Published online May 10, 2014. doi: 10.5306/wjco.v5.i2.71
Revised: February 20, 2014
Accepted: March 13, 2014
Published online: May 10, 2014
Processing time: 139 Days and 15.9 Hours
Breast cancer (BC) is the most frequent type of non skin cancer among women and a major leading cause of cancer-related deaths in Western countries. It is substantial to discover novel biomarkers with diagnostic, prognostic or predictive usefulness as well as therapeutic value for BC. Micro-RNAs (miRNAs) belong to a novel class of endogenous interfering RNAs that play a crucial role in post transcriptional gene silencing through mRNA targeting and, thus, are involved in many biological processes encompassing apoptosis, cell-cycle control, cell proliferation, DNA repair, immunity, metabolism, stress, aging, etc. MiRNAs exert their action mainly in a tumor suppressive or oncogenic manner. The specific aberrant expression patterns of miRNAs in BC that are detected with the use of high-throughput technologies reflect their key role in cancer initiation, progression, migration, invasion and metastasis. The detection of circulating extracellular miRNAs in plasma of BC patients may provide novel, non-invasive biomarkers in favor of BC diagnosis and prognosis and, at the same time, accumulating evidence has underscored the possible contribution of miRNAs as valuable biomarkers to predict response to chemotherapy or radiotherapy. Data from in vitro and in vivo studies on BC have revealed promising therapeutic approaches via miRNA delivery and miRNA inhibition. The purpose of this review is to explore the ontological role of miRNAs in BC etiopathogenesis as well as to highlight their potential, not only as non-invasive circulating biomarkers with diagnostic and prognostic significance, but also as treatment response predictors and therapeutic targets aiding BC management.
Core tip: The specific aberrant expression patterns of micro-RNAs (miRNAs) in breast cancer (BC) that are detected with the use of high-throughput technologies reflect their key role in cancer initiation, progression, migration, invasion and metastasis. The detection of circulating extracellular miRNAs in plasma of BC patients may provide novel, non-invasive biomarkers in favor of BC diagnosis and prognosis and, at the same time, accumulating evidence has underscored the possible contribution of miRNAs as valuable biomarkers to predict response to chemotherapy or radiotherapy. Data from in vitro and in vivo studies on BC have revealed promising therapeutic approaches via miRNA delivery and miRNA inhibition.
- Citation: Christodoulatos GS, Dalamaga M. Micro-RNAs as clinical biomarkers and therapeutic targets in breast cancer: Quo vadis? World J Clin Oncol 2014; 5(2): 71-81
- URL: https://www.wjgnet.com/2218-4333/full/v5/i2/71.htm
- DOI: https://dx.doi.org/10.5306/wjco.v5.i2.71
Breast cancer (BC) is the most frequent type of non skin cancer among women and a major leading cause of cancer-related deaths in women in Western countries[1-3]. One in eight women has a chance of developing BC once in her lifetime[4,5]. Early diagnosis and a followed patient-monitored therapy can lead to successful treatment of BC. However, due to the lack of sensitivity and specificity of known biomarkers, especially in early stage disease, there is no available efficient biomarker up to now for screening or early detection of BC[6,7].
Micro-RNAs (miRNAs or miRs) belong to a novel class of endogenous interfering RNAs that play a crucial role in post transcriptional gene silencing through messenger RNA (mRNA) targeting and are thus involved in many biological processes encompassing apoptosis, cell-cycle control, cell proliferation, DNA repair, immunity, metabolism, stress, aging, etc[8].
Since their initial discovery in 1993 during a study of the gene lin-4 in caenorhabditis elegans[9], more than 2000 molecules have been determined in humans so far, regulating the expression of almost 30% of genes[10]. MiRNAs are short, non-coding RNAs of approximately 20-25 nucleotides in length that are transcribed either from independent genes or from exons or introns of protein-coding genes[8]. MiRNAs present unique nucleotide sequences that are strongly conserved among species[11] and possess a specific critical region of 7 nucleotides long, known as the seed sequence, which is responsible for mRNA base pairing[8]. Their abundant repertoire and the fact that the seed is so short may explain their combinational character in regulation: a given miRNA may target different mRNAs and a given mRNA could similarly be targeted by multiple miRNAs[12].
MiRNA biogenesis is initiated in the cell nucleus with the generation of primary miRNA (pri-miRNA), usually by RNA polymerase II. The next step is the conversion of pri-miRNA into a smaller hairpin precursor miRNA (pre-miRNA) which is arbitrated by the microprocessor complex[8]. The latter consists of the ribonuclease Drosha and the DiGeorge syndrome critical region in gene 8 protein that recognizes the hairpin loop of the pri-miRNA, ensuring unfaulty cleavage by Drosha. Pre-miRNAs are transported to the cytoplasm by the receptor exportin 5, whereas new cleavage occurs by the RNase III enzyme Dicer along with the transactivation response RNA-binding protein, resulting in a double stranded miRNA about 22 nucleotides long[8,13]. One strand of the miRNA/miRNA duplex represents the mature miRNA which in conjunction with the Argonaute (Ago) protein and other proteins form the RNA-induced silencing complex (RISC)[13]. Within RISC, mature miRNA is guided mainly to complementary sequences in 3’ or 5’ untranslated region of mRNA targets, open reading frames and promoter regions[14]. Depending on the perfect or partial base pairing between miRNA and mRNAs molecules, the consequence is mRNA degradation or translational repression at both pre-initiation and post-initiation stages, respectively; leading, thus, to a negative regulation of gene expression[15]. However, studies have also shown a possible miRNAs involvement in positive regulation of their target genes (transcriptional and translational activation)[16].
