Published online May 10, 2014. doi: 10.5306/wjco.v5.i2.48
Revised: January 23, 2014
Accepted: April 16, 2014
Published online: May 10, 2014
Processing time: 136 Days and 14.3 Hours
MicroRNAs (miRNAs) are small non-coding RNAs generated by a two-step complex process and are post transcriptional negative regulators of their target mRNAs. Dysregulation of many of these miRNAs has been associated with tumorigenesis in various cancers including breast cancer. Aberrantly high expression of specific miRNAs in breast cancer cells is demonstrated to be linked with inhibition of tumor suppressor genes and promote tumorigenesis. They are classified as oncogenic miRNAs. However, the tumor suppressor miRNAs are downregulated in breast cancer cells, since their major targets are oncogenic mRNAs. Understanding mechanism of action of specific miRNAs in breast cancer cells can be utilized to develop newer anti-cancer therapies. Recently, newer techniques are also developed to detect abundance of specific miRNA in the blood plasma samples and can be used in early diagnosis or prognosis in breast cancer. In this review article, we have discussed several miRNAs dysregulated in breast cancer and their therapeutic potential.
Core tip: A comprehensive review about the functions and molecular mechanisms of dysregulated microRNAs in breast cancer and their implications in breast cancer diagnosis and treatment.
- Citation: Shah NR, Chen H. MicroRNAs in pathogenesis of breast cancer: Implications in diagnosis and treatment. World J Clin Oncol 2014; 5(2): 48-60
- URL: https://www.wjgnet.com/2218-4333/full/v5/i2/48.htm
- DOI: https://dx.doi.org/10.5306/wjco.v5.i2.48
MicroRNAs (miRNAs) were discovered two decades ago while studying the development of Caenorhabditis elegans. They are conserved, endogenous non-coding RNAs, which are crucial in post transcriptional regulation of several genes involved with various biological functions, such as apoptosis, differentiation, proliferation, etc. Many of these miRNAs have been implicated to take part in various pathological conditions including cancer[1,2].
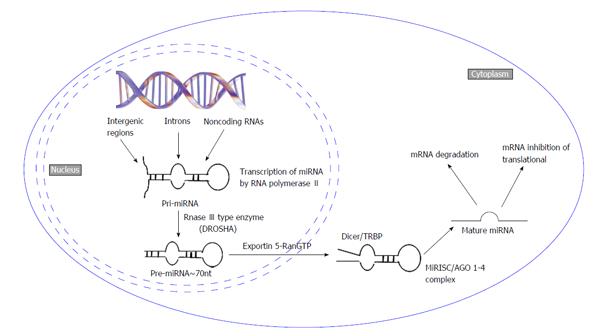
Mature miRNAs are 18-24 nucleotide long single-stranded RNA synthesized from precursor hairpin shaped double-stranded RNAs[3]. Biosynthesis of miRNAs is an extremely complex process. The majority of miRNAs are being transcribed by RNA Pol II into primary miRNA (pri-miRNA) with long stem loop structure[4]. These pri-miRNAs are then cleaved into approximately 70 nt long precursor microRNA (pre-miRNA) by RNAse III endonuclease Drosha with DiGeorge syndrome critical region in gene 8 in humans[5]. This pre-miRNA is transported to the cytoplasm from nucleus by Exportin-5. Once in the cytoplasm, the pre-miRNAs are cleaved into mature 18-24 nucleotide long miRNA by RNAse III type enzyme Dicer. Dicer is a highly specific enzyme which forms a complex with two other proteins TRBP and PACT to form miRNA induced silencing (RISC) complex. The RISC complex is responsible for the degradation of the complementary strand of miRNA and directs miRNA to its mRNA target[4,6]. This results in mRNA degradation or destabilization and subsequently translation inhibition[4,7]. This review summarizes the role of different miRNAs associated with breast cancer progression, breast cancer stem cells (BCSCs) and their implications in cancer diagnosis, prognosis and treatment.
Breast cancer is a leading cause of mortality due to cancer among women. Despite a decrease in mortality due to the advancement of scientific research, it is estimated that approximately 1.3 million females develop breast cancer each year with around 465000 expected to succumb to the disease[8,9]. Early detection and newer treatments are urgently required to inhibit the cancer progression in breast cancer patients. In 2005, a role of miRNA dysregulation in breast cancer was first demonstrated. Since then many studies have found out several different miRNAs which are being deregulated in breast cancer[10]. Some of these miRNAs act as oncogenic miRNAs by suppressing tumor suppressor genes. Whereas, some of the miRNAs exhibit tumor suppressor properties by down-regulating oncogenic genes[7].
Usually, miRNAs promote oncogenesis by either destabilizing or degrading tumor suppressor mRNAs in various types of human cancers[11]. The miRNAs affecting oncogenesis and/or metastasis are classified as oncogenic miRNAs. Several miRNAs have been identified in breast cancer to have such oncogenic potential. Different oncogenic miRNAs exhibit their oncogenic potential by inducing cell proliferation and tumorigenesis and/or metastasis, promoting angiogenesis or inducing epigenetic changes[12-16]. The Table 1 summarizes some of the most commonly deregulated oncogenic miRNA in breast cancer.
