Published online May 24, 2024. doi: 10.5306/wjco.v15.i5.653
Revised: April 1, 2024
Accepted: April 18, 2024
Published online: May 24, 2024
Processing time: 157 Days and 13.1 Hours
Colonization with Helicobacter pylori (H. pylori) has a strong correlation with gastric cancer, and the virulence factor CagA is implicated in carcinogenesis. Studies have been conducted using medicinal plants with the aim of eliminating the pathogen; however, the possibility of blocking H. pylori-induced cell differentiation to prevent the onset and/or progression of tumors has not been addressed. This type of study is expensive and time-consuming, requiring in vitro and/or in vivo tests, which can be solved using bioinformatics. Therefore, prospective com
To perform a computational prospecting of the interactions between phenolic compounds from medicinal plants and the CagA oncoprotein of H. pylori.
In this in silico study, the structures of the phenolic compounds (ligands) kae
The structure model obtained for the CagA oncoprotein exhibited high quality (C-score = 0.09) and was validated using parameters from the MolProbity platform. The GRaSP online platform identified 24 residues (phenylalanine and leucine) as potential binding sites on the CagA oncoprotein. Molecular docking simulations were conducted with the three-dimensional model of the CagA oncoprotein. No complexes were observed in the simulations between the carboxy-terminus region of CagA and the phenolic compounds; however, all phenolic compounds interacted with the central region of the oncoprotein. Phenolic compounds and CagA exhibited significant affinity energy (-7.9 to -9.1 kcal/mol): CagA/kaempferol formed 28 chemical bonds, CagA/myricetin formed 18 chemical bonds, CagA/quercetin formed 16 chemical bonds, CagA/ponciretin formed 13 chemical bonds, and CagA/ch
In silico, the tested phenolic compounds formed stable complexes with CagA. Therefore, they could be tested in vitro and/or in vivo to validate the findings, and to assess interference in CagA/cellular target interactions and in the oncogenic differentiation of gastric cells.
Core Tip: Commonly, studies on the effects of medicinal plants on Helicobacter pylori (H. pylori) infection assess the antimicrobial activity of these plants. However, in this study, the authors conducted a prospective in silico analysis of the activity of certain phenolic compounds from plants used to treat H. pylori infection on stomach cells affected by CagA, aiming to prevent or block the oncogenic differentiation of these cells.
- Citation: Vieira RV, Peiter GC, de Melo FF, Zarpelon-Schutz AC, Teixeira KN. In silico prospective analysis of the medicinal plants activity on the CagA oncoprotein from Helicobacter pylori. World J Clin Oncol 2024; 15(5): 653-663
- URL: https://www.wjgnet.com/2218-4333/full/v15/i5/653.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i5.653
Colonization of gastric epithelial cells by Helicobacter pylori (H. pylori) has a strong positive correlation with gastric diseases such as peptic ulcers and stomach cancer, owing to the virulence factors and evasion capabilities of the bacteria[1]. The standard treatment for eradicating H. pylori is complex and relies on antibiotics[2,3], proton pump inhibitors[4], bismuth salts[5], and H2 blockers[6], typically used in combination for an extended duration[7].
Multiple factors contribute to hindering the eradication of H. pylori, among which drug inefficiency and the bacteria's antibiotic resistance are prominent[7]. In 2017, the World Health Organization classified H. pylori as resistant to clarithromycin, metronidazole, and levofloxacin, emphasizing the urgent need for research into new antibiotics targeting the bacteria[8].
In this context, medicinal plants and their secondary metabolites have emerged as an alternative for managing H. pylori infection. The literature reports several plant species with antimicrobial activity against H. pylori, which act by inhibiting, reducing, and delaying gastric colonization. Plants such as Pistacia lentiscus, Brassica oleracea, Curcuma longa, Coptis chinensis, and Glycyrrhiza glabra were tested in rodents (mice and rats); Vaccinium macrocarpon, Glycyrrhiza glabra, and Nigella sativa were tested in humans, with observed results indicating biological activity against the bacteria and the progression of infection[9]. Dinat et al[10] discuss the use of medicinal plants in treating H. pylori infection and identified antimicrobial activity in several of them, such as Hibiscus sabdariffa and Piper longum.
Commonly, the use of medicinal plants to treat H. pylori infection aims at antimicrobial action to eliminate the path
Although studies have primarily focused on antimicrobial activity, medicinal plants and their metabolites may also interfere with pathogenic cellular processes induced by H. pylori. In cases of infection with CagA-positive H. pylori strains, where the colonization process is advanced, it would be crucial not only to eliminate the pathogen but also to block the cell differentiation induced by CagA to prevent the development of cancer. Therefore, addressing the treatment of H. pylori infection through targeting the CagA would be of interest.
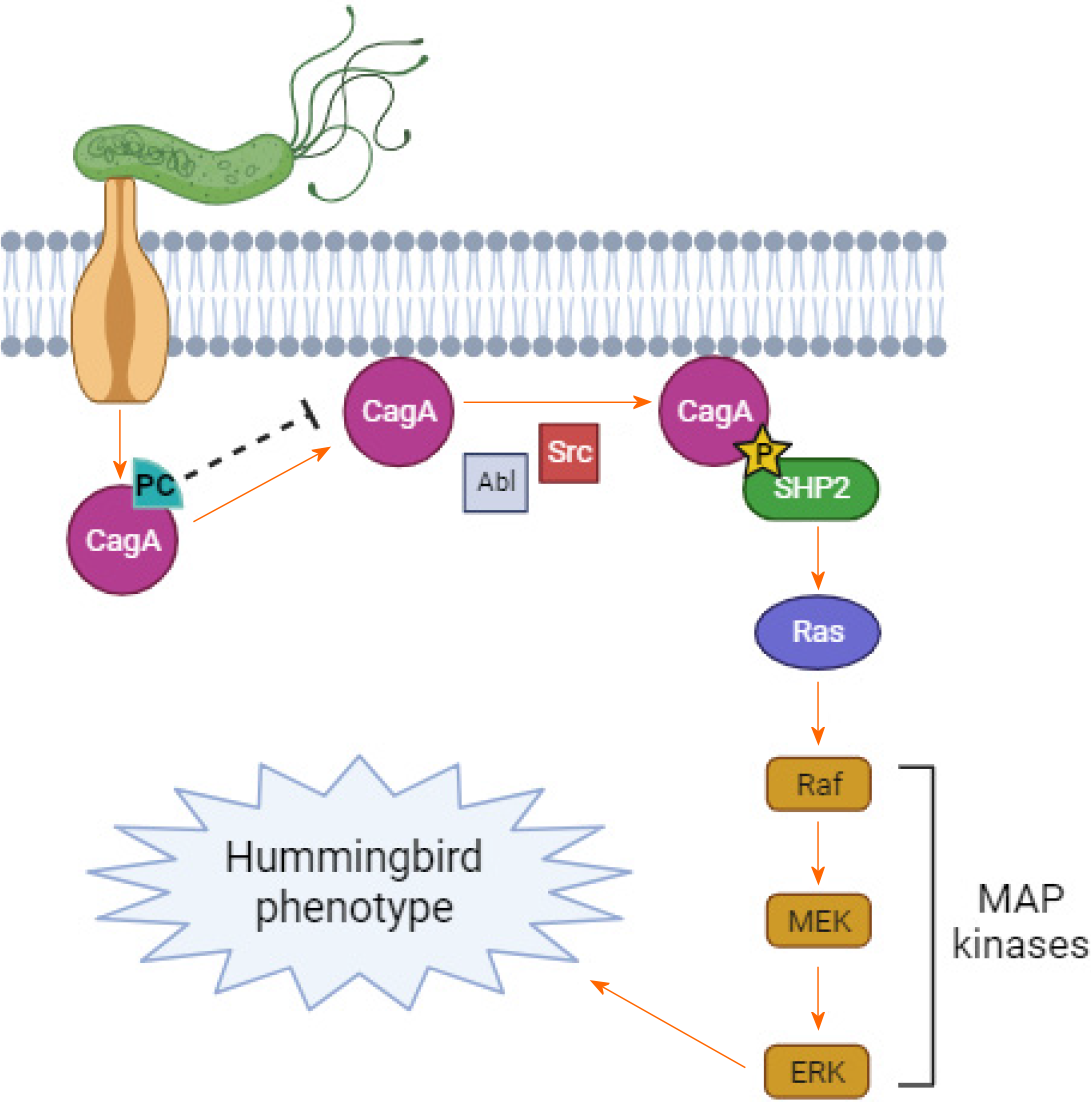
The CagA is an oncoprotein, a virulence factor of H. pylori responsible for inducing genetic mutations and alterations in gastric cells[19]. This oncoprotein is encoded by the pathogenicity island (cag-PAI) and is transported into cells via a type 4 secretion system (T4SS)[20]. Inside cells, CagA attaches to the plasma membrane in two distinct ways: Through the interaction of basic amino acids, including those in the K-Xn-R-X-R binding motif located in the central region of CagA, or through the carboxy-terminus region. Subsequently, CagA undergoes tyrosine phosphorylation on the EPIYA (Glu-Pro-Ile-Tyr-Ala) motif, which is present in multiple copies in the carboxy-terminus polymorphic region, by Src kinase members such as c-Src, Yes, Fyn, and Abl kinase[21]. Phosphorylated CagA interferes with cell signaling pathways, including the MAP kinase pathway, leading to mitogenic imbalance, induction of a pro-inflammatory state, cytoskeleton damage, and disruption of cell-cell junctions[20,22,23].
The carboxy-terminus end of the CagA oncoprotein also contains the CagA-multimerization (CM) motif, which facilitates its dimerization or multimerization. CM motif consists of 16 amino acid residues and is located immediately distal to the last EPIYA segment. Apart from facilitating multimerization, the CM motif enables CagA to interact with regulatory molecules that affect proper cell signaling[21]. The cumulative effect of dysregulated cell signaling, disordered cell growth, endothelial injury, and loss of mucosal integrity caused by the CagA oncoprotein predisposes individuals to precancerous lesions in the stomach, explaining the higher incidence of cancer in individuals colonized with CagA-positive H. pylori[22].
Therefore, approaches could be explored to interfere with the CagA-cellular targets interaction, aiming to reduce or inhibit the activity of this protein and, consequently, its oncogenic potential. Phenolic compounds from plants are capable of crossing the plasma membrane of human cells and interacting with proteins and enzymes of cellular signaling cascades, altering the course of signal transduction[24]. Thus, this study aimed a prospective computational analysis of the action of certain phenolic compounds from medicinal plants, which are reported in the treatment of H. pylori infe
Molecular modeling of the CagA oncoprotein was carried out using the primary sequence BAK52797.1 of the CagA oncoprotein from H. pylori, which contains 1194 residues including the EPIYA-A, B, C, and CM motifs, in FASTA format, selected from GenBank (ncbi.nlm.nih.gov/genbank). The FASTA sequence was utilized to search for X-ray diffraction solved three-dimensional structures deposited in online public databases. However, the structures found in the databases did not include either the amino-terminus or the carboxy-terminus ends of the CagA oncoprotein. Global and local alignment analyses of CagA were performed using the BioEdit software (Informer Technologies, Inc.) and the Basic Local Alignment Search Tool algorithm (National Institutes of Health/United States), respectively. Since no satisfactory hom
The modeled CagA oncoprotein was analyzed using the GRaSP online platform[27] to predict residues that could serve as potential binding sites. This data will be compared with the results obtained from molecular docking.
Chemical compounds selected as ligands were phenolic compounds, and their two-dimensional structures were obtained from the PubChem Database (pubchem.ncbi.nlm.nih.gov)-kaempferol (PubChem: 6325460), myricetin (PubChem: 5281672), quercetin (PubChem: 5280343), ponciretin (PubChem: 25201019), and chlorogenic acid (PubChem: 1794427). The two-dimensional structures were converted to three-dimensional form using PyMol software (Schrödinger, Inc.). The protonation state of each phenolic compound at physiological pH 7.4 was predicted using MarvinSketch software (ChemAxon) before proceeding with docking simulations. All phenolic compounds were selected from studies involving medicinal plants used for the treatment of H. pylori infection[28,29]. AutoDock Tools 4 (ADT) software[30] was used to detect and calculate the points and angles of torsion, respectively. The CagA oncoprotein was prepared using the same software to add missing hydrogen atoms.
Molecular docking was performed using the flexible ligand-rigid receptor methodology[31]. The simulations were conducted by associating ADT with AutoDock Vina v.1.2.0 software[32], enabling the establishment of an algorithm that searches for potential bond combinations, including rotational, translational, and conformational degrees of freedom. This algorithm also establishes scoring criteria to select the best ligand-receptor interactions. Points are assigned according to the molecular force field and the free energy of the bond, with interactions considered stable if the affinity energy is lower than -6.0 kcal/mol[33]. Two sets of molecular docking simulations were carried out: (1) Central region of CagA + phenolic compound; and (2) carboxy-terminus region of CagA + phenolic compound. The membrane binding motif (K-Xn-R-X-R) is located in the central region, while the EPIYA and CM motifs are in the carboxy-terminus region of the CagA oncoprotein. Gridbox dimensions and coordinates were determined separately to allocate the central region and the carboxy-terminus end of the CagA (receptor). Evaluation of the receptor/ligand complexes was performed using PyMol and BIOVIA Discovery Studio (Dassault Systemes) software. Physicochemical analysis of the residues involved in the chemical bonds was carried out using the ProtParam tool[34]. The data was analyzed using descriptive statistics (mean and relative values).
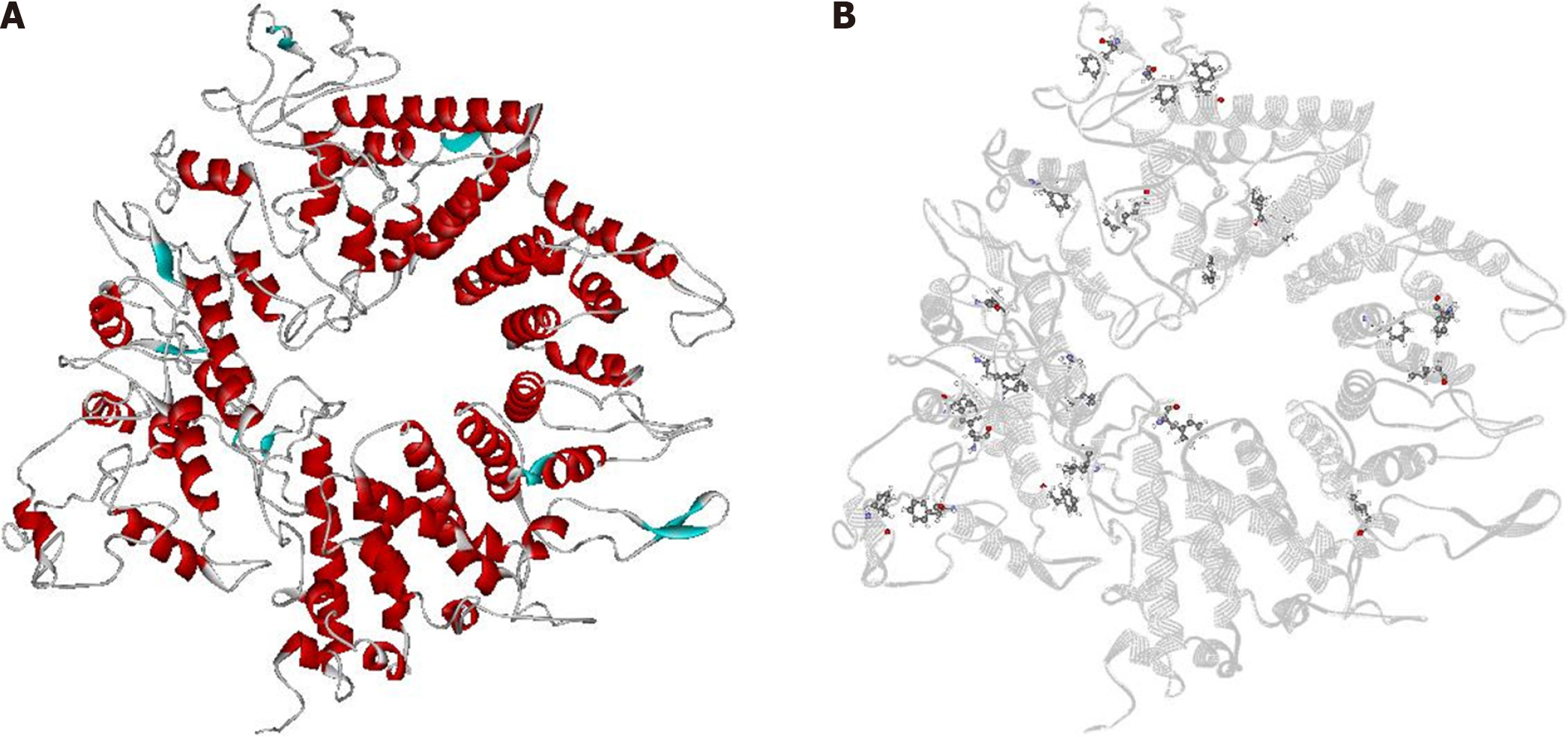
Five structural models were constructed based on 10 three-dimensional structures, and the most satisfactory model, with a C-score value of 0.09, was selected (Figure 1A). The C-score assesses the quality of models predicted by I-Tasser; it typically ranges from -5 to 2, with higher values indicating models with higher reliability. MolProbity indicated that approximately 98% of the amino acid residues in the model were located within the allowed regions of the Rama
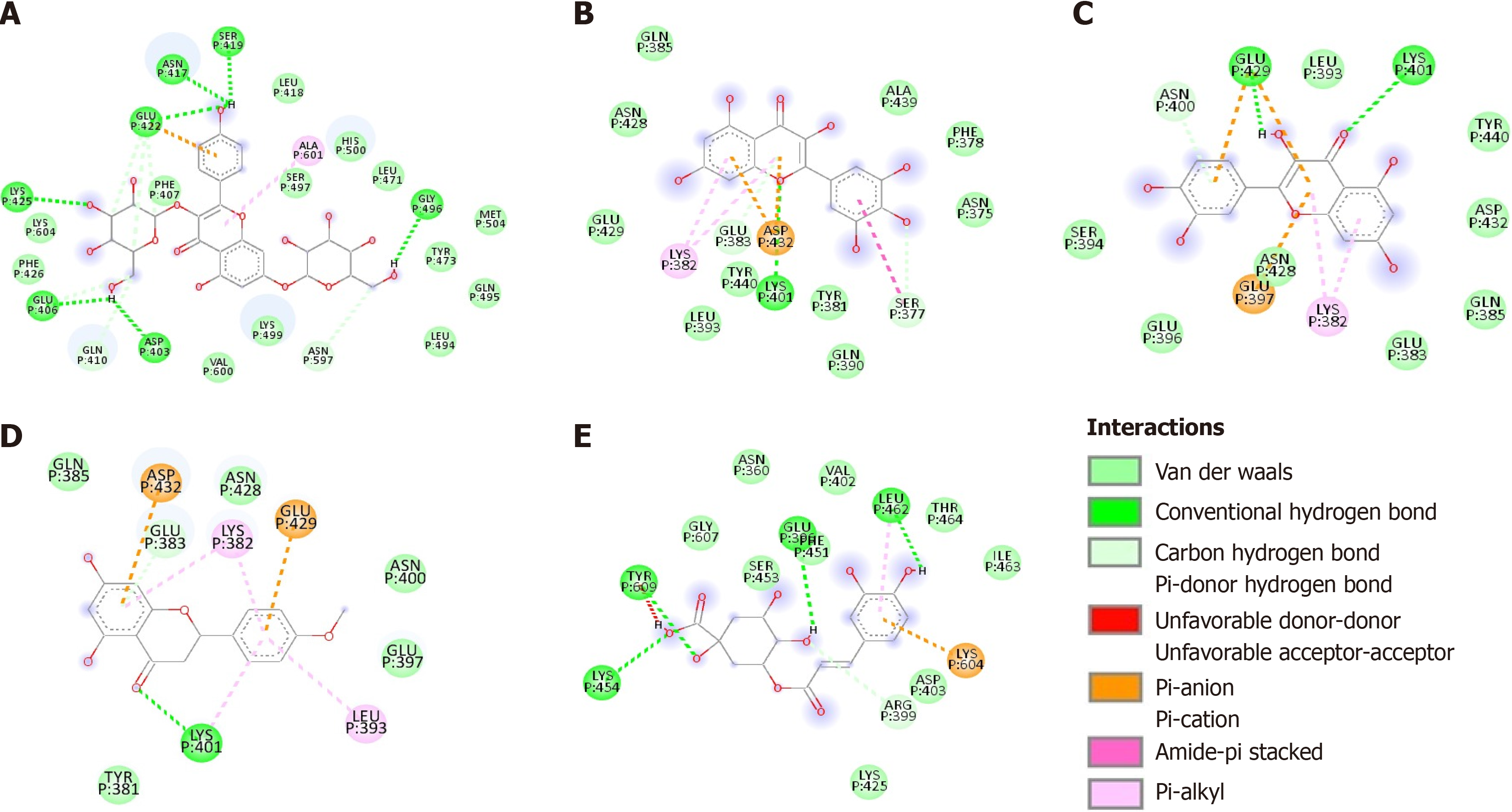
A total of 24 amino acid residues were predicted as possible binding sites for ligands. The algorithm indicated 19 phenylalanine (F) and 5 leucine (L) residues with a probability above 50% of interacting with other molecules. This value represents the algorithm's cut-off point. Among the 24 residues, only one coincided with the molecular docking results (F426) (Table 1). The residues are distributed across the surface of CagA, and no clusters were observed (Figure 1B).
| Molecular docking | Binding sites by GRaSP | |||||
| Phenolic compounds | ||||||
| Kaempferol | Myricetin | Quercetin | Ponciretin | Chlorogenic acid | Residue1/position | |
| Residue1/position | A601 | A439 | D432 | D432 | D403 | F23 |
| D403 | D432 | N400 | E383 | E396 | F31 | |
| E406 | E383 | E383 | E397 | F451 | F41 | |
| E422 | E429 | E396 | E429 | G607 | F60 | |
| F407 | F378 | E397 | K382 | I463 | F88 | |
| F4262 | K382 | E429 | K401 | K425 | F122 | |
| G496 | K401 | K382 | L393 | K604 | F161 | |
| H500 | L393 | K401 | N400 | L454 | F188 | |
| K425 | N375 | L393 | N428 | L462 | F269 | |
| K499 | N428 | N428 | Q385 | N360 | F291 | |
| K604 | Q385 | Q385 | Y381 | R399 | F4262 | |
| L418 | Q390 | S394 | S453 | F537 | ||
| L471 | S377 | Y440 | T464 | F543 | ||
| L494 | Y381 | V402 | F582 | |||
| M504 | Y440 | Y609 | F596 | |||
| N417 | F637 | |||||
| N597 | F702 | |||||
| Q410 | F805 | |||||
| Q495 | F818 | |||||
| S419 | L226 | |||||
| S497 | L260 | |||||
| V600 | L688 | |||||
| Y473 | L835 | |||||
| L964 | ||||||
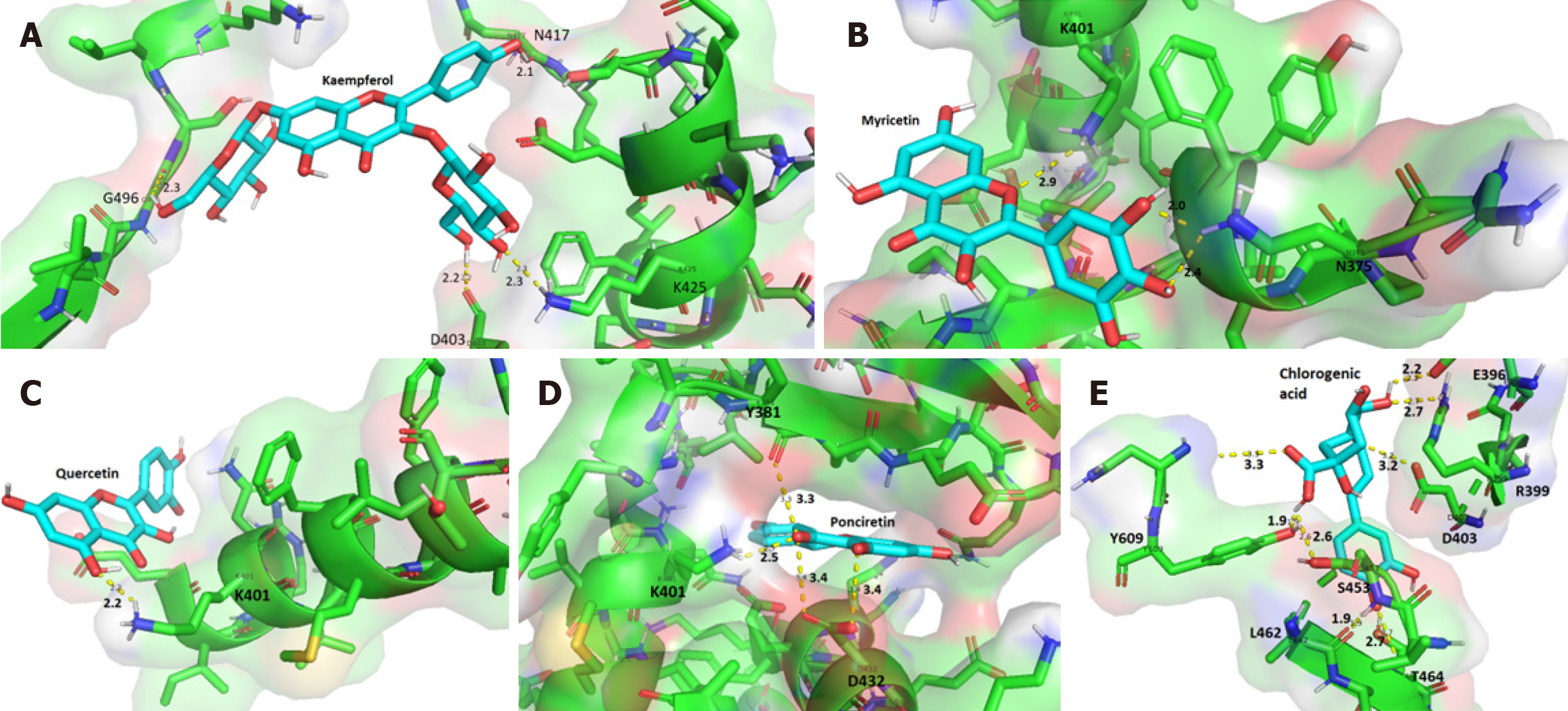
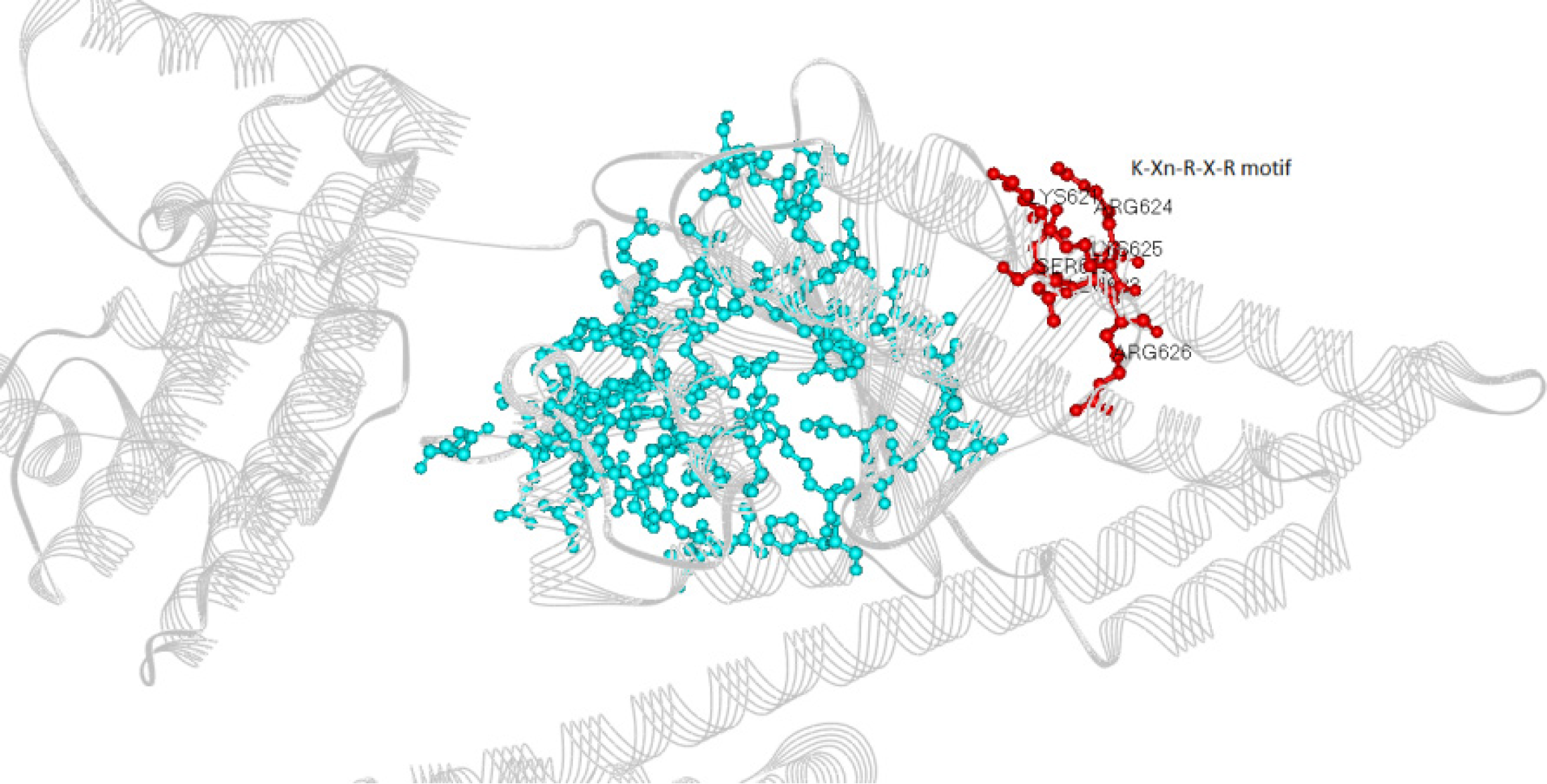
The phenolic compounds analyzed formed complexes with the CagA oncoprotein in silico (Figure 2); all five compounds bound to the central region of the protein close to the spatial region of the membrane-binding motif (residues 621 to 626) (Figure 3). No in silico interactions were observed between the phenolic compounds and the carboxy-terminal region of CagA where the EPIYA motifs are located. The affinity energies were considered satisfactory and ranged from -7.9 kcal/mol (CagA/chlorogenic acid) to -9.1 kcal/mol (CagA/kaempferol). The other complexes showed similar affinity energy values-CagA/quercetin = -8.1 kcal/mol, CagA/myricetin = -8.2 kcal/mol, CagA/ponciretin = -8.4 kcal/mol. In all the complexes, covalent bonds and reversible bonds (hydrogen bond and van der Waals) were observed. The CagA/kaempferol complex showed 28 bonds (2.1Å - 2.3Å) involving 23 amino acid residues; this was the complex with the highest number of chemical bonds. The CagA/myricetin complex showed 18 bonds (2.0Å - 2.9Å) involving 15 residues; CagA/quercetin complex-16 bonds (2.2Å) involving 13 residues; CagA/ponciretin complex-13 bonds (2.5Å - 3.4Å) involving 11 residues; CagA/chlorogenic acid complex-17 bonds (1.9Å - 3.3Å) involving 15 residues. All the chemical bonds present in each complex are shown in Figure 4 in a two-dimensional diagram, and the residues are listed in Table 1. Fifty-three amino acid residues participated in chemical bonds with at least one of the phenolic compounds analyzed in silico. Of these, eight made bonds with two compounds, and eight made bonds with three compounds, most of which were positively (L) or negatively (D/E) charged residues.
H. pylori strains harboring the gene CagA-CagA-positive strains-significantly increase the risk of developing gastric cancer when compared to CagA-negative strains[35]. The cellular modifications triggered by the CagA oncoprotein, after being injected into stomach epithelial cells, involve the EPIYA motifs becoming targets for phosphorylation and recr
The results of this in silico study suggest that, after being injected into the cytoplasm of gastric epithelial cells via T4SS, the CagA oncoprotein can interact with xenobiotics that cross the plasma membrane of these cells, such as phenolic compounds from medicinal plants. The computational search for binding sites in CagA indicated several phenylalanine and leucine residues with this potential. Since these residues did not form clusters, it is possible that they represent the starting point for ligand binding, and the site itself is composed of more residues that become closer together after ligand binding due to a conformational change; in other words, the ligand would actually be an allosteric modulator[36].
Due to the chemical nature of the side chains of these residues, they are expected to form hydrophobic bonds with compounds of a similar chemical nature. This increases the likelihood of secondary metabolites that are more hydr
The phenolic compounds evaluated in this in silico study (ponciretin, kaempferol, quercetin, myricetin, and chlorogenic acid) bound to the central region of the CagA oncoprotein, near the membrane phospholipid-binding motif-K-Xn-R-X-R motif. Due to the proximity of the bonds formed by the phenolic compounds to this motif and the high interaction affinity considered, these compounds could induce conformational changes in the protein, thus destabilizing its previous interactions with cellular targets. Even if the carboxy-terminus region, where the EPIYA and CM motifs are found, does not bind to the analyzed phenolic compounds, the interaction between CagA and the plasma membrane is still necessary for the phosphorylation process. Therefore, interference through the membrane-binding domain could be effective in blocking this process.
Given the consideration of phenolic compounds as an alternative for interfering with CagA oncoprotein activity within epithelial cells, it is important to note that these compounds are absorbed in the human intestine through the action of bile salts, passive diffusion, or transporters[38]. After absorption, phenolic compounds can undergo conjugation with glucuronic acid in enterocytes or hepatocytes, or they can circulate in the bloodstream bound to albumin[39]. Conjugated phenolic compounds can cross cell membranes through carrier proteins and can reach different tissues[38], including the stomach.
Therefore, based on the in silico results and the capacity of phenolic compounds to penetrate cells, these polyphenols have potential for subsequent in vitro and in vivo studies for the treatment of gastric H. pylori infections. They not only exhibit antimicrobial activity, as described for plants containing these compounds, but also have the ability to bind to and destabilize the interaction between CagA and epithelial cells. This interference could potentially prevent the initiation of changes leading to malignant transformation of gastric cells, such as the activation of PAR1/MARK kinases causing loss of cell polarization, and the inactivation of p53 resulting in uncontrolled and disordered cell proliferation[20,29].
A study by Castillo-Juárez et al[40] demonstrated the anti-H. pylori activity of some plants commonly used in trad
Szewczyk et al[41], in a separate study, investigated the antimicrobial properties of plants from the Balsaminaceae family, including the species Impatiens glandulifera, (Himalayan balsam), through an in vitro study. The study revealed that I. glandulifera exhibits high concentrations of phenolic acids, particularly in its aerial parts, and significant antioxidant and antimicrobial activity against Staphylococcus aureus, S. epidermidis, Micrococcus luteus, Bacillus subtilis, B. cereus, Streptococcus pneumoniae, and S. pyogenes. However, direct studies confirming the antimicrobial activity of I. glandulifera against H. pylori are lacking.
Nevertheless, Vieira et al[28] isolated some flavonoids from this plant through chromatography-kaempferol, quercetin, and myricetin-which, according to our findings, demonstrate in silico potential as interferents of the CagA oncoprotein. Among these, due to its binding site, kaempferol appears to be the most effective in destabilizing the interaction between CagA and the epithelial cell membrane. Therefore, I. glandulifera warrants further exploration to evaluate its efficacy in halting precancerous changes induced by CagA-positive H. pylori strains.
The plant Buddleja indica, known for its richness in kaempferol, caffeic acid (a metabolite of chlorogenic acid), and quercetin, is reported to possess anti-diabetic, hepatoprotective, antioxidant, and antimicrobial properties[42,43]. Youssef et al[42] highlighted the bacteriostatic activity of B. indica against H. pylori in an in vitro study. Given that this plant contains two of the compounds discussed in this study, it could be explored in further research on anti-carcinogenic therapy.
Other plants that could be investigated for the presence of the phenolic compounds discussed in this study include Polygonum tinctorium, or indigo (kaempferol, quercetin, and caffeic acid), known for their bactericidal and bacteriostatic activity[44,45]; Rubus ulmifolius, or blackberry (kaempferol, quercetin, and caffeic acid), with bactericidal action[46,47]; Poncirus trifoliata (ponciretin), exhibiting bacteriostatic action[48-50]; Oliveira decumbens (kaempferol), and Hibiscus rosa-sinensis (myricetin, quercetin, kaempferol), with anti-urease and bacteriostatic activity[51-53]. In our study, among the phenolic compounds tested, kaempferol and chlorogenic acid appear to be the most promising candidates for interfering with the interaction between CagA and the phospholipids of the plasma membrane, as they exhibit lower affinity energies, indicating greater stability of the complexes.
The studies correlating flavonoids/phenolic acids and H. pylori primarily focus on the action of these compounds as bactericides, bacteriostats, and anti-urease agents, rather than addressing virulence factors that directly damage host cells, such as CagA. Most research aims to identify alternatives to antibiotics due to the process of bacterial resistance. Indeed, the eradication of H. pylori infection is crucial due to its pathogenic factors, as well as the physiological and biochemical mechanisms that can lead to the malignancy of gastric cells[54].
The utilization of bioinformatics in scientific research is increasingly contributing to the study of molecular interactions[55]. Among these studies, González et al[56] investigated several flavonoids, including kaempferol, quercetin, and myricetin, for their ability to bind to and inactivate the homeostatic stress regulatory protein, yielding promising results. As this protein is crucial for fundamental H. pylori activities such as energy metabolism and genetic material replication, its inactivation results in either bacterial death or reduced multiplication. This study aligns with the same rationale as the present work, wherein the binding of compounds to bacterial components leads to detrimental implications for path
Inhibiting the action of CagA has the potential to halt the progression of gastric H. pylori lesions to malignancy, as this protein disrupts multiple pathways regulating cellular homeostasis. Upon entry into cells through T4SS, phosphorylation of EPIYA motifs occurs, leading to the recruitment of various molecules to the plasma membrane of gastric cells, thereby modulating and altering multiple cell signaling pathways[57]. EPIYA-C motifs are phosphorylated by Src family kinases; H. pylori strains containing higher numbers of EPIYA-C motifs are associated with an increased likelihood of gastric cancer emergence. CagA interacts with the tyrosine phosphatase S H. pylori-2 and potentiates the action of the Erk-MAP kinase, with or without utilizing the Ras protein, while also inactivating the focal adhesion kinase FAK. This pathway results in the hummingbird phenotype, characterized by a rearrangement of the cytoskeleton in gastric cells, leading to enhanced cell motility and elongation[58,59], and is implicated in the malignancy process[60]. Therefore, since the tran
Since the molecular events leading to the hummingbird phenotype are dependent on the action of the Cag oncoprotein, blocking this protein could prevent the cellular oncogenic process. One way to block it would be to prevent CagA from interacting with the plasma membrane of gastric cells, which could be achieved by using a phenolic compound, as sugg
Indeed, a study on the post-translational processing of the CagA oncoprotein revealed that this process may be involved in the pathogenesis of H. pylori infection, as CagA fragmentation alters its functionality, thereby reducing the induction of the hummingbird phenotype[61]. This suggests that interventions targeting CagA may reduce carcinogenic predisposition, similar to the observations made in this in silico study. It is a fact that the tertiary conformation of a protein is determined by the interaction between its amino acid residues, and the binding, whether transient or otherwise, of external molecules to this assembly can lead to a state of inactivity[62]. Thus, by binding to the central region of CagA, the phenolic compounds in this study, especially kaempferol and chlorogenic acid, may induce conformational and functional changes in this virulence factor.
Finally, the present study has some limitations regarding its application in clinical treatments, as the functioning of the human organism is complex, involving a myriad of biochemical and physiological cascades that interact to achieve homeostasis. Therefore, since computational tools simulate only some physiological parameters, the environment in which the interaction between CagA and phenolic compounds was tested does not fully reflect the physiological envir
The in silico data suggest that phenolic compounds (flavonoids and phenolic acids) present in medicinal plants used to treat H. pylori infection bind to the CagA oncoprotein in its central region, close to the membrane anchoring site. Furthermore, none of the amino acid residues of CagA predicted as binding sites are involved in the interaction with the phenolic compounds analyzed in this study. It is possible that these residues interact with other secondary metabolites in medicinal plants, which would increase the chance of interfering with CagA's action. Therefore, medicinal plants have the potential to eliminate H. pylori infection due to their antimicrobial activities already proven in the literature and could also interfere with the action of CagA after being injected into the gastric epithelial cell. This interference could affect the cell differentiation process that culminates in the hummingbird phenotype and, consequently, prevent and/or block the onset of gastric cancer. It is important to emphasize that this is a computational study and, therefore, has limitations; thus, the data obtained must be analyzed in vitro and in vivo to validate the findings.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country of origin: Brazil
Peer-review report’s classification
Scientific Quality: Grade A, Grade B
Novelty: Grade A, Grade B
Creativity or Innovation: Grade B, Grade B
Scientific Significance: Grade A, Grade B
P-Reviewer: Zhu Y, China S-Editor: Qu XL L-Editor: A P-Editor: Zhao YQ
| 1. | Kamboj AK, Cotter TG, Oxentenko AS. Helicobacter pylori: The Past, Present, and Future in Management. Mayo Clin Proc. 2017;92:599-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 2. | Fera MT, Giannone M, Pallio S, Tortora A, Blandino G, Carbone M. Antimicrobial activity and postantibiotic effect of flurithromycin against Helicobacter pylori strains. Int J Antimicrob Agents. 2001;17:151-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 3. | Boyanova L. Comparative evaluation of two methods for testing metronidazole susceptibility of Helicobacter pylori in routine practice. Diagn Microbiol Infect Dis. 1999;35:33-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Park JB, Imamura L, Kobashi K. Kinetic studies of Helicobacter pylori urease inhibition by a novel proton pump inhibitor, rabeprazole. Biol Pharm Bull. 1996;19:182-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 5. | Midolo PD, Norton A, von Itzstein M, Lambert JR. Novel bismuth compounds have in vitro activity against Helicobacter pylori. FEMS Microbiol Lett. 1997;157:229-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Sorba G, Bertinaria M, Di Stilo A, Gasco A, Scaltrito MM, Brenciaglia MI, Dubini F. Anti-Helicobacter pylori agents endowed with H2-antagonist properties. Bioorg Med Chem Lett. 2001;11:403-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 7. | Lamont JT. Treatment regimens for Helicobacter pylori in adults. [cited May 20, 2023]. Available from: https://www.uptodate.com/contents/treatment-regimens-for-helicobacter-pylori-in-adults?search=Helicobacter%20pylori&source=search_result&selectedTitle=1~150&usage_type=default&display_rank=1#H2609998802. |
| 8. | Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology. 2018;155:1372-1382.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 820] [Article Influence: 117.1] [Reference Citation Analysis (0)] |
| 9. | Sathianarayanan S, Ammanath AV, Biswas R, B A, Sukumaran S, Venkidasamy B. A new approach against Helicobacter pylori using plants and its constituents: A review study. Microb Pathog. 2022;168:105594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 10. | Dinat S, Orchard A, Van Vuuren S. A scoping review of African natural products against gastric ulcers and Helicobacter pylori. J Ethnopharmacol. 2023;301:115698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 11. | Wang YC. Medicinal plant activity on Helicobacter pylori related diseases. World J Gastroenterol. 2014;20:10368-10382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 84] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (2)] |
| 12. | Chen AY, Chen YC. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013;138:2099-2107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 537] [Cited by in RCA: 602] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 13. | Singh RP, Agarwal R. Flavonoid antioxidant silymarin and skin cancer. Antioxid Redox Signal. 2002;4:655-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 91] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 14. | Brownson DM, Azios NG, Fuqua BK, Dharmawardhane SF, Mabry TJ. Flavonoid effects relevant to cancer. J Nutr. 2002;132:3482S-3489S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 65] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Chan FL, Choi HL, Chen ZY, Chan PS, Huang Y. Induction of apoptosis in prostate cancer cell lines by a flavonoid, baicalin. Cancer Lett. 2000;160:219-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 166] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 16. | Kuo SM. Dietary flavonoid and cancer prevention: evidence and potential mechanism. Crit Rev Oncog. 1997;8:47-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 115] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 17. | Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: chemistry, bioavailability and effects on health. Nat Prod Rep. 2009;26:1001-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1316] [Cited by in RCA: 1215] [Article Influence: 75.9] [Reference Citation Analysis (0)] |
| 18. | Ullah A, Munir S, Badshah SL, Khan N, Ghani L, Poulson BG, Emwas AH, Jaremko M. Important Flavonoids and Their Role as a Therapeutic Agent. Molecules. 2020;25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 577] [Article Influence: 115.4] [Reference Citation Analysis (0)] |
| 19. | Poorolajal J, Moradi L, Mohammadi Y, Cheraghi Z, Gohari-Ensaf F. Risk factors for stomach cancer: a systematic review and meta-analysis. Epidemiol Health. 2020;42:e2020004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 166] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 20. | Oliveira AKS, Silva LLL, Miguel MP, Blanco AJV, Carneiro LC, Barbosa MS. HELICOBACTER PYLORI cagA VIRULENCE GENE AND SEVERE ESOGASTRODUODENAL DISEASES: IS THERE AN ASSOCIATION? Arq Gastroenterol. 2021;58:468-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Hatakeyama M. Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93:196-219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 160] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 22. | Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015;10:955-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 155] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 23. | Nesić D, Miller MC, Quinkert ZT, Stein M, Chait BT, Stebbins CE. Helicobacter pylori CagA inhibits PAR1-MARK family kinases by mimicking host substrates. Nat Struct Mol Biol. 2010;17:130-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 24. | Rahman MM, Rahaman MS, Islam MR, Rahman F, Mithi FM, Alqahtani T, Almikhlafi MA, Alghamdi SQ, Alruwaili AS, Hossain MS, Ahmed M, Das R, Emran TB, Uddin MS. Role of Phenolic Compounds in Human Disease: Current Knowledge and Future Prospects. Molecules. 2021;27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 489] [Cited by in RCA: 328] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 25. | Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods. 2021;1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 310] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 26. | Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB 3rd, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2746] [Cited by in RCA: 2994] [Article Influence: 427.7] [Reference Citation Analysis (0)] |
| 27. | Santana CA, Silveira SA, Moraes JPA, Izidoro SC, de Melo-Minardi RC, Ribeiro AJM, Tyzack JD, Borkakoti N, Thornton JM. GRaSP: a graph-based residue neighborhood strategy to predict binding sites. Bioinformatics. 2020;36:i726-i734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Vieira MN, Winterhalter P, Jerz G. Flavonoids from the flowers of Impatiens glandulifera Royle isolated by high performance countercurrent chromatography. Phytochem Anal. 2016;27:116-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Gutiérrez-Rebolledo GA, Garduño-Siciliano L, García-Rodríguez RV, Pérez-González MZ, Chávez MI, Bah M, Siordia-Reyes GA, Chamorro-Cevallos GA, Jiménez-Arellanes MA. Anti-inflammatory and toxicological evaluation of Moussonia deppeana (Schldl. & Cham) Hanst and Verbascoside as a main active metabolite. J Ethnopharmacol. 2016;187:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785-2791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19127] [Cited by in RCA: 16216] [Article Influence: 1013.5] [Reference Citation Analysis (0)] |
| 31. | Lengauer T, Rarey M. Computational methods for biomolecular docking. Curr Opin Struct Biol. 1996;6:402-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 417] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 32. | Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J Chem Inf Model. 2021;61:3891-3898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1065] [Cited by in RCA: 2715] [Article Influence: 678.8] [Reference Citation Analysis (0)] |
| 33. | Pantsar T, Poso A. Binding Affinity via Docking: Fact and Fiction. Molecules. 2018;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 318] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 34. | Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 1016] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 35. | Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111-2115. [PubMed] |
| 36. | Saito A, Alvi S, Valant C, Christopoulos A, Carbone SE, Poole DP. Therapeutic potential of allosteric modulators for the treatment of gastrointestinal motility disorders. Br J Pharmacol. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 37. | Paduch R, Kandefer-Szerszeń M, Trytek M, Fiedurek J. Terpenes: substances useful in human healthcare. Arch Immunol Ther Exp (Warsz). 2007;55:315-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 209] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 38. | Meoti FC. Análise dos mecanismos de ação antinociceptiva e antinflamatória do flavonóide miricetina: estudos in vivo e in vitro. UFSM 2006. Available from: https://repositorio.ufsm.br/handle/1/4391. |
| 39. | Murota K, Terao J. Antioxidative flavonoid quercetin: implication of its intestinal absorption and metabolism. Arch Biochem Biophys. 2003;417:12-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 356] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 40. | Castillo-Juárez I, González V, Jaime-Aguilar H, Martínez G, Linares E, Bye R, Romero I. Anti-Helicobacter pylori activity of plants used in Mexican traditional medicine for gastrointestinal disorders. J Ethnopharmacol. 2009;122:402-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 41. | Szewczyk K, Zidorn C, Biernasiuk A, Komsta Ł, Granica S. Polyphenols from Impatiens (Balsaminaceae) and their antioxidant and antimicrobial activities. Ind Crops Prod. 2016;86:262-272. [DOI] [Full Text] |
| 42. | Youssef FS, Altyar AE, Omar AM, Ashour ML. Phytoconstituents, In Vitro Anti-Infective Activity of Buddleja indica Lam., and In Silico Evaluation of its SARS-CoV-2 Inhibitory Potential. Front Pharmacol. 2021;12:619373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 43. | Elhady SS, Youssef FS, Alahdal AM, Almasri DM, Ashour ML. Anti-Hyperglycaemic Evaluation of Buddleia indica Leaves Using In Vitro, In Vivo and In Silico Studies and Its Correlation with the Major Phytoconstituents. Plants (Basel). 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Jang HG, Heo BG, Park YS, Namiesnik J, Barasch D, Katrich E, Vearasilp K, Trakhtenberg S, Gorinstein S. Chemical composition, antioxidant and anticancer effects of the seeds and leaves of indigo (Polygonum tinctorium Ait.) plant. Appl Biochem Biotechnol. 2012;167:1986-2004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 45. | Kataoka M, Hirata K, Kunikata T, Ushio S, Iwaki K, Ohashi K, Ikeda M, Kurimoto M. Antibacterial action of tryptanthrin and kaempferol, isolated from the indigo plant (Polygonum tinctorium Lour.), against Helicobacter pylori-infected Mongolian gerbils. J Gastroenterol. 2001;36:5-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 104] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 46. | da Silva LP, Pereira E, Pires TCSP, Alves MJ, Pereira OR, Barros L, Ferreira ICFR. Rubus ulmifolius Schott fruits: A detailed study of its nutritional, chemical and bioactive properties. Food Res Int. 2019;119:34-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 47. | Martini S, D'Addario C, Colacevich A, Focardi S, Borghini F, Santucci A, Figura N, Rossi C. Antimicrobial activity against Helicobacter pylori strains and antioxidant properties of blackberry leaves (Rubus ulmifolius) and isolated compounds. Int J Antimicrob Agents. 2009;34:50-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 48. | Bae EA, Han MJ, Kim DH. In vitro anti-Helicobacter pylori activity of some flavonoids and their metabolites. Planta Med. 1999;65:442-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 86] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 49. | Kim MJ, Kim BG, Ahn JH. Biosynthesis of bioactive O-methylated flavonoids in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:7195-7204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 50. | Kang GD, Kim DH. Ponciretin attenuates ethanol-induced gastric damage in mice by inhibiting inflammatory responses. Int Immunopharmacol. 2017;43:179-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 51. | Eftekhari M, Shams Ardekani MR, Amin M, Mansourian M, Saeedi M, Akbarzadeh T, Khanavi M. Anti-Helicobacter pylori Compounds from Oliveria decumbens Vent. through Urease Inhibitory In-vitro and In-silico Studies. Iran J Pharm Res. 2021;20:476-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 52. | Tran Trung H, Truong Thi Huynh H, Nguyen Thi Thuy L, Nguyen Van Minh H, Thi Nguyen MN, Luong Thi MN. Growth-Inhibiting, Bactericidal, Antibiofilm, and Urease Inhibitory Activities of Hibiscus rosa sinensis L. Flower Constituents toward Antibiotic Sensitive- and Resistant-Strains of Helicobacter pylori. ACS Omega. 2020;5:20080-20089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 53. | Mejía JJ, Sierra LJ, Ceballos JG, Martínez JR, Stashenko EE. Color, Antioxidant Capacity and Flavonoid Composition in Hibiscus rosa-sinensis Cultivars. Molecules. 2023;28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 54. | Stein M, Rappuoli R, Covacci A. The cag Pathogenicity Island. In: Helicobacter pylori: Physiology and Genetics. 2001. [PubMed] |
| 55. | Verli H. Bioinformática: da biologia à flexibilidade molecular. 2014; 1-12 Available from: https://lume.ufrgs.br/bitstream/handle/10183/166105/001012172.pdf?sequence=1&isAllowed=y. |
| 56. | González A, Salillas S, Velázquez-Campoy A, Espinosa Angarica V, Fillat MF, Sancho J, Lanas Á. Identifying potential novel drugs against Helicobacter pylori by targeting the essential response regulator HsrA. Sci Rep. 2019;9:11294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 57. | Tegtmeyer N, Backert S. Role of Abl and Src family kinases in actin-cytoskeletal rearrangements induced by the Helicobacter pylori CagA protein. Eur J Cell Biol. 2011;90:880-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 58. | Bessède E, Mégraud F. Microbiota and gastric cancer. Semin Cancer Biol. 2022;86:11-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 57] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 59. | Tohidpour A. CagA-mediated pathogenesis of Helicobacter pylori. Microb Pathog. 2016;93:44-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 60. | Fidler R, Sokolov AV, Verbitskiĭ MSh, Papazov IP. [Monoclonal antibodies against the beta subunit of human chorionic gonadotropin]. Biull Eksp Biol Med. 1988;106:693-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 388] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 61. | Chang CC, Kuo WS, Chen YC, Perng CL, Lin HJ, Ou YH. Fragmentation of CagA Reduces Hummingbird Phenotype Induction by Helicobactor pylori. PLoS One. 2016;11:e0150061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 62. | Nelson DL. Princípios de bioquímica de Lehninger. 6ed. 2014; 115-151. |