. 2011;13:R116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 37. | Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: MicroRNAs in Cancer. Cell. 2009;137:586-586.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 192] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 38. | Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 278] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 39. | Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 726] [Cited by in RCA: 752] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 40. | Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348-2360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 825] [Cited by in RCA: 899] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 41. | Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Mu L, Yu H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 194] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 42. | Walter BA, Gómez-Macias G, Valera VA, Sobel M, Merino MJ. miR-21 Expression in Pregnancy-Associated Breast Cancer: A Possible Marker of Poor Prognosis. J Cancer. 2011;2:67-75. [PubMed] |
| 43. | Ota D, Mimori K, Yokobori T, Iwatsuki M, Kataoka A, Masuda N, Ishii H, Ohno S, Mori M. Identification of recurrence-related microRNAs in the bone marrow of breast cancer patients. Int J Oncol. 2011;38:955-962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 44. | He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-833. [PubMed] |
| 45. | Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806-2811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 396] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 46. | Liu S, Goldstein RH, Scepansky EM, Rosenblatt M. Inhibition of rho-associated kinase signaling prevents breast cancer metastasis to human bone. Cancer Res. 2009;69:8742-8751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 208] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 47. | Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191-8201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 389] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 48. | Li H, Bian C, Liao L, Li J, Zhao RC. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 2011;126:565-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 162] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 49. | Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA. 2009;106:15732-15737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 282] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 50. | Kim K, Chadalapaka G, Lee SO, Yamada D, Sastre-Garau X, Defossez PA, Park YY, Lee JS, Safe S. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene. 2012;31:1034-1044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 51. | Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, McCue PA, Quong AA, Lisanti MP, Pestell RG. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci USA. 2010;107:8231-8236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 195] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 52. | Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001-11011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 370] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 53. | Banerjee N, Talcott S, Safe S, Mertens-Talcott SU. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo: potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res Treat. 2012;136:21-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 54. | Mertens-Talcott SU, Noratto GD, Li X, Angel-Morales G, Bertoldi MC, Safe S. Betulinic acid decreases ER-negative breast cancer cell growth in vitro and in vivo: role of Sp transcription factors and microRNA-27a: ZBTB10. Mol Carcinog. 2013;52:591-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 55. | Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-7070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2930] [Cited by in RCA: 3052] [Article Influence: 152.6] [Reference Citation Analysis (0)] |
| 56. | Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, Klijn JG, Wiemer EA, Martens JW. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021-13026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 299] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 57. | Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 601] [Cited by in RCA: 623] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 58. | Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 252] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 59. | Shah MY, Calin GA. MicroRNAs miR-221 and miR-222: a new level of regulation in aggressive breast cancer. Genome Med. 2011;3:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 60. | Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O’Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:pt5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 61. | Lambertini E, Lolli A, Vezzali F, Penolazzi L, Gambari R, Piva R. Correlation between Slug transcription factor and miR-221 in MDA-MB-231 breast cancer cells. BMC Cancer. 2012;12:445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 62. | Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247-256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1073] [Cited by in RCA: 1074] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 63. | Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 427] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 64. | Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147-152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1524] [Cited by in RCA: 1485] [Article Influence: 87.4] [Reference Citation Analysis (0)] |
| 65. | Zhang S, Lei P, Liu X, Li X, Walker K, Kotha L, Rowlands C, Safe S. The aryl hydrocarbon receptor as a target for estrogen receptor-negative breast cancer chemotherapy. Endocr Relat Cancer. 2009;16:835-844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 66. | Zhang S, Kim K, Jin UH, Pfent C, Cao H, Amendt B, Liu X, Wilson-Robles H, Safe S. Aryl hydrocarbon receptor agonists induce microRNA-335 expression and inhibit lung metastasis of estrogen receptor negative breast cancer cells. Mol Cancer Ther. 2012;11:108-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 67. | Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, Chen H, Ding F, Wang X, Liu Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32:4294-4303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 68. | Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 372] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 69. | Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 613] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 70. | Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081-18086. [PubMed] |
