INTRODUCTION
Miniature chromosome maintenance (MCM) proteins were initially identified from autonomously replicating sequences in Saccharomyces cerevisiae. Mutations of some of these proteins such as MCM7 or MCM3 in yeast result in loss of many yeast chromosomes. MCM7 cDNA encodes a 543-amino-acid protein and is ubiquitously expressed in all tissues. Initiation of DNA replication is a complex process that involves the concerted action of many proteins. Many studies have indicated that MCM7 is a crucial component of the DNA replication licensing complex in yeast and Xenopus[1-4]. Some studies have suggested that MCM4, 6 and 7 complexes contain DNA helicase activity[5,6]. The DNA replication licensing complex is multimeric and phase specific. In yeast, DNA replication licensing proteins such as MCM2-7 and several replication origin binding proteins such as Cdc6, geminin and Cdt1 form a DNA replication licensing complex in G1 phase, to enable DNA replication and promote cell cycle entry into S phase. Such complexes, however, dissipate in the S, G2 and M phases to prevent re-firing of DNA replication, and thus protect the integrity of the genome. There was little interest in the MCM complex as a target for oncogenic or tumor suppressor pathways until a link was found between MCM7 overexpression and amplification and several human malignancies[7-10].
MCM7 TRANSFORMING ONCOGENIC ACTIVITY
The initial implication of MCM7 involvement in human malignancies was positive immunostaining of MCM7 in several cancers, including endometrial carcinoma[11], melanoma[12], esophageal adenocarcinoma[9], colorectal adenocarcinoma[13], oral squamous cell carcinoma[14], glioblastoma[15], and thyroid cancer[16]. Most of these studies used MCM7 as a proliferation marker to compare with the existing markers such as Ki-67 or proliferating cell nuclear antigen. The first study to address the oncogenic role of MCM7 came from genome analysis of prostate cancer. By performing a genome-wide copy number analysis using biotin-labeled genome DNA on Affymetrix U133 2.0 chip[10], the DNA copy number of MCM7 was found to increase several-fold, accompanied by a concomitant increase in MCM7 mRNA level. Subsequent validation analyses have suggested that MCM7 copy number and/or protein level increase is associated with prostate cancer relapse and metastasis. Amplification of MCM7 has also been found in esophageal carcinoma[9]. The magnitude of MCM7 amplification correlates with the expression of MCM7, tumor grade, and aggressiveness of esophageal cancer[9]. It is presumed that amplification of MCM7 is the driving force of MCM7 overexpression in primary human malignancies. The oncogenic role of MCM7 was initially demonstrated by higher levels of cell growth and more invasiveness of MCM7-transformed prostate cancer cells (DU145) and in an animal xenograft model[10]. When MCM7 was transgened into murine skin basal cells utilizing a keratin promoter, the animals developed squamous cell carcinoma upon 7,12-dimethyl-benz[a]anthracene/phorbol ester 12-O-tetradecanoylphorbol-13 acetate (DMBA/TPA) challenge, compared with completely negative results from the wild-type controls[8]. However, MCM7 does not play an initiator role in cancer development, because organ-specific MCM7 transgenic mice develop no spontaneous cancer in skin or prostate cancer models[8,17]. The inability of MCM7 expression alone to initiate carcinogenesis could result from other negative feedback mechanisms that neutralize MCM DNA replication licensing in the cell, such as retinoblastoma (Rb) or integrin-linked kinase (ILK) signaling discussed later. Even though the transforming activity of MCM7 has been clearly observed, strictly speaking, it does not fall into the category of a typical proto-oncogene because it does not have a viral counterpart nor sequence mutations that confer its oncogenic potential. MCM7 probably falls into the broadly defined oncogene category whose gain of oncogenic function is generated by epigenome or chromosomal numerical alterations.
MCM7 AS A TARGET OF ONCOGENIC OR TUMOR SUPPRESSOR SIGNALING PATHWAYS
The first significant signaling pathway that targets MCM7 was found by yeast two-hybrid screening analysis in which the N terminus of Rb bait probe bound with MCM7[18]. Additional analysis has suggested that other Rb homologs p107 and p130 also bind with MCM7[18]. In vitro analysis has indicated that the binding of MCM7 and Rb inhibits DNA replication. However, the biological significance of such interaction was not elucidated until 11 years later, when it was shown that interaction of Rb and MCM7 is essential for transforming growth factor (TGF)-β-induced blockade of entry into S phase[19].
These studies suggest that MCM7 could be the main target of Rb in controlling the S phase check point (Figure 1). This is because overexpression of MCM7 can reverse the Rb inhibitory effect, and a peptide that interferes with MCM7/Rb binding but not other activity of Rb also reverses the Rb check point blockade. Another salient example of MCM7 as a target of a signaling pathway is androgen receptor (AR) signaling. It is well known that AR regulates cell growth and proliferation. However, most of the studies have focused on gene expression regulation, which might play a secondary role in controlling cell cycle progression. It was found later that AR interacts with MCM7 directly, and inactivates or activates MCM DNA replication licensing, depending on the nature of the ligands or their concentrations[20]. Mutation of MCM7 that abrogates its interaction with AR, but not its DNA replication licensing activity, eliminates the pleiotropic effect of testosterone. Furthermore, an AR mutant that does not bind with MCM7 can translocate into the nucleus upon androgen stimulation, but it fails to induce cell proliferation or to enhance transcription of androgen-dependent genes. It appears that DNA replication activity and transcription activity of AR are dependent on its binding with MCM7. It is likely that AR serves as a co-replication factor that directs the MCM complex DNA replication licensing through its interaction with MCM7 (Figure 1). One surprising finding is that MCM7 also serves as a co-transcription factor for AR. Several studies have shown that transcription activity enhances DNA replication, and that transcription activity is dependent on DNA replication in eukaryotic cells[21,22]. It remains to be seen whether MCM7-dependent transcription activity is maintained with other MCM7-interacting transcription factors. The MCM7 dependency of AR transcription activity suggests that androgen-dependent gene expression can only occur in actively proliferating cells, because MCM7 is excluded from the nucleus during S, G2 and M phases, and it only re-enters the nucleus in G1 phase.
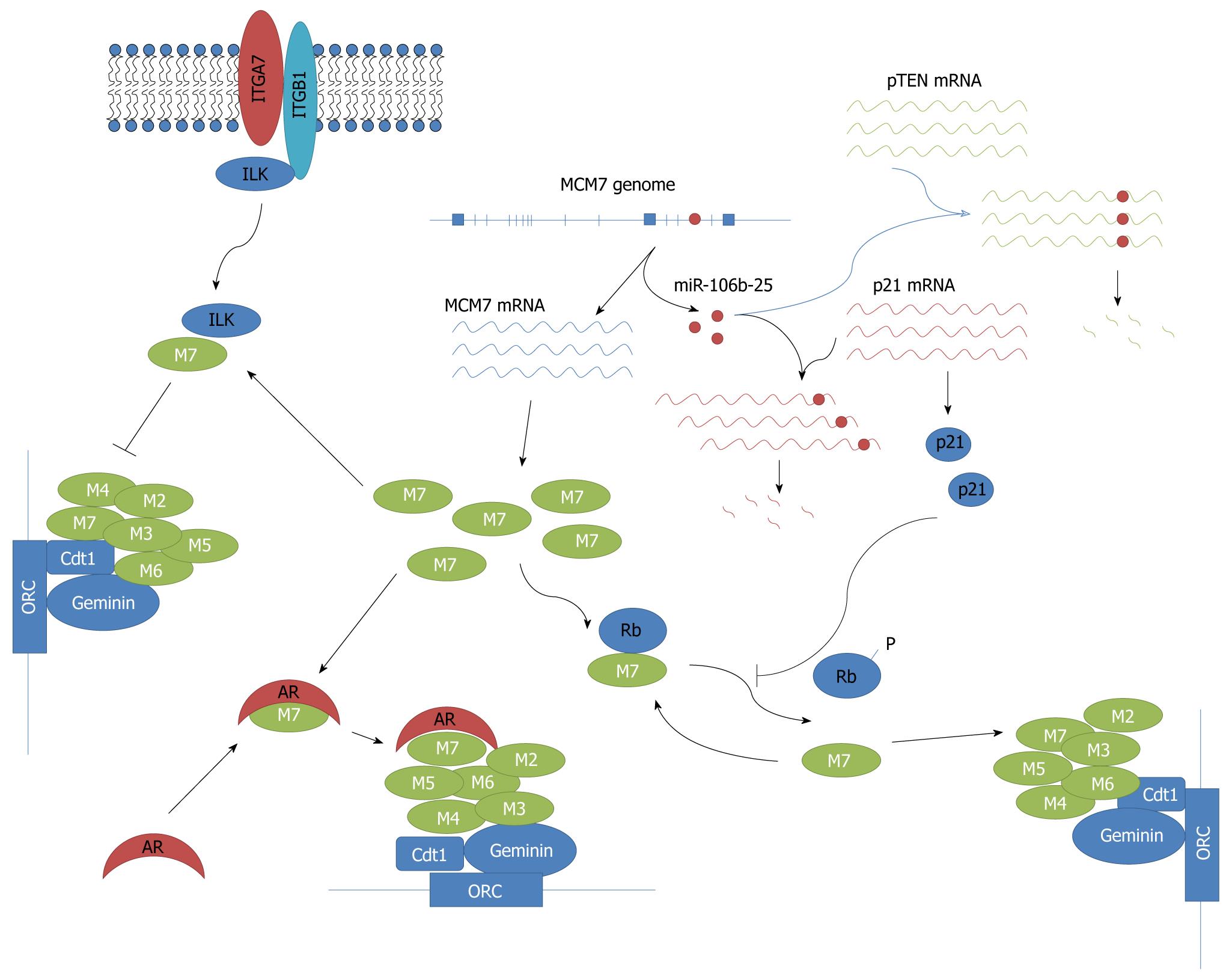
Figure 1 Diagram of miniature chromosome maintenance 7 oncogenic signaling.
Fifteen exons and miR106b-25 oncogenic miRNA cluster of miniature chromosome maintenance (MCM) 7 genome were transcribed into MCM7 mRNA and miRNA 106b, 93 and 25. Two-prong pathways were directed at promoting DNA replication: degradation of crucial tumor suppressor genes p21 and pTEN mRNA by the miR106b-25 cluster, and increased DNA replication licensing by MCM7 protein. Androgen receptor (AR) promotes DNA replication by enhancing MCM7 replication licensing activity. Integrin α7 inhibits DNA replication through inhibition of MCM7 activity. M7: MCM7; M6: MCM6; M5: MCM5; M4: MCM4; M3: MCM3; M2: MCM2; ITGA7: integrin α7; ILK: Integrin-linked kinase; ORC: Origin of replication complex; Rb: Retinoblastoma.
MCM7 also plays a crucial role in mediating the function of cell membrane receptors. This is demonstrated in its interaction with ILK (Figure 1). It appears that MCM7 is a substrate of ILK. The binding and phosphorylation of MCM7 N terminus by ILK reduce the binding of MCM7 with other DNA replication licensing factors, and lead to slower cell growth[23]. The phosphorylation of MCM7 by ILK has been proven to be a crucial link with the tumor suppressor activity of integrin α7. A dominant negative mutant of ILK interrupts the ILK/MCM7 interaction, and partly blocks the tumor suppression activity of integrin α7. An MCM7 mutant that lacks the ILK binding motif activates DNA replication licensing similar to that of wild-type, but is unresponsive to integrin α7 signaling. These findings suggest that MCM7 could be the end target of many oncogenic or tumor-suppressing signaling pathways. Depending on the nature of these interactions or modification of MCM7, it might lead to increased or decreased DNA replication licensing activity of the MCM complex, and guide the cells into a higher level of proliferation or cell growth arrest.
TRANSFORMING miRNA CLUSTER IN MCM7 GENOME
A unique feature of the MCM7 genome is that it contains an intronic miRNA miR-106b-25 cluster in intron 13, which includes miR-106b, miR93 and miR-25. miR-25 is highly homologous to miR-32, a onco-miRNA, whereas miR-106b and miR-93 belong to the miR-17 family. All three members of the miR-106b-25 cluster are abundantly expressed in most prostate cancer cell lines. Similar to their host gene MCM7, amplification and upregulation of all members of the miR-106b-25 cluster are found in several human malignancies, including esophageal and prostate cancers[9,17,24-26]. These miRNAs target multiple tumor suppressor genes, and shut down their expression levels (Figure 1). One of the most notable examples is pTEN gene expression. miR-25 and miR-93 expression decrease pTEN protein levels, and result in activation of the Akt pathway[17]. The expression of the miR106-25 cluster increases tumorigenesis in anchorage-independent assays and animal tumor xenograft models. Mice with knock-in prostate-specific MCM7 and miR-106-25 cluster develop cancer like dysplasia, whereas mice with pure MCM7 knock-in do not appear to develop dysplasia without carcinogen challenge. One could interpret such observations as showing that the miR-106-25 cluster functions as a tumor initiator by knocking down pTEN in mice, whereas overexpression of MCM7 serves as a mechanism for the development of invasive phenotype. The miR-106b-25 cluster appears more versatile than knocking down just one tumor suppressor gene. Several studies have suggested that miR-106 and miR-25 target p21, Bim and E2F1[24,25,27]. These targets and their relationship have been clearly demonstrated in prostate cancer, esophageal adenocarcinoma and gastric cancer. Inhibition of E2F1 by miR-106b and miR-93 in gastric cancer cell lines partially blocks TGF-β-induced apoptosis[27]. E2F1 is a strong stimulator of MCM7 locus transcription. As a result, a negative feedback loop of MCM7/miR-106b-25 is formed. Such a feedback loop might serve to limit the excessive cell death induced by TGF-β under physiological conditions, and thus achieve a balance of cell growth and cell death. In the event of malignancy, amplification of the MCM7 locus might bypass such a negative feedback loop, and tip the balance towards cell survival. Targeting of p21waf and bim by miR-106b and miR-25 has been demonstrated in esophageal and prostate cancers. Inhibitors of miR-106, miR-93 or miR-25 inhibit tumorigenesis in vitro and in vivo. With these targets that all appear crucial for the oncogenic activity of the miR-106b-25 cluster, it is not easy to analyze the contribution of each of these pathways to carcinogenesis, because there has been a lack of experiments to block off each of the pathways to offset the oncogenic activity of the miR-106b-25 cluster. The miR-25/93-pTEN pathway is compelling because of the inverse correlation of pTEN expression and miR-106b-25 miRNA levels in primary and mouse tumor samples. The animal model initiates prostate cancer with the MCM7/miR-106b-25 cluster transgene, which shows clear downregulation of pTEN expression and a concomitant increase in miR-106b-25 cluster expression, and consequent Akt pathway activation. It would be of interest to see how much oncogenic phenotype is reversed if these animals are transgened with pTEN, p21, BIM or E2F1 gene constructs that lack the miR-106b-25 target sequences in their 3’ untranslated regions.
POTENTIAL THERAPEUTIC TARGET
MCM7 is essential for any cell that undergoes proliferation. This poses a dilemma for MCM7-gene-targeted therapy. Nevertheless, one study has indicated that shRNA that targets MCM7 in PC3 and DU145 tumor xenografts in mice dramatically reduces tumor volume, rate of metastasis and fatality[28]. The drawback of this analysis is that the shRNA target might not recognize the MCM7 sequence from mice because the target sequence is intended for human MCM7. Nevertheless, the study is a proof of principle that MCM7 knockdown is a potential effective approach in combating prostate cancer, particularly that with MCM7 amplification and overexpression. In light of the double oncogenic effect of the MCM7 genome cluster, it might be more effective if a shRNA is designed to neutralize the effect of miR-106b, miR-93 and miR-25 onco-miRNA. This can be accomplished with a large dosage of morpholino oligonucleotide that is specific for these onco-miRNAs. These RNase-resistant oligonucleotides have a clear advantage over genomic approaches in that they do not interfere with the genome structure of a cell, but they do have a long-lasting presence once they have been taken. Furthermore, there is no evidence to suggest that knocking down these miRNAs adversely affects the survival of normal cells.
Peer reviewers: Shiaw-Yih Lin, PhD, Associate Professor, Department of Systems Biology, The University of Texas MD Anderson Cancer Center, South Campus Research Building II, 7435 Fannin Street, Unit 950, Houston, TX 77054, United States; Melanie H Kucherlapati, PhD, Instructor, Associate Geneticist, Department of Medicine, Division of Genetics, Harvard Medical School, Brigham and Women’s Hospital, 77 Avenue Louis Pasteur, NRB 160B, Boston, MA 02115, United States
S- Editor Cheng JX L- Editor Kerr C E- Editor Ma WH