MiRNAs, as major gene-expression regulators, are implicated in the pathogenesis of many diseases, including cancer[17]. The specific aberrant expression patterns of miRNAs in several cancer types, such as BC, that are detected with the use of high-throughput technologies reflect their key role in cancer initiation, progression, migration, invasion and metastasis[18]. MiRNAs exert their action mainly in a tumor suppressive or oncogenic manner[19].
The purpose of this review is to explore the ontological role of miRNAs in BC etiopathogenesis as well as to highlight their potential, not only as non-invasive circulating biomarkers with diagnostic and prognostic significance, but also as treatment response predictors and therapeutic targets aiding BC management.
MiRNA expression profiling studies have detected aberrations with specific signatures of miRNA expression in breast carcinoma[20]. Certain miRNA expression signatures have been associated with tumor classification, stage and prognosis, while others have been useful in detecting the primary site of tumors of unknown origin[21].
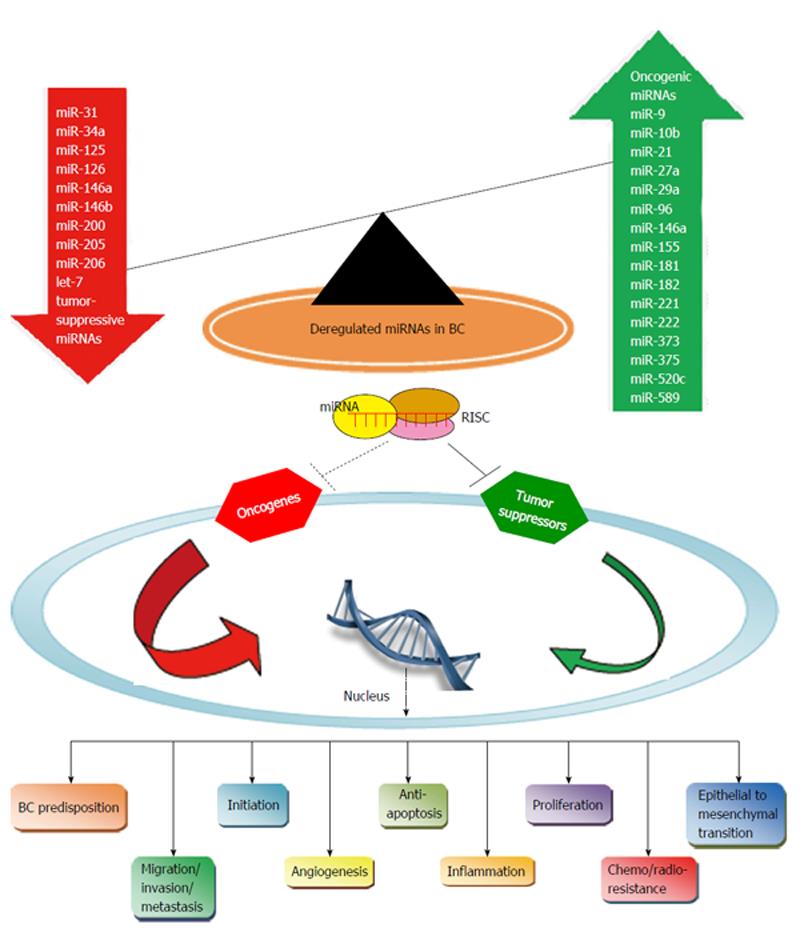
Accumulating evidence has revealed that miRNAs may act either as oncogenes, commonly named oncomirs, by suppressing the expression of tumor suppressor genes or genes responsible for apoptosis; or as tumor suppressors or oncosuppressors by inhibiting genes that promote carcinogenesis and therefore controlling apoptosis and differentiation. Nevertheless, this miRNA categorization in oncogenes with an upregulated profile and tumor suppressors with a downregulated profile may be inaccurate, as many studies have shown that miRNAs may present a dual function with oncogenic or tumor suppressive properties based on tumor type and cellular context[22].
In Figure 1, we have included a list of the most important miRNAs acting as oncogenes or tumor suppressors in BC. MiR-21 appears as a significant BC-related intracellular and extra-cellular biomarker and a therapeutic target with upregulated expression detected in human BC tissues and cell lines, playing a key role in all phases of BC pathogenesis[23,24]. In BC clinical specimens, miR-21 increased expression was associated with advanced clinical stage, lymph-node positivity and shorter survival[20]. Another example of a miRNA associated with an oncogenic potential in BC is miR-155, whereas the upregulation of miR-155 was correlated with advanced grade, clinical stage, estrogen receptor (ER) negative tumors, lymph node invasion, metastasis and poor prognosis[25]. However, miR-21 and miR-155 increased expression do not characterize only BC but also colorectal and lung cancer as well as leukemia[25,26].
Examples of overexpressed miRNAs in BC include miR-9/10b/21/27a/29a/96/146a/155/181/221/222/373/375/520c/589, where some of these have been validated in BC clinical specimens, highlighting their potential role in BC diagnosis, prognosis and therapeutics[18,23]. Concerning downregulated miRNAs, miR-30a/31/34a/125/126/146a/146b/200/205/206 and let-7 emerge in BC pathogenesis through the loss of their tumor suppressor properties[18,23]. Generally, it is important to mention that miRNA downregulation represents a more frequent event in BC pathogenesis, allowing oncogenes to be activated during BC development.
Aberrant expression of miRNAs in cancer-associated genomic regions: MiRNA genes are often situated in cancer-associated genomic regions and are subject to deletions, rearrangements, breakpoints and loss of heterozygosity[27]. Approximately half of all annotated human miRNA genes are situated in fragile sites or genomic areas that have been associated with cancer[27,28]. For example, miR-15a and miR-16-1, which are often downregulated in cancer, occupy the most frequently deleted genomic region and may harbor germline mutations in familial cases of B-cell chronic lymphocytic leukemia and BC[22,28].
Single nucleotide polymorphisms in miRNA genes or miRNA target genes and genetic susceptibility to BC: Single nucleotide polymorphisms (SNPs) in miRNA genes, their processing machinery and their target binding sites could also increase the susceptibility to BC and affect patient prognosis and treatment efficacy[29]. Despite the fact that SNPs are rare in miRNA genes, they may alter miRNA biogenesis and function as well as miRNA binding sites[30]. Many SNPs in miRNA genes or in miRNA target genes in germline cells, independently from their possible/putative functional effects, have been analyzed in association (case-control) studies. For example, the SNP rs11614913 in pre-miR-196a-2 has been linked to an elevated BC risk[31] and the SNP rs895819 in pre-miR-27a has been associated with a decreased BC risk[32]. Therefore, miRNAs may be used as potential biomarkers for BC predisposition in populations at risk.
Aberrant expression of miRNAs in BC initiation: Cancer stem cells (CSCs) or tumor-initiating cells lead tumor promotion, progression and heterogeneity by proliferating and forming some differentiated tumor cells. CSCs present a self-renewal (symmetric and asymmetric division) potential and the capacity to generate all types of cancer cells within the tumor[33]. Targeting these initial CSCs may be an effective therapeutic strategy as CSCs are responsible for tumor growth and propagation of cancer and are considered more resistant to chemotherapy and radiotherapy. Breast CSCs were first reported by Al-Hajj et al[34] and are characterized by the expression of the surface biomarkers CD44+, CD24-/low, the epithelial specific antigen and aldehyde dehydrogenase 1[33]. Under non-adherent conditions for human mammary epithelial cells, only breast CSCs are capable of surviving, proliferating and building mammospheres, which are multicellular formations containing a large number of mammary stem cells[33]. The Hedgehog signaling pathway activates the self-renewal of breast CSCs via the polycomb ring finger oncogene B lymphoma Mo-MLV insertion region 1 homolog (Bmi-1)[35]. The aberrant expression of miRNAs may play a role in carcinogenesis and breast CSCs self-renewal by acting as oncogenic or tumor suppressive miRNAs and also regulating the stem cell-like phenotype of breast CSCs[33]. The miRNA triad let-7/miR-200c/miR-30 suppresses the self-renewal of breast CSCs and the spontaneous conversion of immortalized mammary epithelial cells to a stem-like phenotype with less differentiated and mesenchymal properties by targeting Ras, Bmi-1, ubiquitin-conjugating enzyme 9 and integrin b3 respectively[24,33]. On the contrary, the upregulation of miR-181 family members and miR-495 plays a significant role in modulating breast CSCs and maintaining a stem cell-like phenotype in BC by targeting respectively the tumor suppressor serine/threonine kinase ataxia telangiectasia mutated and E-cadherin and short for regulated in development and DNA damage responses. In particular, over-expression of miR-495 in human BC cells enhances colony formation in vitro and carcinogenesis in vivo[36].
Aberrant expression of miRNAs in BC progression: MiRNAs may be involved in the cell cycle by controlling critical components of the regulatory pathways. In particular, miRNAs could regulate the cyclin/cyclin dependent kinase (CDK) pathway which constitutes a significant pathway in the cell cycle control. The pair miR-17-5p/miR-20a has been shown to attenuate the synthesis of cyclin D1 encoded by the gene CCND1 in BC MCF-7 cell line, blocking S-phase entry and inhibiting cell proliferation[37]. MiR-27a, which is also associated with the cyclin/CDK pathway, targets zinc finger and BTB domain containing 10 and Myt-1 which halter BC cell proliferation by suppressing cyclin D1 and cyclin B respectively[38].
The estradiol (E2)/ERα/Sp1 is another important cell cycle regulatory pathway in BC that aids proliferation by activating cyclin D1, leading to the G1/S phase transition. In ER positive BC, E2 increases the expression of miR-21 and let-7 family members[24]. The upregulation of let-7 family members in ER positive BC may result in diminished ERα activity and cell proliferation[18]. BC cells at the stage of ductal carcinoma in situ and invasive ductal carcinoma are characterized by let-7 downregulation in comparison to benign lesions[18]. In contrast to ER positive BC, the E2/ER signaling pathway is inhibited and miR-21 is downregulated in ER negative BC, whereas miR-18a, miR-18b, miR-206, miR-221 and miR-222 are upregulated leading to inhibition of ER expression and induction of other signaling pathways regulating cell growth and proliferation[39]. The upregulation of the pair miR-221/222 in ER negative BC may lead to reduced p27kip1 levels and continuous BC proliferation[40].
In BC, miR-31 acts as a tumor suppressor targeting the human frizzled transmembrane receptor frizzled-3 and is downregulated in all interactions with the Wnt signaling transduction pathway[41]. The oncosuppressor miR-34a, which is downregulated in triple negative and mesenchymal-type BC cell lines, has been shown to inhibit BC proliferation and migration via downregulation of B-cell lymphoma 2 (Bcl-2) and sirtuin 1[18]. The tumor suppressor miR-205 targets human epidermal growth factor receptor 3 (HER3) directly, a receptor tyrosine kinase of the epidermal growth factor receptor (EGFR) family, inactivating thereby the downstream mediator Akt, suppressing the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway and inhibiting BC proliferation with improved response to targeted therapies[42]. Finally, the overexpression of the oncogenic miR-146a may be responsible for the altered expression of the breast cancer type 1 susceptibility protein (BRCA1), which is a negative regulator of BC growth associated with the hypophosphorylated form of Rb, and the BC metastasis suppressor 1 (BRMS1), which reduces the metastatic potential of BC cells[43].
MiRNAs also interfere with the apoptotic process in BC cells. MiR-21 has been shown to play an anti-apoptotic role by indirectly targeting Bcl-2 in MCF-7 BC cells[44]. On the contrary, the oncosuppressor miR-145 targets Rhotekin (RTKN), the gene coding for the Rho effector, which activates B-cell lymphoma extra large via the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling pathway[45]; hence, promoting apoptosis. The oncogenic miR-155 is an effective suppressor of the execution phase of apoptosis via its actions in caspase 3 and its negative regulation of the tumor suppressor gene suppressor of cytokine signaling 1[18]. The members of forkhead box protein O family (FOXO) which are often the target of miRNAs, represent transcription factors characterized by a distinctive forkhead DNA binding domain and play an important role in promoting the cell cycle arrest at the G1/S checkpoint, apoptotic responses via the pro-apoptotic factor Bcl-2 homology domain 3-only molecule Bcl-2-interacting mediator (Bim) of cell death and cellular metabolism[43]. In BC, the up-regulation of the oncogenic triad of miR-27a, miR-96 and miR-182 which targets FOXO1 may contribute to BC progression and maintenance of the oncogenic state[46]. In addition, the oncogenic miR-155 may induce BC cell survival and anti-apoptosis by blocking FOXO3a[47].
Aberrant expression of miRNAs in BC migration, invasion and metastasis: After initiation and progression, tumor cells proceed to invasion and metastasis, which are enabled by the epithelial to mesenchymal transition (EMT). BC cells in carcinoma in situ lose cell adhesion mediated by E-cadherin repression, become more motile and break through the basement membrane with increased invasive properties, progressing to invasive BC. EMT, which is essential in cancer invasion, metastatic dissemination and resistance acquisition to cancer therapy, is characterized by the loss of the epithelial phenotype marker E-cadherin[18,43]. EMT is activated by tumor necrosis factor-α, hepatocyte growth factor and transforming growth factor-β (TGF-β), while many transcription factors, including zinc finger E-box binding homeobox 1 (ZEB1) and ZEB2, can repress E-cadherin directly or indirectly[43]. In BC, the expression of the miR-200 family is downregulated, resulting in the overexpression of ZEB1 and ZEB2, which are crucial EMT activators by inhibiting E-cadherin expression[48]. MiR-9 directly targets the E-cadherin coding gene CDH1, leading to an enhanced cell motility and invasiveness of SUM149 human BC cells[49]. Ras homolog gene family member (Rho) A, a prometastatic gene regulating EMT in a multiphasic manner, is the target of both oncogenic miRNAs such as miR-155, which mediates the TGF-β induced EMT, and tumor suppressor miRNAs such as miR-31[41].
For the invasion and metastatic processes, attachment of BC cells to matrix components must take place. Metalloproteinases (MMP) degrade the extra-cellular matrix (ECM) which mediates cell attachment, while the tissue inhibitor of metalloproteinases suppresses the MMP activities[50]. MiRNAs may induce cell migration and invasion through ECM destruction or disruption of recognition between ECM and cells. Later, circulating BC cells exit the bloodstream to form micro-metastases, undergoing mesenchymal to epithelial transition for clonal outgrowth at the metastatic sites.
MiR-21 inhibits TIMP3 expression in BC, promoting ECM disruption, BC invasion in multiple cell lines in vitro and metastasis[51]. Furthermore, the overexpressed miR-21 directly blocks the tumor suppressor programmed cell death protein 4 (Pdcd4), tropomyosin 1 and maspin, enhancing the promotion of invasion and metastasis[52]. The prometastatic miRNAs miR-373 and miR-520c may promote cell migration and invasion in vitro and in vivo by inhibiting the expression of CD44, which is a cell surface receptor for ECM components and cell to cell interactions with ECM[53]. The basal-like subtype-specific miRNAs miR-221 and miR-222 are associated with increased cell migration and invasion, aiding in the progression of the clinically aggressive basal-like BC[39]. Interestingly, miR-223 transferred from macrophages to BC cells through exosomes may lead to enhanced invasiveness of BC cells; highlighting the important role of exosomal communication between BC cells and macrophages[54]. The oncogenic miR-10b, which is overexpressed in metastatic BC, may initiate tumor invasion and metastasis in vivo and in vitro by interrupting the homeobox D10 (HOXD10) expression (a transcription factor that maintains a differentiated phenotype in epithelial cells), resulting in an increased expression of Ras homolog gene family member C which leads to BC cell invasion and metastasis[55]. The upregulation of miR-375 contributes to breast lobular neoplasia and invasive lobular breast carcinoma progression[18].
The miRNA pair miR-126 and miR-335 has been associated with the capacity of BC cells to metastasize to bones and lungs through blocking the expression of tenascin c, a ECM component[56]. The downregulation of the tumor suppressive let-7 contributes to BC metastasis. Let-7 modulates the repressive action of Raf kinase inhibitory protein, a BC suppressor gene inhibiting NF-κΒ, mitogen-activated protein kinases and G protein-coupled receptor kinase-2 signaling pathways in BC metastatic cells[57]. The tumor suppressive miR-31, which is undetectable in metastatic BC cells, has been shown to inhibit the expression of multiple prometastatic genes blocking BC metastasis[18]. The tumor suppressor miR-146a/b diminishes the expression of EGFR, inhibiting metastasis[58].
Distant BC metastases need tumor-induced formation of new blood vessels (angiogenesis) in order to allow expansion of the primary breast tumor and to obtain sufficient oxygen and nutrients. The angiogenic factor vascular endothelial growth factor (VEGF) represents the most important inducer of angiogenesis and may be regulated by several miRNAs. In particular, miR-126 has been shown to target VEGF expression in BC whereas the VEGF/PI3K/Akt signaling cascade is activated[59]. On the contrary, miR-9 may promote angiogenesis by enhancing VEGF-A expression in BC and downregulating E-cadherin[49]. In hypoxic conditions within the tumor microenvironment, hypoxia inducible factor-1 may also mediate the expression of VEGF in BC cells in a miR-20b-dependent way[60]. Also, the downregulation of the oncosuppressor miR-125a is associated with the overexpression of a stress-induced HuR protein in the cytoplasm, which in turn could increase the invasiveness of BC cells and angiogenesis via VEGF-Α expression[61]. In addition to angiogenesis, miRNAs may constitute feedback mechanisms that link inflammation to BC. In particular, the upregulation of miR-155 in BC could lead to stimulation of signal transducer and activator of transcription 3 via the Janus kinase pathway and activation of BC cells by interleukin-6, interferon γ and lipopolysaccharide[62].
Finally, miRNAs may also represent key regulators of the epigenetic interaction that takes place in BC cells with DNA methylation and histone modifications. The expression of the oncosuppressor miR-200 was shown to be epigenetically modulated by DNA promoter methylation and histone modifications[33]. The downregulation of the pro-apoptotic and tumor suppressor miR-34c, which inhibits invasion, occurs through hypermethylation of the promoter region and may lead to enhanced self-renewal and EMT of breast CSCs[63].
Several blood-based profiling studies have tried to elucidate the role of extracellular miRNAs in BC biology and pathogenesis[64]. It should be also noted that miRNAs have been detected in breast milk[22]. MiRNAs could be used as promising diagnostic, prognostic and predictive biomarkers in BC, presenting the following advantages: (1) their remarkable stability in plasma due to their association not only with RNA binding proteins [Ago2 protein, high density lipoprotein or nucleophosmin 1 (NPM1)] but also exosomal vesicular transportation[65,66]; (2) miRNAs represent a non-invasive diagnostic approach as a liquid biopsy in contrast to the existing tissue-dependent biopsy; and (3) miRNAs may be regarded as tumor-derived molecules that have been present early into circulation, therefore reflecting tumor status.
Genome-wide expression profiling studies of extracellular miRNAs have investigated whether serum samples could be used to identify differentiated miRNA expression levels between BC patients and healthy individuals; thus, distinguishing normal from diseased state. Wu et al[67] showed that serum miR-29a and miR-21 levels were significantly increased in 20 BC patients compared to healthy controls. Kumar et al[68] also demonstrated an overexpression of miR-21 and miR-146a in plasma samples of BC patients. Using microarray-based expression profiling followed by real time quantitative polymerase chain reaction (RT-qPCR), Zhao et al[69] found deregulated expression levels of 49 miRNAs in plasma from 20 women with early stage BC compared to 20 matched controls. Furthermore, the authors showed that both upregulated (n = 26) and downregulated (n = 23) miRNAs could discriminate patients from controls with acceptable specificity and sensitivity scores. Let-7c and miR-589 were significantly decreased and increased respectively in BC patients. In a study by Chan et al[70], 4 (miR-1, miR-92a, miR-133a and miR-133b) of the 7 miRNAs that were differentially expressed in a set of serum samples from a cohort of 132 Asian BC patients and 101 healthy controls were validated and identified as the most significant diagnostic markers. Interestingly, only 7 miRNAs out of the total 20 were overexpressed in both tumor and serum of BC patients, indicating that miRNAs could be released into serum selectively. Profiling results of another study have indicated that the combination of circulating miR-145 and miR-451 seems capable of predicting BC patients from normal individuals[71].
Other investigations have shown a correlation between systemic miRNA levels and various clinicopathological features of BC. MiR-10b and miR-373 were related to lymph node status[72], while the upregulated miR-21 was associated significantly with visceral metastasis[73]. MiR-21, miR-126, miR-155, miR-199a and miR-335 levels were closely correlated with histological grade and hormone receptor expression. A significantly higher relationship of miRNA expression levels between BC tumor tissues and sera was also found[74]. Roth et al[75] revealed that circulating miR-10b, miR-34a and miR-155 levels were significantly related to the presence of overt metastases. Serum levels of miR-182 in ER and progesterone receptor (PR) positive BC patients were lower when compared with patients suffering from ER-PR negative BC[76]. On the other hand, miR-155 expression levels were higher in serum of women with hormone sensitive BC (PR positive)[77]. Similarly, in a prospective study, levels of miR-195 and let-7a were significantly correlated with ER and lymph nodal status and decreased interestingly in BC patients postoperatively following curative tumor resection[78]. Cuk et al[79] showed that a panel of 7 circulating miRNAs, including miR-127-3p, miR-148b, miR-409-3p, miR-652 and miR-801, presents a substantial diagnostic potential, not only as a screening method for benign and malignant breast tumors, but also for the detection of early BC stage. Notably, another study has linked exosomal miRNAs to poor prognosis in BC through the maintenance of dormant BC cells in the bone marrow stroma[80]. Sieuwerts et al[81] have highlighted the diagnostic potential of detecting tumor specific miRNAs in circulating tumor cells (CTCs) in the bloodstream in an attempt to discriminate BC patients with CTCs from patients with no detectable CTCs and healthy volunteers. Accumulating evidence has underscored the possible contribution of miRNAs as valuable biomarkers to predict response to chemotherapy or radiotherapy. For example, downregulated miR-34, miR-17 and let-7a were associated with chemosensitivity to fluorouracil, adriamycin and cyclophosphamide, respectively[82]. Studies in BC cell lines have also related targeted miR-21 downregulation with increased sensitivity to topotecan and taxol[44,83], whereas other investigations have indicated that restoration of the oncosuppressor miR-205 expression levels was associated with improved response to tyrosine-kinase inhibitors gefitinib and lapatinib through abrogating the HER3-mediated resistance[42]. A growing number of studies have demonstrated a correlation between circulating miRNA expression levels and patterns of chemoresistance or chemosensitivity. Zhao et al[84] have found that plasma miR-221 could be a predictive biomarker for neoadjuvant chemotherapy sensitivity in BC patients. In other studies, circulating miR-210 and miR-125b were associated with sensitivity to trastuzumab and neoadjuvant chemotherapeutic resistance respectively; underlining the possibility of using them as indicators of treatment response[85,86].
The pivotal role of miRNA as oncogenes or tumor suppressors has stimulated scientists to manipulate their expression; an effort that indicates their emerging role as therapeutic targets and replacement therapies for BC treatment. Depending on a given miRNA that is up or downregulated, various methods exist in order to inhibit or increase its expression, including miRNA inhibition via antisense targeting with oligonucleotides (anti-miRs) or miRNA replacement via viral or liposomal delivery (miRNA mimics), respectively[87,88]. Functional analyses using knockdown mouse models and BC cell lines have revealed great therapeutic potential for the studied miRNA molecules. Potentially, every miRNA could serve as a possible therapeutic target. Si et al[44] showed that inhibition of miR-21 expression using anti-miR-21 oligonucleotides resulted in reduced MCF-7 BC cell growth and tumor growth in the xenograft mouse model due to decreased proliferation and increased apoptosis. In agreement with these findings, Yan et al[89] showed that knockdown of miR-21 inhibited growth and migration of MCF-7 and MDA-MB-231 BC cell lines in vitro and tumor growth in nude mice in vivo. MiR-21’s potential therapeutic relevance is also supported by its capacity to sensitize BC cells to anticancer therapy. MiR-21 suppression has been reported to increase sensitivity of BC cells to topotecan and taxol[44,83], whereas its tumor suppressive gene target phosphatase and tensin homolog has been shown to be a regulator of sensitivity to trastuzumab[90]. These findings suggest that the combination of anti-miR-21 with classical chemotherapy may result in overcoming drug resistance and in individualizing therapy in BC patients. Furthermore, Kong et al[47] demonstrated that knockdown of miR-155 led to apoptosis and increased chemosensitivity by upregulation of its direct target FOXO3a, suggesting that miR-155 inhibition could present a promising therapeutic potential for BC. Additionally, let-7 could contribute to cancer therapeutics due to its association with self-renewal ability and tumorigenicity of BC cells[91]. Let-7 also regulates apoptosis and CSC differentiation[92]; thus, targeting let-7 in BC could serve as an effective treatment option.
Recent studies have targeted miR-205 for inhibiting the metastatic nature of BC. Wu et al[93] demonstrated that ectopic expression of the downregulated miR-205 effectively hinders cell proliferation, anchorage-independent growth and cell invasion, supporting its use as a possible therapeutic target. MiRNA delivery via nanoparticles is also a promising technique. Jin et al[94] have recently used nanoparticles to deliver anti-miR-10b for targeting the overexpressed miR-10b which is related to BC cell migration and invasion through inhibition of HOXD10 target synthesis. A RNA poly L-lysine complex has been developed; it releases concentrations of anti-miR-10b into the cytoplasm of BC cells with sustainable effectiveness.
Further therapeutic potential is likely via targeting breast CSCs with miRNA manipulation. Certain miRNAs seem to be responsible for breast CSCs behavior, including self-renewal characteristics, increased chemotherapeutic resistance and EMT[33]. Thus, anti-CSC therapy with miRNAs could combat breast CSCs’ positive effect on tumorigenesis. Additionally, a potential mesenchymal stem cell-mediated anti-miR delivery directly to the tumor area was proposed based on mesenchymal stem cells ability to migrate[95].
Unmasking the precise role of miRNAs in the regulation of breast CSC renewal and the potential for combination of stem cell and novel miRNA-associated targeted therapies may represent effective therapeutic strategies of significant clinical benefit, probably when combined with the classic anticancer agents.
MiRNAs constitute a novel class of dysregulated molecules that could provide new avenues for diagnosing and classifying tumor-specific malignancies such as BC but also different phases of BC development from initiation to progression, migration, invasion and metastasis. The scientific interest is mainly concentrated on two basic aspects in regard to the clinical utility of miRNAs: their extracellular presence in body fluids, particularly blood, and their potential therapeutic applications either by miRNA replacement or miRNA inhibition. In BC, both aspects appear promising, with miRNAs being used as potential circulating non-invasive biomarkers detected in serum and plasma samples, and as therapeutic targets for cancer under current investigation, respectively. Although the available recent data are almost exclusively pre-clinical evidence, the application of miRNAs in BC therapy as adjuvant tools or targets appears exciting and very promising. A better understanding of the complex network of genes and cellular signaling transduction pathways regulated by miRNAs would enrich our knowledge of BC etiopathogenesis and hence would improve the therapeutic outcome of BC patients.
The diagnostic potential of circulating miRNAs as BC biomarkers is based mainly on their non-invasive detection in serum and plasma and on their high resistance and stability under difficult environmental conditions that could degrade the majority of RNAs, such as extended storage, frequent freeze-thaw cycles, extreme PH variations, boiling, preservation in archived human blood samples for several years, transport, etc. Several methodologies are available for establishing miRNA signatures in BC, such as RT-PCR, miRNA microarrays and next-generation sequencing, with several limitations regarding their cross-comparison, various reference genes used to normalize miRNA levels and differences in blood collection.
Nonetheless, crucial issues need to be resolved before establishing extracellular miRNAs as biomarkers and therapeutic tools for BC. The lack of larger prospective clinical trials with robust and standardized analyzing methods, the necessity for clarifying the real origin of circulating miRNAs as they may be confused with by-products of normal tissues or dead cells, and the validation of a well-characterized BC-specific signature of circulating miRNAs are important limitations that need to be overcome when bringing miRNAs from bench to bedside. However, taking into account the important pace of evolution in understanding the main ways of miRNA effects on BC pathogenesis, these small molecules will amaze the scientific world with more revelations in the near future.
P- Reviewers: Groisman I, Peterlongo P S- Editor: Zhai HH L- Editor: Roemmele A E- Editor: Liu SQ
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25541] [Article Influence: 1824.4] [Reference Citation Analysis (7)] |
| 2. | Vogel VG. Epidemiology, genetics, and risk evaluation of postmenopausal women at risk of breast cancer. Menopause. 2008;15:782-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Dalamaga M. Nicotinamide phosphoribosyl-transferase/visfatin: a missing link between overweight/obesity and postmenopausal breast cancer? Potential preventive and therapeutic perspectives and challenges. Med Hypotheses. 2012;79:617-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9215] [Cited by in RCA: 9856] [Article Influence: 821.3] [Reference Citation Analysis (4)] |
| 5. | Dalamaga M, Sotiropoulos G, Karmaniolas K, Pelekanos N, Papadavid E, Lekka A. Serum resistin: a biomarker of breast cancer in postmenopausal women? Association with clinicopathological characteristics, tumor markers, inflammatory and metabolic parameters. Clin Biochem. 2013;46:584-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 6. | Duffy MJ. Serum tumor markers in breast cancer: are they of clinical value? Clin Chem. 2006;52:345-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 309] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 7. | Dalamaga M, Archondakis S, Sotiropoulos G, Karmaniolas K, Pelekanos N, Papadavid E, Lekka A. Could serum visfatin be a potential biomarker for postmenopausal breast cancer? Maturitas. 2012;71:301-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 8. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27845] [Article Influence: 1326.0] [Reference Citation Analysis (0)] |
| 9. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8672] [Cited by in RCA: 8876] [Article Influence: 277.4] [Reference Citation Analysis (0)] |
| 10. | Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152-D157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2680] [Cited by in RCA: 2787] [Article Influence: 185.8] [Reference Citation Analysis (0)] |
| 11. | Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 946] [Cited by in RCA: 982] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 12. | Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3534] [Cited by in RCA: 3632] [Article Influence: 181.6] [Reference Citation Analysis (0)] |
| 13. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2105] [Cited by in RCA: 2359] [Article Influence: 131.1] [Reference Citation Analysis (0)] |
| 14. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16085] [Article Influence: 1005.3] [Reference Citation Analysis (2)] |
| 15. | He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4964] [Cited by in RCA: 5320] [Article Influence: 253.3] [Reference Citation Analysis (0)] |
| 16. | Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931-1934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1948] [Cited by in RCA: 2051] [Article Influence: 113.9] [Reference Citation Analysis (0)] |
| 17. | Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390-7394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 787] [Cited by in RCA: 812] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 18. | Tang J, Ahmad A, Sarkar FH. The Role of MicroRNAs in Breast Cancer Migration, Invasion and Metastasis. Int J Mol Sci. 2012;13:13414-13437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 19. | Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2468] [Cited by in RCA: 2464] [Article Influence: 154.0] [Reference Citation Analysis (0)] |
| 20. | Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-7070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2930] [Cited by in RCA: 3055] [Article Influence: 152.8] [Reference Citation Analysis (0)] |
| 21. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7370] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 22. | Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1009] [Cited by in RCA: 1130] [Article Influence: 80.7] [Reference Citation Analysis (0)] |
| 23. | Corcoran C, Friel AM, Duffy MJ, Crown J, O’Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 24. | Zhang ZJ, Ma SL. miRNAs in breast cancer tumorigenesis (Review). Oncol Rep. 2012;27:903-910. [PubMed] |
| 25. | Mattiske S, Suetani RJ, Neilsen PM, Callen DF. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:1236-1243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 218] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 26. | Faltejskova P, Besse A, Sevcikova S, Kubiczkova L, Svoboda M, Smarda J, Kiss I, Vyzula R, Slaby O. Clinical correlations of miR-21 expression in colorectal cancer patients and effects of its inhibition on DLD1 colon cancer cells. Int J Colorectal Dis. 2012;27:1401-1408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 28. | Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524-15529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3675] [Cited by in RCA: 3795] [Article Influence: 165.0] [Reference Citation Analysis (0)] |
| 29. | Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1029] [Cited by in RCA: 1054] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 30. | Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300-3305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 535] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 31. | Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, Zhang Y, Paranjape T, Zhu Y. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970-5977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 285] [Cited by in RCA: 296] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 32. | Kontorovich T, Levy A, Korostishevsky M, Nir U, Friedman E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int J Cancer. 2010;127:589-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 33. | Schwarzenbacher D, Balic M, Pichler M. The role of microRNAs in breast cancer stem cells. Int J Mol Sci. 2013;14:14712-14723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 34. | Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274-7282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 639] [Cited by in RCA: 625] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 35. | Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063-6071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 894] [Cited by in RCA: 936] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 36. | Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY, Lee WH. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463-2474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 37. | Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 292] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 38. | Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001-11011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 371] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 39. | Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079-31086. [PubMed] |
| 40. | le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699-3708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 627] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 41. | Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032-1046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 663] [Cited by in RCA: 660] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 42. | Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Ménard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195-2200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 277] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 43. | Kayani Mu, Kayani MA, Malik FA, Faryal R. Role of miRNAs in breast cancer. Asian Pac J Cancer Prev. 2011;12:3175-3180. [PubMed] |
| 44. | Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799-2803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1136] [Cited by in RCA: 1205] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 45. | Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L, Zhou H, Zhao RC. miR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol. 2009;34:1461-1466. [PubMed] |
| 46. | Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204-23216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 482] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 47. | Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869-17879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 285] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 48. | Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4:e7181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 259] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 49. | Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247-256. [PubMed] |
| 50. | Baker AH, George SJ, Zaltsman AB, Murphy G, Newby AC. Inhibition of invasion and induction of apoptotic cell death of cancer cell lines by overexpression of TIMP-3. Br J Cancer. 1999;79:1347-1355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 215] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 51. | Song B, Wang C, Liu J, Wang X, Lv L, Wei L, Xie L, Zheng Y, Song X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res. 2010;29:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 200] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 52. | Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 862] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 53. | Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 745] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 54. | Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 491] [Cited by in RCA: 557] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 55. | Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1936] [Cited by in RCA: 1999] [Article Influence: 111.1] [Reference Citation Analysis (0)] |
| 56. | Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147-152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1524] [Cited by in RCA: 1485] [Article Influence: 87.4] [Reference Citation Analysis (0)] |
| 57. | Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 295] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 58. | Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279-1283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 320] [Cited by in RCA: 326] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 59. | Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 165] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 60. | Cascio S, D’Andrea A, Ferla R, Surmacz E, Gulotta E, Amodeo V, Bazan V, Gebbia N, Russo A. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol. 2010;224:242-249. [PubMed] |
| 61. | Gubin MM, Calaluce R, Davis JW, Magee JD, Strouse CS, Shaw DP, Ma L, Brown A, Hoffman T, Rold TL. Overexpression of the RNA binding protein HuR impairs tumor growth in triple negative breast cancer associated with deficient angiogenesis. Cell Cycle. 2010;9:3337-3346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 62. | Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119-3127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 542] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 63. | Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 874] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 64. | Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1258] [Cited by in RCA: 1333] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 65. | Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 289] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 66. | Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 67. | Wu Q, Lu Z, Li H, Lu J, Guo L, Ge Q. Next-generation sequencing of microRNAs for breast cancer detection. J Biomed Biotechnol. 2011;2011:597145. [PubMed] |
| 68. | Kumar S, Keerthana R, Pazhanimuthu A, Perumal P. Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J Biochem Biophys. 2013;50:210-214. [PubMed] |
| 69. | Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5:e13735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 292] [Cited by in RCA: 300] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 70. | Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA, Tan PH, Ho GH, Lee AS. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19:4477-4487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 215] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 71. | Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 197] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 72. | Chen W, Cai F, Zhang B, Barekati Z, Zhong XY. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumour Biol. 2013;34:455-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 73. | Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 362] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 74. | Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 241] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 75. | Roth C, Rack B, Müller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 324] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 76. | Wang PY, Gong HT, Li BF, Lv CL, Wang HT, Zhou HH, Li XX, Xie SY, Jiang BF. Higher expression of circulating miR-182 as a novel biomarker for breast cancer. Oncol Lett. 2013;6:1681-1686. [PubMed] |
| 77. | Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 279] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 78. | Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 523] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 79. | Cuk K, Zucknick M, Madhavan D, Schott S, Golatta M, Heil J, Marmé F, Turchinovich A, Sinn P, Sohn C. Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS One. 2013;8:e76729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 80. | Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, Greco SJ, Bryan M, Patel PS, Rameshwar P. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 2011;71:1550-1560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 329] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 81. | Sieuwerts AM, Mostert B, Bolt-de Vries J, Peeters D, de Jongh FE, Stouthard JM, Dirix LY, van Dam PA, Van Galen A, de Weerd V. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin Cancer Res. 2011;17:3600-3618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 172] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 82. | Salter KH, Acharya CR, Walters KS, Redman R, Anguiano A, Garman KS, Anders CK, Mukherjee S, Dressman HK, Barry WT. An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer. PLoS One. 2008;3:e1908. [PubMed] |
| 83. | Mei M, Ren Y, Zhou X, Yuan XB, Han L, Wang GX, Jia Z, Pu PY, Kang CS, Yao Z. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol Cancer Res Treat. 2010;9:77-86. [PubMed] |
| 84. | Zhao R, Wu J, Jia W, Gong C, Yu F, Ren Z, Chen K, He J, Su F. Plasma miR-221 as a predictive biomarker for chemoresistance in breast cancer patients who previously received neoadjuvant chemotherapy. Onkologie. 2011;34:675-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 85. | Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC, Park ST. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603-2614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 86. | Wang H, Tan G, Dong L, Cheng L, Li K, Wang Z, Luo H. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One. 2012;7:e34210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 206] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 87. | Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294-2304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 318] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 88. | Liu Z, Sall A, Yang D. MicroRNA: An emerging therapeutic target and intervention tool. Int J Mol Sci. 2008;9:978-999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 118] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 89. | Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, Fu J, Zeng MS, Yun JP, Wu QL. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011;13:R2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 262] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 90. | Crowder RJ, Lombardi DP, Ellis MJ. Successful targeting of ErbB2 receptors-is PTEN the key? Cancer Cell. 2004;6:103-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 91. | Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1416] [Cited by in RCA: 1474] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 92. | Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111-1115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 633] [Cited by in RCA: 641] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 93. | Wu H, Mo YY. Targeting miR-205 in breast cancer. Expert Opin Ther Targets. 2009;13:1439-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 94. | Jin H, Yu Y, Chrisler WB, Xiong Y, Hu D, Lei C. Delivery of MicroRNA-10b with Polylysine Nanoparticles for Inhibition of Breast Cancer Cell Wound Healing. Breast Cancer (Auckl). 2012;6:9-19. [PubMed] |
| 95. | Lowery AJ, Miller N, McNeill RE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin Cancer Res. 2008;14:360-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 117] [Article Influence: 6.9] [Reference Citation Analysis (0)] |