| 71. | Mulrane L, McGee SF, Gallagher WM, O’Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554-6562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 212] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 72. | You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 794] [Cited by in RCA: 802] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 73. | Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, Kim H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410-3421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 74. | Leslie EM, Deeley RG, Cole SP. Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol Appl Pharmacol. 2005;204:216-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 996] [Cited by in RCA: 1034] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 75. | Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75:1374-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 76. | Chen GQ, Zhao ZW, Zhou HY, Liu YJ, Yang HJ. Systematic analysis of microRNA involved in resistance of the MCF-7 human breast cancer cell to doxorubicin. Med Oncol. 2010;27:406-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 72] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 77. | Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010;127:1785-1794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 271] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 78. | Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, Wagar N, Yoon Y, Cho HT, Scala S. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010;79:817-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 266] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 79. | Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, Gong C, Chen J, Su F, Zhang Y. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17:7105-7115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 203] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 80. | Takwi AA, Wang YM, Wu J, Michaelis M, Cinatl J, Chen T. miR-137 regulates the constitutive androstane receptor and modulates doxorubicin sensitivity in parental and doxorubicin-resistant neuroblastoma cells. Oncogene. 2014;33:3717-3729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 81. | Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284:26533-26546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 254] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 82. | Niu J, Shi Y, Tan G, Yang CH, Fan M, Pfeffer LM, Wu ZH. DNA damage induces NF-κB-dependent microRNA-21 up-regulation and promotes breast cancer cell invasion. J Biol Chem. 2012;287:21783-21795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 126] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 83. | Kastl L, Brown I, Schofield AC. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat. 2012;131:445-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 145] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 84. | Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, Xiong W, Li G, Lu J, Fodstad O. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496-21507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 338] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 85. | Wang H, Tan G, Dong L, Cheng L, Li K, Wang Z, Luo H. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One. 2012;7:e34210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 206] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 86. | Wang ZX, Lu BB, Wang H, Cheng ZX, Yin YM. MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch Med Res. 2011;42:281-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 87. | Tryndyak VP, Beland FA, Pogribny IP. E-cadherin transcriptional down-regulation by epigenetic and microRNA-200 family alterations is related to mesenchymal and drug-resistant phenotypes in human breast cancer cells. Int J Cancer. 2010;126:2575-2583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 88. | Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687-1717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5655] [Cited by in RCA: 5738] [Article Influence: 286.9] [Reference Citation Analysis (0)] |
| 89. | Cittelly DM, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31:2049-2057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 90. | McCafferty MP, McNeill RE, Miller N, Kerin MJ. Interactions between the estrogen receptor, its cofactors and microRNAs in breast cancer. Breast Cancer Res Treat. 2009;116:425-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 91. | Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897-29903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 603] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 92. | Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079-31086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 402] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 93. | Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, Burow ME, Ivan M, Croce CM, Nephew KP. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30:1082-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 282] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 94. | Lu Y, Roy S, Nuovo G, Ramaswamy B, Miller T, Shapiro C, Jacob ST, Majumder S. Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein and modulating mitogenic signal. J Biol Chem. 2011;286:42292-42302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 95. | Zhao R, Wu J, Jia W, Gong C, Yu F, Ren Z, Chen K, He J, Su F. Plasma miR-221 as a predictive biomarker for chemoresistance in breast cancer patients who previously received neoadjuvant chemotherapy. Onkologie. 2011;34:675-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 96. | Rodríguez-González FG, Sieuwerts AM, Smid M, Look MP, Meijer-van Gelder ME, de Weerd V, Sleijfer S, Martens JW, Foekens JA. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2011;127:43-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 97. | Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M, Hui AB, Sykes J, P’ng C, Miller N. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926-2937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 202] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 98. | Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395-402. [PubMed] |