| MiRNA | Known target mRNA | Function | Ref. |
| MiR-10b | HOXD10 | Promotes cell proliferation, metastasis and angiogenesis | [108] |
| MiR-126 | IGFBP2, MERTK, PITPNC1 | Promotes angiogenesis | [18] |
| MiR-155 | SOCS1, TP53INP1, FOXO3, RhoA | Promotes cell proliferation | [12,20,21,62] |
| MiR-21 | PTEN, TPM1, PDCD4, Maspin | Promotes cell proliferation | [22-24,109] |
| MiR-375 | RASD1 | Epigenetic modification of tumor suppressor genes | [110,111] |
| MiR-221/222 | TRPS1 | Induce metastasis | [28,29,65] |
| MiR-373 | CD-44 | Induce metastasis | [14,26] |
| MiR-520c | CD-44 | Induce metastasis | [14,26] |
| MiR-9 | SOCS5, E-cadherin | Induce metastasis | [27,112,113] |
| MiR-632 | DNAJB6 | Induce metastasis | [30] |
| MiR-196b | HOXD10 (indirect) | Promotes angiogenesis | [15] |
| MiR-7 | HOXB3 | Epigenetic modification of tumor suppressor genes | [16] |
| MiR-218 | HOXB3 | Epigenetic modification of tumor suppressor genes | [16] |
| MiR-203 | SOCS3 | Promotes cell proliferation | [25] |
The miR-10b is shown to be upregulated in breast cancer cells and correlated with increased cell migration and metastasis[11]. Overexpression of miR-10b disrupts the homeobox D10 (HOXD10) mRNA, which leads to the increased expression of RhoC [a Rat Sarcoma (RAS) family member] known to promote proliferation and metastasis[17,18]. Several studies have suggested that miR-155 is an oncogenic miR by demonstrating that, its upregulation results in inhibition of tumor suppressor genes in breast cancer cells[12,19-21]. The miR-155 exhibits its oncogenic ability by suppressing the expression of protein Suppressor of cytokine expression 1 (SOCS1) both in vitro and in vivo[21]. Inhibition of SOCS1 is inversely correlated to pro-tumorigenesis. The miR-155 also represses tumor suppressor forkhead box O3a expression in breast cancer[19,20]. Similarly, miR-21 is also demonstrated to be upregulated in breast cancer cells by multiple studies and also considered as oncogenic miRNA. Research thus far have shown that miR-21 inhibits the expression of various tumor suppressor proteins such as TIMP3, PDCD4 and tropomyosin 1 (alpha)[22-24]. Recently, miR-203 also has been shown to inhibit SOCS3 expression in breast cancer cells[25]. Inhibition of miR-203 leads to the activation of several tumor suppressor proteins including p53, Bax and p21[25].
Oncogenic miRNAs such as miR-520c, miR-373, miR-221 and 222, miR-9 and others are known to promote metastasis in breast cancer and are sometimes referred as MetastamiRs[11,26,27]. MiRNAs 520c and 373 have been shown to increase migration and invasion both in vitro and in vivo by targeting the expression of CD44[26]. miR-221 and 222 induce metastasis by targeting trichorhinophalangeal syndrome 1[28,29]. The miR-9 is also shown to be significantly upregulated in breast cancer, by targeting E-cadherin to promote metastasis[27]. A recent study has also revealed that miR-632 stimulates metastasis by down regulating the HSP40 family member: DNAJB6 in breast cancer[30].
Several oncogenic miRNAs are also known to deregulate angiogenesis. The expression of miR-126 has been seen to be upregulated in many breast cancer cells[31]. Recent studies suggests that miR-126 affects angiogenesis by inhibiting the protein synthesis of insulin-like growth factor binding protein 2, c-Mer tyrosine kinase and phosphatidylinositol transfer protein cytoplasmic 1. MiRNAs 10b and 196b have also been shown to regulate angiogenesis targeting vascular endothelial growth factor (VEGF) signaling through HOXD10[16].
Some miRNAs are known to inhibit tumor suppressor genes by affecting epigenetic changes. In breast cancer cells MDA-MB-231 and MCF-7 miRNAs miR-7 and miR-218 affects histone modification and DNA methylation by targeting HOXB3. This results in inhibition of RASSF1A and Claudin 6 expression[16].
Tumor suppressor miRNAs target mRNAs of various oncogenes and their dysregulation is critical in carcinogenesis[11]. The most commonly deregulated tumor suppressor miRNAs in breast cancer are compiled in the Table 2. The Lethal-7 (let-7) family of miRNA due to their abundance was among first to be identified. This family of miRNA contains 12 members[32,33]. Various studies have shown that, expression of let-7 family members is downregulated in malignant breast cells, compared to the healthy tissues[1,33,34]. Oncogenes RAS and High-Mobility Group AT-hook 2 (HMGA2) are found to be the direct targets of let-7[33]. Increased expression of let-7 reduces cell proliferation and mammosphere formation by breast cancer initiating cells and also decreases metastasis in vivo[33]. Another study revealed that let-7a directly binds to the 3’-Untranslated Regions (UTR) region of C-C chemokine receptor type-7 (CCR7) gene. Signaling of CCR7 and its ligand CCL21 has been demonstrated to play key role in cancer progression and metastasis[35]. It was shown that expression of let-7a inhibits CCR7 expression and suppresses migration and metastasis in Zebrafish[36].
| MiRNA | Known target mRNA | Function | Ref. |
| Let-7 family | RAS, HMGA2 | Inhibit cell proliferation and mammosphere formation | [11,114] |
| MiR-125 | HuR, HER2, ETS1, Cyclin J, MEGF9 | Inhibit cell proliferation and invasion | [38,39,115,116] |
| MiR-205 | ZEB1 and ZEB2 | Reduces EMT and metastasis | [1,40] |
| MiR-200 family | ZEB1/2 | Reduces EMT and metastasis | [117] |
| MiR-206 | Cyclin D2 | Inhibits Cyclin D2 in MCF-7 cells | [118] |
| MiR-34a | Bcl2, SIRT1 | Inhibits migration, invasion and metastasis | [69] |
| MiR-335 | SOX-4, TNC | Inhibits metastasis | [41,42] |
| MiR-342 | HER2 | Increases cell proliferation | [119] |
| MiR-15a/16 | HER2 | Increases cell proliferation | [68] |
| MiR-302 | RAD52 and AKT1 | Affects DNA repair | [78] |
| MiR-31 | RhoA, ITGA5, RDX | Reduces invasion and metastasis | [44] |
| MiR-519c | HIF-1α | Inhibits angiogenesis | [45] |
In many breast cancer cell lines and breast cancer patient samples, the levels of miR-125a and miR-125b are often found to be greatly downregulated. miR-125b directly targets ETS: an oncogenic transcription factor and functions as a tumor suppressor miRNA[37]. Further, both miR-125a and miR-125b are shown to be downregulated in human epidermal growth factor receptor (HER2) overexpressing cells. Expression of miR-125 in SKBR3 cells lead to the suppression of HER2 transcripts, which eventually lead to the slower cell growth and decreased invasiveness[38]. Recently, several novel targets such as cyclin J (CCNJ) and multiple EGF-like-domains 9 (MEGF9) were found to be the direct targets of miR-125b[39]. Both CCNJ and MEGF9 are potential oncogenes and their roles in tumorigenesis are only recently emerging[39].
Another frequently down regulated tumor suppressor miRNA in breast cancer is miR-205, which is suggested to be the negative regulator of epithelial to mesenchymal transition (EMT) and metastasis[1]. The miR-205 like the miR-200 family members targets Zeb1 and Zeb2 to prevent cells from undergoing EMT. Even though miR-200 and miR-205 share similar functionality, their expression levels differ in normal mammary gland and BCSCs. The expression of miR-205 is found to be elevated, whereas expression of miR-200 family members is decreased in normal mammary gland stem cells and BCSCs[40]. This indicates the unique functionality of miR-205 in breast cancer.
The miR-335 act as a tumor suppressor miRNA in breast cancer cells because its expression suppressed the migration, invasion and metastasis[41]. It was discovered that miR-335 targets transcription factor SOX4 and matrix protein Tenascin-C (TNC)[42]. Both SOX4 and TNC were recently discovered to promote metastasis in pancreatic and liver cancers[42].
miR-31 also recently came into light when it was demonstrated that multiple genes involved in the metastatic pathway of breast cancer were being targeted by it, including radixin, RhoA and integrin-α-5. When miR-31 is overexpressed expression of all three target genes is reduced and cells become less invasive and metastatic[43]. In another study it was also seen that the ectopic expression of miR-31 in MDA-MB-231 and SUM-159 cell lines both in vivo and in vitro suppressed invasion and metastatic ability[44].
The anti-tumor activity of miR-519c is attributed to its ability in regulating angiogenesis. In a study miR-519c directly targeted the hypoxia inducible factor 1α (HIF-1α), which regulates the angiogenesis by activating VEGF, interlukin-8 (IL-8) and basic fibroblast growth factor. Ectopic expression of miR-519c significantly suppressed HIF-1α and reduced angiogenesis in a nude mouse model[45]. Also, miR-519a/b/c expression is reduced in ovarian, kidney and lung tumor samples as compared to the healthy samples. Expression of miR-519 is found to be inversely correlated to the RNA binding protein HuR expression[46].
Around a decade ago a concept was proposed that a small subset of cancer cells with stem-like characteristics might be the key factor in tumor development and metastasis in various types of cancers[47]. Cancer stem cells (CSCs) gained more attention when its role was suggested in providing chemoresistance[48]. In breast cancer, CD44+/CD24-/low or high aldehyde dehydrogenase 1 (ALDH1) expression are typical characteristics of BCSCs. To enrich BCSCs, breast cancer cells are stained with fluorescently labelled antibodies for these markers and then sorted using Fluorescence-activated cell sorting[49]. However, it should be noted that even after sorting, it is virtually impossible to get a pure cancer stem cell population. Recent researches have shown that the expression profile of specific miRNAs in BCSCs is distinct compared to the normal breast cells[33,50]. The dysregulation of miRNA might contribute to the self-renewal of BCSCs and cancer progression[33]. Here we have summarized the several miRNAs identified to be deregulated in BCSCs and their mechanism of action (Figure 1).
One of the first group of miRNAs discovered to be dysregualted in BCSCs were the let-7 family members. It was noticed that expression of let-7 miRNA was significantly downregulated in SKBR-3 tumor-initiating cells than non-self-renewing population[33]. These two population of cells were separated using CD44+ CD24-/low phenotype. Let-7 miRNAs act as tumor suppressors mainly by targeting RAS oncogene as described earlier[33]. Upon induced expression of let-7 miRNAs led to the decreased BCSCs and decreased mammosphere formation[33]. Another study have shown that decreased expression of let-7 is attributed to the RNA binding protein Lin28[51]. It is also demonstrated that activation of STAT3 via inflammatory cytokines activates Lin28 expression and this results in let-7 downregulation. Consequently, target of let-7, HMGA2 is increased, which in turn enhances the EMT in CSCs.
The BCSCs can be enriched also by cultivating cancer cells as mammospheres. The 3D mammosphere is believed to be enriched with BCSCs. In a recent finding, miRNAs belong to miR-30 family are found to be downregulated in BCSCs enriched via mammosphere. The miR-30e downregulation increases the ubiquitin-conjugating enzyme 9 and integrinb 3 expression. Increasing the expression of miR-30e reduced self-renewal ability of CSCs and tumorigenesis[52]. Overexpression of miR-30a reduces total number of mammospheres in MCF-7 cells and its downregulation leads to the increase in the number of mammospheres[53].
MiR-200 family members (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) have been shown to regulate the BCSC population. The miRNAs belong to the miR-200 family are significantly downregulated in BCSCs (CD44+ CD24-/low) when compared to non-cancerous cells. Expression of tumor suppressor miR-200c decreased the self-renewal ability of BCSCs in vitro and tumor formation ability in vivo[40]. The decreased expression of miR-200b lead to the reduction of E-cadherin expression, which results in increase EMT[54]. Another group has also demonstrated that re-expressing the miR-200 family members reprograms BCSCs to a more non-stem like cells and also promotes mesenchymal to epithelial transition[55].
The miR-128 was also found to be downregulated in BCSCs (in both CD44+ CD24-/low and mammospheres) compared to the non-cancerous cells. The miR-128 targets polycomb ring finger oncogene (Bmi-1) and ATP binding cassette sub-family C member 5 (ABCC5). Both of these genes are known to induce chemoresistance in breast tumor initiating cells. Induced expression of miR-128 reduces the levels of both Bmi-1 and ABCC5 and increases the drug efficacy[56]. Also, similar to other miRNAs mentioned in this section, reduction in miR-128 expression results in the increase number of mammospheres[57].
Another miRNA recently shown to be downregulated in BCSCs (high ALDH1) is miR-93. Ectopic expression of miR-93 prevents tumor growth in xenografts. In this study it was demonstrated that miR-93 regulates BCSCs by reducing the expression of several stem cell regulatory genes such as SOX4, STAT3 and AKT3[58].
BCSCs enriched from MCF-7 and SK-3rd (CD44+ CD24-/low) cells were found to have lower levels of miR-34c. Decreased expression of miR-34c promotes self-renewal and EMT. When re-expressed, miR-34c inhibited the expression of Notch4, reduced the number of mammospheres[59].
Another tumor suppressor miRNA often downregulated in BCSCs (enriched using mammosphere) is miR-16. It targets the oncogene Wip1 and ectopic expression of miR-16 inhibits Wip1 expression in MCF-7 cells and also increases the sensitivity to chemotherapeutic drugs[60].
In the mammospheres of human breast cancer cell lines MDA361, MCF-7 and BT474, miR-181 levels are found to be elevated compared to non-cancerous cells. Potential target of miR-181 seems to be the tumor suppressor gene: Ataxia telangiectasia mutated (ATM). ATM levels are usually reduced in mammospheres treated with transforming growth factor-β.
Another miR-found to be significantly upregulated in BCSCs is miR-495. The miR-495 is found to be upregulated in two distinct subpopulations of BCSCs based on two surface markers: (1) commonly used CD44+ CD24-/low; and (2) novel surface marker PROCR+/ESA+ (PROCR stands for protein C receptor). Also, overexpression of miR-495 increased tumor formation in vivo. Moreover, expression of E-cadherin was lowered with overexpression of miR-495 which enhances the stem like phenotype in BCSCs[61].
So far it has been established that, miRNAs play critical role in regulation of several genes associated with various cancers including breast cancer[4,11,62]. Many recent studies have implicated that several miRNAs can confer resistance against common chemotherapeutic drugs in breast cancer[63,64]. The proposed mechanisms to explain this drug resistance in breast cancer are (1) intracellular drug depletion via transporters and enzymes; (2) impairing cellular functions through cell cycle arrest, DNA damage or apoptosis; (3) Inducing signaling cascade which promote transformation; and (4) Inducing epigenetic changes such as DNA methylation[63-65].
The miR-7, miR-27, miR-326, miR-328, miR-451, miR-489 confer resistance to chemotherapeutic drugs such as Doxorubicin, Cisplatin or taxol via drug depletion by targeting transporters and enzymes in breast cancer cells[63,66]. Some of these miRNAs target the ABC drug transporters and affect drug availability in the cell. The decrease in miR-451 leads to the increase in its target P-glycoprotein, a member of the ABC transporter family[67]. Another family of genes associated with the drug resistance is multidrug resistance associated proteins (MRP-1). Various independent studies have revealed that miR-7 and miR-345 directly bind at the 3’-UTR region of MRP-1 mRNA and decrease their expression levels correlated with resistance to Cisplatin in MCF-7 cells[64]. Similarly, miR-489 targets MRP-2 and affects the drug efflux in breast cancer cells[63].
Dysregulation of tumor suppressor miRNAs miR-342 and miR-15a/16 are related to the tamoxifen resistance in HER2 overexpressing tumors. HER2 overexpression is found in approximately 30% of breast cancers and is one of the major factors responsible for the chemoresistance[11]. A new study revealed that a splice variant of HER2, HER2Δ16 is linked to metastatic breast cancer and chemoresistance[68]. It was demonstrated that decreased levels of miR-342 and miR-15a/16 expression contribute to tamoxifen resistance by increasing anti-apoptotic protein B-cell lymphoma 2 (Bcl-2) expression in cell lines overexpressing HER2Δ16[68].
Similarly, expression of miR-34a is also found to be downregulated in breast cancer. It is suggested that the miR-34a is regulated by p53. Breast cancer cell line MDA-MB-231 with low levels of miR-34a was more resistant to radiation therapy compared to cells with elevated levels of miR-34a[69]. In a separate report, miR-34a was found to be downregulated in multiple breast cancer cell lines. Here it was shown that miR-34a suppresses proliferation and migration by inhibiting Bcl-2 and sirtuin[70].
Several chemotherapeutic drugs used in the treatment of breast cancer interfere with cellular functions such as DNA repair, cell cycle and apoptosis. Over activation of DNA repair pathways are associated with resistance to chemotherapy[71]. MiR-373 down regulation is shown to increase DNA repair efficiency and resistance to drugs[72]. Similarly, miR-21 and miR-141 are also associated with drug resistance, which target the mismatch repair pathway[73,74]. One of the most studied genes in breast cancer, BRCA1 also achieves drug resistance by inducing repair in double strand DNA breaks generated by anticancer drugs[75]. The miR-17, miR-182, miR-146 and miR-28 have a potential binding site at the 3’-UTR of BRCA1 transcript[63,76,77]. Another, miRNA affecting dsDNA repair is miR-302. The expression of miR-302 is associated with radioresistant breast cancer cells. The miR-302 directly inhibits the expression of RAD52 and AKT that are known to provide radioresistance in many cancers including breast cancer[78] to provide radioresistance in many cancers including breast cancer. The PI3K/AKT is found to be one of the major pathways in various mechanisms to confer radioresistance in breast cancer. RAD52 is important in dsDNA repair mechanism and its activation leads to radioresistance upon down regulation of miR-302[79].
As mentioned above, miRNA dysregulation can trigger several signaling pathways to alter the levels of various receptors or hormones. The estrogen receptor (ER) is expressed in the vast majority of breast cancers and induces cell proliferation. Anticancer drugs like Tamoxifen inhibit the expression of steroid receptors to prevent ER-driven cell proliferation. Increased expression of miR-221/222 and miR-101 decreases the sensitivity to Tamoxifen in ER-positive tumors[65]. Overexpression of HER2 (or HER2) is found in approximately 30% of malignant breast cancer patients and known for poor prognosis[80]. The downregulation of miR-125 and miR-331 is suggested to be responsible for reduced efficacy of HER2 targeted therapy such as trastuzumab. Both miR-125 and miR-331 can decrease the expression of HER2[63].
Several genes responsible for induction of breast cancer can be regulated epigenetically, and several of them are known to induce drug resistance in breast cancer cells. Abnormal DNA methylation is one of the major characteristic of cancer cells. DNA methyl transfrases DNMT1, 2 and 3 are crucial for DNA methylation. Often levels of DNMTs have been found to be elevated in breast cancer cells. Several recent studies have suggested that miR-148, miR-152, miR-29, miR-194 and miR-143 regulate the expression of various DNMTs in breast cancer, thereby inducing drug resistance[81-84]. Moreover, it is shown that upregulation of DNMTs leads to the hypermethylation of miRNAs such as miR-148 and represses their expression. Increasing the miR-148 expression resulted in decreased tumor growth and metastasis[85]. Recent studies clearly demonstrated that miR-143 is downregulated in breast cancer cells and regulates the expression of DNMT3A. Restoring the expression of miR-143 can decrease the phosphatase and tensin homolog (PTEN) hypermethylation[81]. Other than DNA methylation, aberrant change in chromatin structure can induce drugresistance in cancer cells[86]. The miR-101 and miR-221/222 upregulation leads to aberrant histone H3 modification and is associated with chemoresistance[87,88]. Similarly, miR-342 downregulation affects histone demethylation and is associated with Cisplatin resistance[64].
Chances of survival of a breast cancer patient is significantly more when detected at an early stage over late detection[89]. Moreover, biomarkers which can predict the treatment outcome are also very important in managing breast cancer therapy. Various approaches have been used to identify different biomarkers to diagnose breast cancer at an early stage. In recent years miRNAs have emerged as important diagnostic and prognostic tool in breast cancer research.
Other than early detection of breast cancer, prediction of treatment response is also very critical in better outcome of the treatment and patient survival. It was hypothesized that dysregulation of specific miRNAs in breast tumors can be used in determining prognosis of drug treatment in breast cancer patients. Data from various findings with breast tumor samples have supported this hypothesis. In this section role of different miRNAs in breast cancer prognosis is briefly discussed.
A recent study showed that expression levels of miR-10B, miR-21, and miR-335 were higher in 112 breast tumor samples compared to the healthy tissues and directly correlated with disease free survival[13]. Also, upregulation of miR-21 can be used as prognostic tool for the lung metastasis of breast cancer[90]. In this study it was also reported that down regulation of tumor suppressor miR-205 correlates with disease free interval and overall survival[13]. Recent computational analysis indicated that association of miR-330-3p with MAF mRNA directly correlates with lower survival rates of breast cancer patients[91]. Further validation of these miRNAs can be useful in identifying reliable prognostic biomarker of breast cancer.
Among all breast cancer patients around 30% patients are HER2 positive and found to have poor prognosis than PR+ or ER+[92]. Moreover, around 10% breast cancer patients acquire triple negative (PR-/ER-/HER2-) phenotype with worse prognosis[77,92]. Finding a better prognostic marker is urgently required for these types of breast cancers. In a study RNA samples of 49 HER2 positive and 48 triple negative breast cancer (TNBC) patients were subjected to the deep sequencing to identify potential prognostic markers of these relatively resistant breast cancers to chemotherapy. This study revealed that patients who developed metastasis, had increased levels of miR-184 and decreased levels of miR-375 and miR-423[10]. This data suggest that these miRNAs can be used as prognostic markers for HER2+ or TNBC breast cancer. Further, research is required to confirm these findings with bigger sample size.
Specific miRNAs are very stable in serum of various breast cancer patients, making them very valuable targets in early diagnosis of the disease. Recent advances in high-throughput techniques provided an instrumental platform to detect dysregulation of miRNA in serum samples of breast cancer patients. Various studies have identified several miRNA specifically deregulated in the blood plasma of breast cancer patients compared to a healthy individual[6,9,32]. Moreover, these findings also demonstrated that dysregulation of miRNA in plasma is attributed to the primary tumor site[6,9,32]. In this section, several of these miRNAs identified as biomarkers of breast cancer diagnosis are discussed.
A recent study has identified miR-571, miR-139-3p, miR-206, miR-193a, miR-526b, miR-519 to be more than 1.5 fold downregulated in breast cancer patients of age 50-53[9]. In the same study they also found elevated levels of miR--376c, miR-801, miR-148b, miR-424, miR-184, miR-409, miR-376a, miR-190 and miR-127-3p in the blood plasma of breast cancer patients[9]. It is important to determine that the dysregulation of miRNA found in the serum samples of patients is tumor-derived. To test this, Ng et al[93] developed a method, in which miRNA levels of serum samples of patients are compared with their tumor samples. During the profiling they found that 8 miRNAs (miR-16, miR-21, miR-27a, miR-150, miR-191, miR-200c, miR-210, miR-451) up-regulated and miR-145 down-regulated in both plasma and tumor tissues in breast cancer patients. Further it was demonstrated that miR-451, miR-21 and miR-16 levels were significantly elevated in the serum of breast cancer patients compared to the healthy individuals[93]. Interestingly miRNA levels measured in the postoperative samples were drastically lowered when compared with preoperative samples. These findings indicate that miRNAs from serum samples can be used as diagnostic tool in breast cancer[9,93].
Another recent study found that miR-18a, miR-181a and miR-222 showed the highest percentage difference in the serum samples of 205 women who eventually developed cancer and 205 women who remained cancer free[94]. This study shed light on detecting miRNAs which can predict increased risk of getting breast cancer. Together all these results clearly demonstrates that using the high-throughput methods, dysregulation of miRNA can be used as a diagnostic tool in various cancers including breast cancer to predict disease progression, risk of metastasis and/or treatment outcome[9,93,94].
Few reports have suggested that a limited number of specific miRNAs detected in blood plasma can also be used as both diagnostic and prognostic marker of breast cancer. One study demonstrated that miR-155 levels are significantly higher in 81.9% breast cancer patient samples and is an excellent diagnostic marker. After surgical removal of breast tumor and four rounds of chemotherapy, 79% of patients exhibited reduced levels of miR-155 and sustained treatment response[95]. In the blood plasma elevated levels of miR-200a/b/c, miR-203 and miR-210 were also shown to have prognostic significance in breast cancer patients[96].
So far we established that several miRNAs are play critical role in breast cancer initiation, progression and metastasis, and thus become attractive targets for therapy [31,49,79]. Various research groups have demonstrated different approaches to regulate these target miRNA expression to improve therapy. Two distinct ways miRNA based therapy can be utilized as anticancer treatment: (1) by antagonizing oncogenic miRNA; or (2) by enhancing expression of tumor suppressor miRNAs.
There are various approaches being utilized to inhibit oncogenic and metastatic functions of miRNA. One of the most obvious strategies is to silence these oncogenic miRNAs using an anti-microRNA oligonucleotide (AMO) to prevent interaction with their target proteins. The AMO competes with the target mRNA of oncogenic miRNAs and inhibits their function[17]. These anti-sense miRNAs are chemically modified to increase their stability in the system. These chemical modifications also make them more stable in the hybridization state[7].
Use of anti-sense RNA of miR-10b in mouse reduced mammary tumor metastasis[17]. It was clearly observed that upon treatment with AMO against miR-10b decreased the levels of miR-10b significantly and by simultaneously increased the levels of its target, HOXD10. Also, it was observed that anti-sense miR-10b did not reduce the size of primary tumor but prevented lung metastasis. These results suggest that anti-miR-10b can be a good candidate as an anti-metastatic agent but might not be useful in reducing primary tumor burden in breast cancer[17].
Another strategy used in miR-therapeutics is modification of 2’-hydroxyl to 2’O-methyl in ribonucleotide. This chemical modification also prevents the degradation of these miRNAs and improves their stability. The anti-sense of oncogenic miR-21 is used in the xenograft carcinoma model using MCF7 breast cancer cells. It was observed that mice treated with anti-miR-21 had half the size of tumors when compared to the control group[97]. The miR-is an oncogenic miRNA and known to induce cell proliferation by targeting PTEN. Using 2’-O-methyl antisense oligonucleotide only destabilizes the mRNA and does not degrade them. Therefore, any change in their levels can be measured only by measuring the levels of their targets[98].
Another novel method to inhibit miRNAs was introduced as an alternative to the chemically modified AMO. Here, miRNA-inhibitors are directly expressed in cells under the strong promoter which contains multiple sites complementary to the target miRNA and are known as “miRNA sponges”[99]. Utilizing this novel approach a very recent investigation have revealed that inhibition of miR-22 by providing complementary binding site via reporter gene in 3’UTR reduced the cell migration and metastatic phenotype of breast cancer cell line LM2. Oncogenic miR-22 is known to increase breast cancer metastasis and stemness. Moreover, using same approach inhibition of miR-22 also led to the inhibition of breast cancer metastasis in vivo[100].
Several studies have made it apparent that dysregulation of multiple miRNAs seems to be responsible for oncogenesis and/or metastasis. Thus targeting single miRNA might not be sufficient to achieve the optimal result in inhibiting cancer progression. To address this issue, a novel method has been proposed where AMO is designed to target more than one miRNA. In a study it was shown that a longer multiple target AMO (MTg-AMO21/155/17) decreased the cell viability of MCF-7 cancer cells to 18% at 10 nmol/L concentration. To achieve the similar levels of cell death by individual AMOs against miR-21, miR-155 or miR-17 required 10 times higher concentration of AMOs. These results indicated that targeting multiple miRNAs by longer AMOs can be a better approach to inhibit the disease progression[101].
Recently, libraries of active clinical small molecule compounds have been tested for their ability to inhibit specific miRNA of interest[102,103]. A recent finding demonstrated that levels of miR-21 were effectively reduced by the small molecule inhibitor “azobenzene”[102]. Similarly, another study identified two small molecule inhibitors polylysine and trypaflavine as effective inhibitors of various oncogenic miRNAs and attenuation of tumorigenesis[103].
Another novel approach in miRNA antagonism is by using peptide nucleic acid (PNA) as a miRNA inhibitor. Chemically in PNA the sugar-phosphate backbone is replaced by N-(2-aminoethyl) glycine, which makes them efficient hybridization agent resistant to DNAses and proteases. In aggressive breast cancer cells PNAs targeting miR-221 and miR-210 reduced the levels of miR-221 and miR-210 respectively. Also, inhibition of these miRNAs resulted in the elevated levels of a major apoptosis player p27Kip1[104]. Treatment with PNA targeting miR-21 showed inhibition of tumorigenesis of MCF-7 breast cancer cells in female nude mice compared to the control group of mice[105].
Several tumor suppressor miRNAs are shown to be downregulated in various types of cancers. It is proposed that restoring their levels should be beneficial in inhibiting cancer progression. One approach to restore the tumor suppressor miRNA in cancer cells is by using the miRNA mimics. A study demonstrated that in aggressive breast cancer cell MDA-MB-231 restoring the levels of miR-200c using the miR-200c mimic results in reduced cell proliferation, invasion and migration[106]. Similarly, in breast cancer cells increasing the levels of let-7 miRNA by let-7-lentivirus infection reduced the cell proliferation and mammospheres formation[33].
Accumulating evidences supports that dysregulation of specific miRNAs is crucial in breast cancer progression. Targeting these miRNAs can have tremendous potential in breast cancer diagnosis and treatment. This review summarizes the impact of various deregulated miRNAs in breast cancer progression, metastasis and angiogenesis. Understanding the molecular mechanisms of these miRNAs with their target genes is critical in utilizing them as a therapeutic tool. To manage more aggressive types of breast cancers, other than development of novel therapies, early diagnosis and prognosis markers are urgently required. Recently, various online databases of miRNA targets are made available to predict the role of specific miRNA in regulating biological pathways. Better use of these databases can save a lot of time and efforts for clinical research in finding better target miRNA for cancer therapeutics or diagnosis. Also, for future a better collaboration among clinicians and researchers is required to develop novel miRNA based anti-cancer therapies.
Several publications have indicated that differences in specific miRNA levels among blood plasma samples of breast cancer patients compare to the healthy individuals can be used as diagnostic markers of breast cancer. However, it is not fully understood that, what is the source of these miRNAs? And how are they released in the blood stream? Future research is necessary to find this mechanism to develop better diagnostic tools using miRNAs.
Despite of all the advances, miRNA based therapy faces some stiff challenges. One of them is specificity. Also, therapeutic miRNAs are subjected to degradation. This results in inefficient treatment in cancer cells. Further research is required to address these issues to improve their specificity and enhancing efficiency of treatment. Moreover, future research warranted to improve delivery of these miRNA to prevent their degradation[1,80].
Studying the function of various miRNAs in different biological pathways have improved our understanding of their role in cancer development. Also, several studies have shown very encouraging data in breast cancer treatment using miRNA both in vitro and in vivo. Despite of these successes it is believed that, it will take several years for miRNA based therapy to enter clinics to treat human cancers because of the limitations we discussed above[1,107].
We would like to thank Sara H Johnson for her assistance with the manuscript.
P- Reviewers: de Andrade Urban C, Hernanz F, Khajehei M S- Editor: Gou SX L- Editor: A E- Editor: Liu SQ
| 1. | Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-7070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2930] [Cited by in RCA: 3052] [Article Influence: 152.6] [Reference Citation Analysis (0)] |
| 2. | Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 706] [Cited by in RCA: 738] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 3. | Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M. A uniform system for microRNA annotation. RNA. 2003;9:277-279. [PubMed] |
| 4. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 5. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3628] [Article Influence: 164.9] [Reference Citation Analysis (0)] |
| 6. | Cookson VJ, Bentley MA, Hogan BV, Horgan K, Hayward BE, Hazelwood LD, Hughes TA. Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cell Oncol (Dordr). 2012;35:301-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 7. | Corsini LR, Bronte G, Terrasi M, Amodeo V, Fanale D, Fiorentino E, Cicero G, Bazan V, Russo A. The role of microRNAs in cancer: diagnostic and prognostic biomarkers and targets of therapies. Expert Opin Ther Targets. 2012;16 Suppl 2:S103-S109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 8. | Herranz M, Ruibal A. Optical imaging in breast cancer diagnosis: the next evolution. J Oncol. 2012;2012:863747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Cuk K, Zucknick M, Heil J, Madhavan D, Schott S, Turchinovich A, Arlt D, Rath M, Sohn C, Benner A. Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer. 2013;132:1602-1612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 199] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 10. | Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443-4453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 289] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 11. | Tang J, Ahmad A, Sarkar FH. The Role of MicroRNAs in Breast Cancer Migration, Invasion and Metastasis. Int J Mol Sci. 2012;13:13414-13437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 12. | Zhang CM, Zhao J, Deng HY. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci. 2013;20:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 13. | Markou A, Yousef GM, Stathopoulos E, Georgoulias V, Lianidou E. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem. 2014;60:197-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 14. | Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 744] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 15. | Plummer PN, Freeman R, Taft RJ, Vider J, Sax M, Umer BA, Gao D, Johns C, Mattick JS, Wilton SD. MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Cancer Res. 2013;73:341-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 99] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 16. | Li Q, Zhu F, Chen P. miR-7 and miR-218 epigenetically control tumor suppressor genes RASSF1A and Claudin-6 by targeting HoxB3 in breast cancer. Biochem Biophys Res Commun. 2012;424:28-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 17. | Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1936] [Cited by in RCA: 1996] [Article Influence: 110.9] [Reference Citation Analysis (0)] |
| 18. | Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 427] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 19. | Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773-6784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 545] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 20. | Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869-17879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 21. | Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119-3127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 540] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 22. | Qi L, Bart J, Tan LP, Platteel I, Sluis Tv, Huitema S, Harms G, Fu L, Hollema H, Berg Av. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 168] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 23. | Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Mu L, Yu H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 194] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 24. | Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348-2360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 825] [Cited by in RCA: 899] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 25. | Ru P, Steele R, Hsueh EC, Ray RB. Anti-miR-203 Upregulates SOCS3 Expression in Breast Cancer Cells and Enhances Cisplatin Chemosensitivity. Genes Cancer. 2011;2:720-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 26. | Yang K, Handorean AM, Iczkowski KA. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int J Clin Exp Pathol. 2009;2:361-369. [PubMed] |
| 27. | Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247-256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1073] [Cited by in RCA: 1074] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 28. | Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897-29903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 603] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 29. | Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O’Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:pt5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 30. | Mitra A, Rostas JW, Dyess DL, Shevde LA, Samant RS. Micro-RNA-632 downregulates DNAJB6 in breast cancer. Lab Invest. 2012;92:1310-1317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | Harquail J, Benzina S, Robichaud GA. MicroRNAs and breast cancer malignancy: an overview of miRNA-regulated cancer processes leading to metastasis. Cancer Biomark. 2012;11:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Ali AS, Ali S, Ahmad A, Bao B, Philip PA, Sarkar FH. Expression of microRNAs: potential molecular link between obesity, diabetes and cancer. Obes Rev. 2011;12:1050-1062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 33. | Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1416] [Cited by in RCA: 1471] [Article Influence: 86.5] [Reference Citation Analysis (0)] |
| 34. | Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612-11620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 446] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 35. | Cunningham HD, Shannon LA, Calloway PA, Fassold BC, Dunwiddie I, Vielhauer G, Zhang M, Vines CM. Expression of the C-C chemokine receptor 7 mediates metastasis of breast cancer to the lymph nodes in mice. Transl Oncol. 2010;3:354-361. [PubMed] |
| 36. | Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW, Nam SJ, Chun KH. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012;14:R14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 37. | Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552-3562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 234] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 38. | Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479-1486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 487] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 39. | Feliciano A, Castellvi J, Artero-Castro A, Leal JA, Romagosa C, Hernández-Losa J, Peg V, Fabra A, Vidal F, Kondoh H. miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS One. 2013;8:e76247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 40. | Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592-603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 972] [Cited by in RCA: 955] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 41. | Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147-152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1524] [Cited by in RCA: 1485] [Article Influence: 87.4] [Reference Citation Analysis (0)] |
| 42. | Png KJ, Yoshida M, Zhang XH, Shu W, Lee H, Rimner A, Chan TA, Comen E, Andrade VP, Kim SW. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011;25:226-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 174] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 43. | Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 2010;70:5147-5154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 44. | Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032-1046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 663] [Cited by in RCA: 660] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 45. | Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, Yu SL, Chen JS, Chang KJ, Jee SH. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010;70:2675-2685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 46. | Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, Cabo Rd, Gorospe M. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle. 2010;9:1354-1359. [PubMed] |
| 47. | Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 874] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 48. | Abdullah LN, Chow EK. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med. 2013;2:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 477] [Cited by in RCA: 558] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 49. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7715] [Article Influence: 350.7] [Reference Citation Analysis (0)] |
| 50. | Schwarzenbacher D, Balic M, Pichler M. The role of microRNAs in breast cancer stem cells. Int J Mol Sci. 2013;14:14712-14723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 51. | Sakurai M, Miki Y, Masuda M, Hata S, Shibahara Y, Hirakawa H, Suzuki T, Sasano H. LIN28: a regulator of tumor-suppressing activity of let-7 microRNA in human breast cancer. J Steroid Biochem Mol Biol. 2012;131:101-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 52. | Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194-4204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 53. | Ouzounova M, Vuong T, Ancey PB, Ferrand M, Durand G, Le-Calvez Kelm F, Croce C, Matar C, Herceg Z, Hernandez-Vargas H. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genomics. 2013;14:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 54. | Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761-772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 322] [Cited by in RCA: 329] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 55. | Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, Thomas D, Lopez AF, Drew PA, Khew-Goodall Y. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J Cell Sci. 2013;126:2256-2266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 147] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 56. | Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, Gong C, Chen J, Su F, Zhang Y. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17:7105-7115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 203] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 57. | Qian P, Banerjee A, Wu ZS, Zhang X, Wang H, Pandey V, Zhang WJ, Lv XF, Tan S, Lobie PE. Loss of SNAIL regulated miR-128-2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res. 2012;72:6036-6050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 58. | Liu S, Patel SH, Ginestier C, Ibarra I, Martin-Trevino R, Bai S, McDermott SP, Shang L, Ke J, Ou SJ. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8:e1002751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 135] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 59. | Yu F, Jiao Y, Zhu Y, Wang Y, Zhu J, Cui X, Liu Y, He Y, Park EY, Zhang H. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J Biol Chem. 2012;287:465-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 60. | Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70:7176-7186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 61. | Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY, Lee WH. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463-2474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 62. | Corcoran C, Friel AM, Duffy MJ, Crown J, O’Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 63. | Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152-2159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 500] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 64. | Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010;127:1785-1794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 271] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 65. | Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079-31086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 402] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 66. | Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, Xiong W, Li G, Lu J, Fodstad O. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496-21507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 338] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 67. | Gottesman MM, Ling V. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett. 2006;580:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 387] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 68. | Cittelly DM, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE. Oncogenic HER2{Delta}16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31:2049-2057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 69. | Kato M, Paranjape T, Müller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419-2424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 181] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 70. | Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 250] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 71. | Reardon JT, Vaisman A, Chaney SG, Sancar A. Efficient nucleotide excision repair of cisplatin, oxaliplatin, and Bis-aceto-ammine-dichloro-cyclohexylamine-platinum(IV) (JM216) platinum intrastrand DNA diadducts. Cancer Res. 1999;59:3968-3971. [PubMed] |
| 72. | Xin F, Li M, Balch C, Thomson M, Fan M, Liu Y, Hammond SM, Kim S, Nephew KP. Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics. 2009;25:430-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 73. | Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 616] [Cited by in RCA: 683] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 74. | Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci USA. 2010;107:21098-21103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 296] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 75. | Thangaraju M, Kaufmann SH, Couch FJ. BRCA1 facilitates stress-induced apoptosis in breast and ovarian cancer cell lines. J Biol Chem. 2000;275:33487-33496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 115] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 76. | Yao E, Ventura A. A new role for miR-182 in DNA repair. Mol Cell. 2011;41:135-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 77. | Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, Lidereau R, Mikaélian I, Mazoyer S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3:279-290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 210] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 78. | Liang Z, Ahn J, Guo D, Votaw JR, Shim H. MicroRNA-302 replacement therapy sensitizes breast cancer cells to ionizing radiation. Pharm Res. 2013;30:1008-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 79. | Jameel JK, Rao VS, Cawkwell L, Drew PJ. Radioresistance in carcinoma of the breast. Breast. 2004;13:452-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 80. | Piva R, Spandidos DA, Gambari R. From microRNA functions to microRNA therapeutics: novel targets and novel drugs in breast cancer research and treatment (Review). Int J Oncol. 2013;43:985-994. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 81. | Ng EK, Li R, Shin VY, Siu JM, Ma ES, Kwong A. MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumour Biol. 2014;35:2591-2598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 82. | Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 438] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 83. | Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805-15810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1300] [Cited by in RCA: 1278] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 84. | Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove R, Xu R, Huang W. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148-2157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 177] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 85. | Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556-13561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 821] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 86. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 46962] [Article Influence: 3354.4] [Reference Citation Analysis (5)] |
| 87. | Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, Burow ME, Ivan M, Croce CM, Nephew KP. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30:1082-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 282] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 88. | Sachdeva M, Wu H, Ru P, Hwang L, Trieu V, Mo YY. MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene. 2011;30:822-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 89. | Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007;12:20-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 680] [Cited by in RCA: 728] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 90. | Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 330] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 91. | Ben-Hamo R, Efroni S. MicroRNA-gene association as a prognostic biomarker in cancer exposes disease mechanisms. PLoS Comput Biol. 2013;9:e1003351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 92. | Singh R, Mo YY. Role of microRNAs in breast cancer. Cancer Biol Ther. 2013;14:201-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 93. | Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 197] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 94. | Godfrey AC, Xu Z, Weinberg CR, Getts RC, Wade PA, Deroo LA, Sandler DP, Taylor JA. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 2013;15:R42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 95. | Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, Li J. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 2012;7:e47003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 96. | Madhavan D, Zucknick M, Wallwiener M, Cuk K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res. 2012;18:5972-5982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 203] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 97. | Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799-2803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1136] [Cited by in RCA: 1205] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 98. | Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 705] [Cited by in RCA: 798] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 99. | Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721-726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1744] [Cited by in RCA: 1650] [Article Influence: 91.7] [Reference Citation Analysis (0)] |
| 100. | Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 372] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 101. | Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z, Yang B. A single anti-microRNA antisense oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers an improved approach for microRNA interference. Nucleic Acids Res. 2009;37:e24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 102. | Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482-7484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 343] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 103. | Watashi K, Yeung ML, Starost MF, Hosmane RS, Jeang KT. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J Biol Chem. 2010;285:24707-24716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 104. | Brognara E, Fabbri E, Aimi F, Manicardi A, Bianchi N, Finotti A, Breveglieri G, Borgatti M, Corradini R, Marchelli R. Peptide nucleic acids targeting miR-221 modulate p27Kip1 expression in breast cancer MDA-MB-231 cells. Int J Oncol. 2012;41:2119-2127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 105. | Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, Fu J, Zeng MS, Yun JP, Wu QL. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011;13:R2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 262] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 106. | Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, Zhang JD, Wiemann S, Sahin Ö. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. 2012;32:633-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 199] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 107. | Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 278] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 108. | Gabriely G, Teplyuk NM, Krichevsky AM. Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy. 2011;7:1384-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 109. | Huang GL, Zhang XH, Guo GL, Huang KT, Yang KY, Shen X, You J, Hu XQ. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009;21:673-679. [PubMed] |
| 110. | Ward A, Balwierz A, Zhang JD, Küblbeck M, Pawitan Y, Hielscher T, Wiemann S, Sahin Ö. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32:1173-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 211] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 111. | de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, Malekpour F, Volinia S, Croce CM, Najmabadi H. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175-9184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 221] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 112. | Hildebrandt MA, Gu J, Lin J, Ye Y, Tan W, Tamboli P, Wood CG, Wu X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29:5724-5728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 164] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 113. | Khew-Goodall Y, Goodall GJ. Myc-modulated miR-9 makes more metastases. Nat Cell Biol. 2010;12:209-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 114. | Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 315] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 115. | Guo Y, Liu Y, Li Z, Wang D, Du Y, Chen J, Jin Z. EUS-guided implantation of radioactive iodine-125 seeds in retroperitoneal metastatic adenocarcinoma. Endoscopy. 2009;41 Suppl 2:E301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 116. | Guo L, Chen C, Shi M, Wang F, Chen X, Diao D, Hu M, Yu M, Qian L, Guo N. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene. 2013;32:5272-5282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 174] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 117. | Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1717] [Cited by in RCA: 1845] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 118. | Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, Kong R, Luo Y, Shi Y, Wang K. miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun. 2013;433:207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 119. | Cittelly DM, Das PM, Spoelstra NS, Edgerton SM, Richer JK, Thor AD, Jones FE. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9:317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 144] [Article Influence: 9.6] [Reference Citation Analysis (0)] |