| 99. | Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, Su F, Yao H, Song E. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127-19137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 248] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 100. | Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Ménard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195-2200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 277] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 101. | Casalini P, Iorio MV, Galmozzi E, Ménard S. Role of HER receptors family in development and differentiation. J Cell Physiol. 2004;200:343-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 160] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 102. | Ménard S, Casalini P, Campiglio M, Pupa SM, Tagliabue E. Role of HER2/neu in tumor progression and therapy. Cell Mol Life Sci. 2004;61:2965-2978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 84] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 103. | Campbell MR, Amin D, Moasser MM. HER3 comes of age: new insights into its functions and role in signaling, tumor biology, and cancer therapy. Clin Cancer Res. 2010;16:1373-1383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 155] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 104. | Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC, Park ST. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603-2614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 105. | Banerjee S, Kaye SB, Ashworth A. Making the best of PARP inhibitors in ovarian cancer. Nat Rev Clin Oncol. 2010;7:508-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 106. | Anders CK, Winer EP, Ford JM, Dent R, Silver DP, Sledge GW, Carey LA. Poly(ADP-Ribose) polymerase inhibition: “targeted” therapy for triple-negative breast cancer. Clin Cancer Res. 2010;16:4702-4710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 107. | Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41:210-220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 359] [Cited by in RCA: 355] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 108. | Chen J, Wang BC, Tang JH. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106:260-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 82] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 109. | Anastasov N, Höfig I, Vasconcellos IG, Rappl K, Braselmann H, Ludyga N, Auer G, Aubele M, Atkinson MJ. Radiation resistance due to high expression of miR-21 and G2/M checkpoint arrest in breast cancer cells. Radiat Oncol. 2012;7:206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 110. | Kato M, Paranjape T, Müller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419-2424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 181] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 111. | He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819-822. [PubMed] |
| 112. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1665] [Cited by in RCA: 1698] [Article Influence: 141.5] [Reference Citation Analysis (0)] |
| 113. | Uhlmann S, Mannsperger H, Zhang JD, Horvat EÁ, Schmidt C, Küblbeck M, Henjes F, Ward A, Tschulena U, Zweig K. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 2012;8:570. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 161] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 114. | Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493-507. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 547] [Cited by in RCA: 531] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 115. | Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341-347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 638] [Cited by in RCA: 599] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 116. | Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13:464-477. [PubMed] |
| 117. | Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011;18:1104-1110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 325] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 118. | Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev. 2014;66:74-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 119. | Melo S, Villanueva A, Moutinho C, Davalos V, Spizzo R, Ivan C, Rossi S, Setien F, Casanovas O, Simo-Riudalbas L. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci USA. 2011;108:4394-4399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 120. | Bao B, Ali S, Banerjee S, Wang Z, Logna F, Azmi AS, Kong D, Ahmad A, Li Y, Padhye S. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72:335-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 228] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 121. | Terao M, Fratelli M, Kurosaki M, Zanetti A, Guarnaccia V, Paroni G, Tsykin A, Lupi M, Gianni M, Goodall GJ. Induction of miR-21 by retinoic acid in estrogen receptor-positive breast carcinoma cells: biological correlates and molecular targets. J Biol Chem. 2011;286:4027-4042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 122. | Watashi K, Yeung ML, Starost MF, Hosmane RS, Jeang KT. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J Biol Chem. 2010;285:24707-24716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 123. | Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 199] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 124. | Kim SJ, Oh JS, Shin JY, Lee KD, Sung KW, Nam SJ, Chun KH. Development of microRNA-145 for therapeutic application in breast cancer. J Control Release. 2011;155:427-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 125. | Huang JW, Wang Y, Dhillon KK, Calses P, Villegas E, Mitchell PS, Tewari M, Kemp CJ, Taniguchi T. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol Cancer Res. 2013;11:1564-1573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 126. | Cittelly DM, Das PM, Spoelstra NS, Edgerton SM, Richer JK, Thor AD, Jones FE. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9:317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 144] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 127. | Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene. 2012;31:39-47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 128. | Zhao Y, Deng C, Lu W, Xiao J, Ma D, Guo M, Recker RR, Gatalica Z, Wang Z, Xiao GG. let-7 microRNAs induce tamoxifen sensitivity by downregulation of estrogen receptor α signaling in breast cancer. Mol Med. 2011;17:1233-1241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 129. | Masri S, Liu Z, Phung S, Wang E, Yuan YC, Chen S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res Treat. 2010;124:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |