Published online Mar 24, 2024. doi: 10.5306/wjco.v15.i3.391
Peer-review started: October 17, 2023
First decision: December 31, 2023
Revised: January 14, 2024
Accepted: February 3, 2024
Article in press: February 3, 2024
Published online: March 24, 2024
Processing time: 156 Days and 20 Hours
Ferroptosis has recently been associated with multiple degenerative diseases. Ferroptosis induction in cancer cells is a feasible method for treating neoplastic diseases. However, the association of iron proliferation-related genes with prognosis in HER2+ breast cancer (BC) patients is unclear.
To identify and evaluate fresh ferroptosis-related biomarkers for HER2+ BC.
First, we obtained the mRNA expression profiles and clinical information of HER2+ BC patients from the TCGA and METABRIC public databases. A four-gene prediction model comprising PROM2, SLC7A11, FANCD2, and FH was subsequently developed in the TCGA cohort and confirmed in the METABRIC cohort. Patients were stratified into high-risk and low-risk groups based on their median risk score, an independent predictor of overall survival (OS). Based on these findings, immune infiltration, mutations, and medication sensitivity were analyzed in various risk groupings. Additionally, we assessed patient prognosis by combining the tumor mutation burden (TMB) with risk score. Finally, we evaluated the expression of critical genes by analyzing single-cell RNA sequencing (scRNA-seq) data from malignant vs normal epithelial cells.
We found that the higher the risk score was, the worse the prognosis was (P < 0.05). We also found that the immune cell infiltration, mutation, and drug sensitivity were different between the different risk groups. The high-risk subgroup was associated with lower immune scores and high TMB. Moreover, we found that the combination of the TMB and risk score could stratify patients into three groups with distinct prognoses. HRisk-HTMB patients had the worst prognosis, whereas LRisk-LTMB patients had the best prognosis (P < 0.0001). Analysis of the scRNA-seq data showed that PROM2, SLC7A11, and FANCD2 were significantly differentially expressed, whereas FH was not, suggesting that these genes are expressed mainly in cancer epithelial cells (P < 0.01).
Our model helps guide the prognosis of HER2+ breast cancer patients, and its combination with the TMB can aid in more accurate assessment of patient prognosis and provide new ideas for further diagnosis and treatment.
Core Tip: A prognostic model constructed with four ferroptosis-related genes (PROM2, SLC7A11, FANCD2, and FH) combined with tumor mutation burden can be used to evaluate the prognosis of patients with HER2-positive breast cancer more accurately.
- Citation: Shi JY, Che X, Wen R, Hou SJ, Xi YJ, Feng YQ, Wang LX, Liu SJ, Lv WH, Zhang YF. Ferroptosis biomarkers predict tumor mutation burden's impact on prognosis in HER2-positive breast cancer. World J Clin Oncol 2024; 15(3): 391-410
- URL: https://www.wjgnet.com/2218-4333/full/v15/i3/391.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i3.391
Breast cancer (BC) is the most prevalent malignancy in the world and the primary cause of cancer-related deaths in women[1]. As a highly heterogeneous disease, BC has four molecular subtypes: Basal/triple-negative, luminal A, luminal B, and HER2-positive[2]. HER2 is an orphan tyrosine kinase receptor that regulates cell proliferation and survival when activated. Located at chromosome 17q12, the HER2 oncogene is amplified in 15-20% of all BCs[3]. The primary and essential mechanism of HER2 receptor overexpression is amplification[4,5]. Due to its role in cell proliferation, invasion, and survival, this mechanism confers a poor prognosis. Standard treatment modalities include surgery combined with chemotherapy, radiotherapy, endocrine therapy, and HER2-targeted therapy which are widely used in clinical practice[5]. For the HER2-positive subtype, HER2-targeted treatments, such as trastuzumab, pertuzumab, T-DM1, DS8201, and tyrosine kinase inhibitors (TKIs), can significantly improve disease-free survival and overall survival (OS)[6,7]. Notably, not all patients derive equal benefits from existing anti-HER2 therapies, and HER2-positive BC is inherently heterogeneous. Although numerous studies have focused on investigating the prognostic significance of ferroptosis-related genes in BC[8-10], analyses specific to BC subtypes are lacking. A more comprehensive understanding of tumor biology and the HER2 signaling pathway is essential for advancing novel strategies to improve patient outcomes.
A new type of controlled cell death known as ferroptosis differs from apoptosis, necrosis, and autophagy in morphology, biochemistry, and genetics[11]. It is characterized by disruption of the intracellular redox balance and nonapoptotic cell death. Previous studies revealed that the NAD (P)H/FSP1/CoQ10 and cyst (e)ine/GSH/glutathione peroxidase 4 (GPX4) signaling pathways control ferroptosis. Ferroptosis is caused by the buildup of lipid peroxidation products and reactive oxygen species generated from iron metabolism. Increasing evidence suggests that ferroptosis is closely related to many diseases, especially HER2+ BC[12]. Thus, ferroptosis has gained popularity as a potential therapeutic strategy to promote cancer cell death. Various studies have reported ferroptosis induction by afatinib and lapatinib[13]. However, the association between iron proliferation-related genes and prognosis in HER2+ BC patients has yet to be determined, hindering practical clinical assessment before treatment.
This research systematically analyzed HER2+ BC expression data and clinical information from the TCGA and METABRIC cohorts. Furthermore, we identified genes associated with ferroptosis that were differentially expressed in patient tissues compared with normal tissues, screened four signatures related to survival, and constructed a consistent prediction model. We also explored the associations of ferroptosis with immune cell infiltration, mutations, and immune checkpoints in HER2+ BC patients. These results provide a foundation for developing comprehensive therapeutic strategies for HER2+ BC patients.
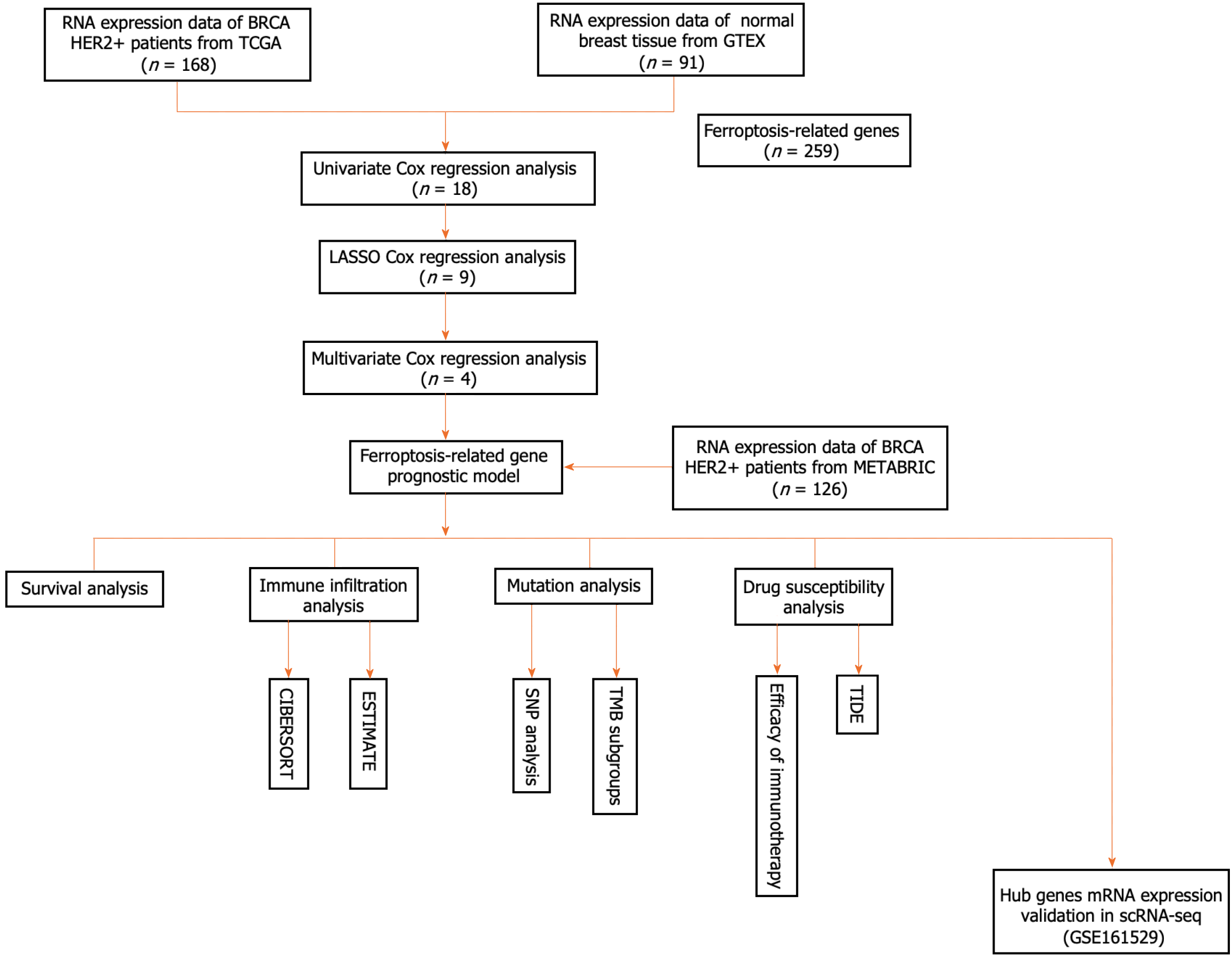
We used HER2+ BC datasets downloaded from the TCGA (https://protal.gdc.cancer.gov/repository) and the METABRIC databases (www.cbioportal.org/). The downloaded data were filtered using the following criteria: (1) Histologically diagnosed with malignant BC; (2) complete corresponding clinical data; and (3) available OS data for more than 90 d. Additional average breast tissue mRNA expression data (n = 91) were obtained from GTEx (https://gtexportal.org/home/datasets). The final sample included 168 patients from the TCGA cohort and 126 patients from the METABRIC cohort with complete follow-up information. A total of 259 genes associated with ferroptosis were retrieved from the FerrDb website (http://www.zhounan.org/ferrdb/current/) and are reported in Supplementary Table 1 (marker: 111; Driver: 108; suppressor: 69). The single-cell RNA sequencing (scRNA-seq) dataset was assessed from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo).
The TCGA and METABRIC cohorts were selected as training and validation sets, respectively. First, the log2 transformation approach was used after the raw count data had been normalized using the transcripts per million method. The number of DEGs was calculated using the “DESeq2” package (P < 0.05) in R. Venn was used to identify DEGs associated with ferroptosis. Univariate Cox regression analysis was performed to identify ferroptosis-related prognostic genes, and relevant genes were selected using a cutoff of P < 0.05. In total, 18 genes were chosen for the minor absolute shrinkage and selection operator (LASSO) Cox regression. Multivariate Cox regression analysis was subsequently applied to further assess the significant factors. We used lasso-penalized Cox regression analysis with the "glmnet" package in R to choose prognostic ferroptosis-related genes and construct a predictive model. The following formula was used to generate the risk score: Risk score = sum (corresponding coefficient × expression level of the gene). The expression levels of the genes were normalized, and the regression coefficients were calculated from the training set. Patients were then categorized into high- or low-risk groups according to the median risk score. The "Rtsne" package was used to run t-SNE to investigate the distribution of various groups. Using the "survminer" package in R, Kaplan-Meier (K-M) curves were created to predict OS. The "survival receiver operating characteristic (ROC)" package in R was used to run a time-dependent ROC curve analysis to evaluate the ability of the signature genes to predict survival.
To determine the relevant immune cell infiltration patterns and immunological characteristics, the CIBERSORT algorithm was used. The "ESTIMATE" program was applied to estimate the tumor purity scores. We examined the expression of immune-related signal transduction pathway components in various risk groups. We compared the estimated immune and stromal scores between the high- and low-risk groups. We also examined the underlying mechanisms in two risk subgroups using TCGA gene mutation data. The "MAFtools" package of R was used to evaluate SNP mutations and visualize the results. The tumor mutation burden (TMB) was subsequently determined. Patients were separated into two groups based on the median TMB value: The high TMB group and the low TMB group. Subsequently, the TMB score was combined with the risk score to form a new subgroup.
Our study included specific well-known immune checkpoint genes to evaluate gene expression levels across various risk score groups. Drug susceptibility was predicted using information from the GDSC database (https://www.cancerrxgene.org/celllines). The half-maximal inhibitory concentration (IC50) indicated the patient’s drug response and was calculated using the "pRRophetic" package.
scRNA-seq data (GSE161529) from 6 HER2-positive BC patients and 13 healthy controls were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Subsequently, low-quality cells were removed using the following criteria: (1) Had a number of expressed genes lower than 500; (2) Had a number of expressed genes higher than 2500; or (3) Had a proportion of mitochondria larger than 10%. The R "Seurat" package was used for cell cluster analysis. Cellular regions were manually annotated based on marker gene expression patterns and cell subset grouping patterns. The expression levels of the hub genes were subsequently displayed in each cell subset. Additionally, we validated the differential expression profiles of prognostic genes in the epithelial cells of patients and healthy controls using the tool “FindMarkers.”
R 4.1.0 was used to perform all the statistical analyses. A log-rank test and K-M analysis were used to compare OS among various risk subgroups. The primary prognostic variables connected to OS were identified using univariate and multivariate Cox regression analyses. Continuous and categorical variables were compared in the training and verification sets using Spearman correlation analysis. Unless otherwise stated, all the statistical tests were two-sided, and values with P < 0.05 were considered to indicate statistical significance.
A flowchart of our research is shown below (Figure 1). In this study, 168 patients with the HER2+ subtype of BC from the TCGA database served as the training cohort, whereas 126 patients from the METABRIC cohort were enrolled as the validation cohort. Supplementary Table 2 summarizes the clinical features of the two cohorts.
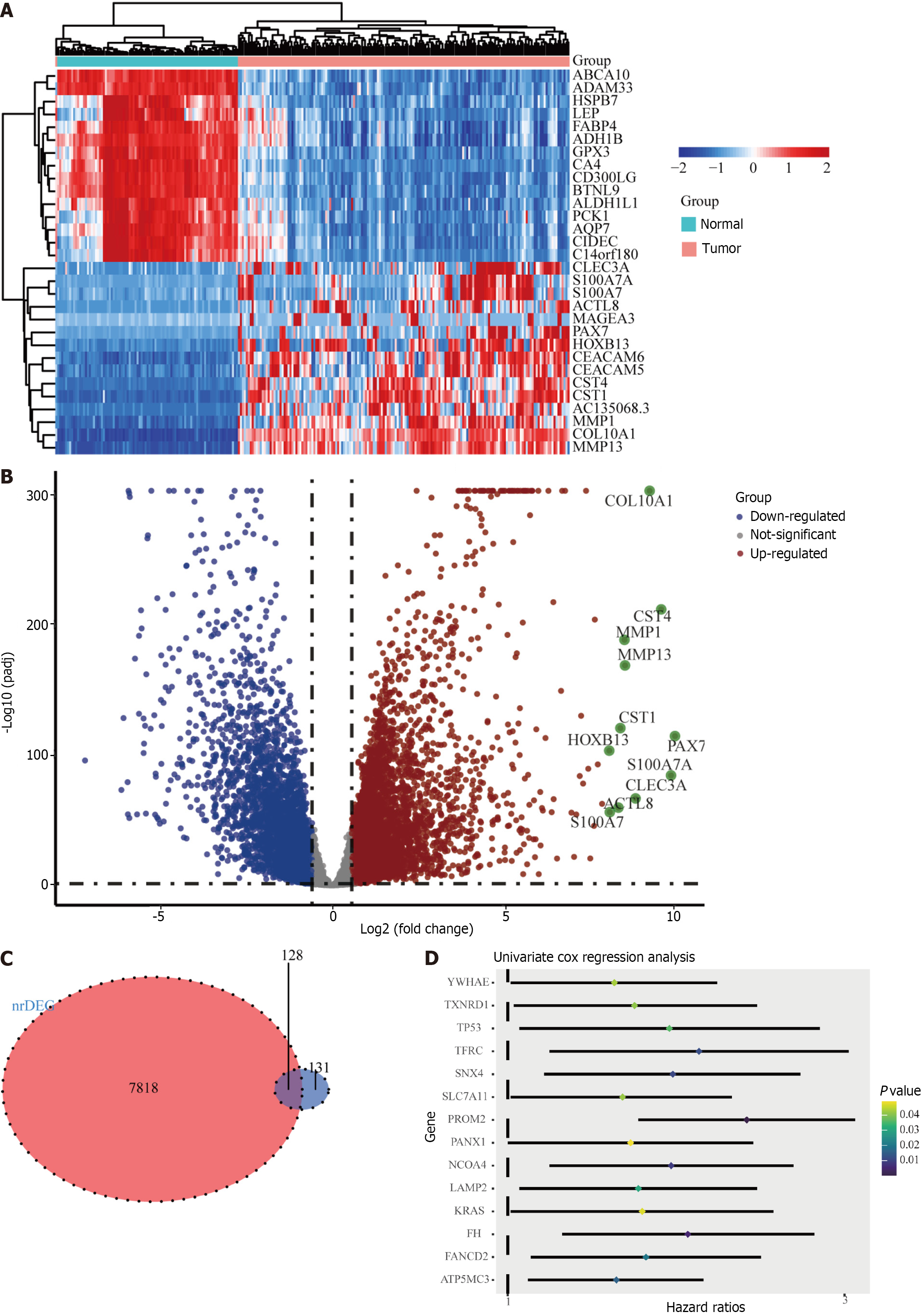
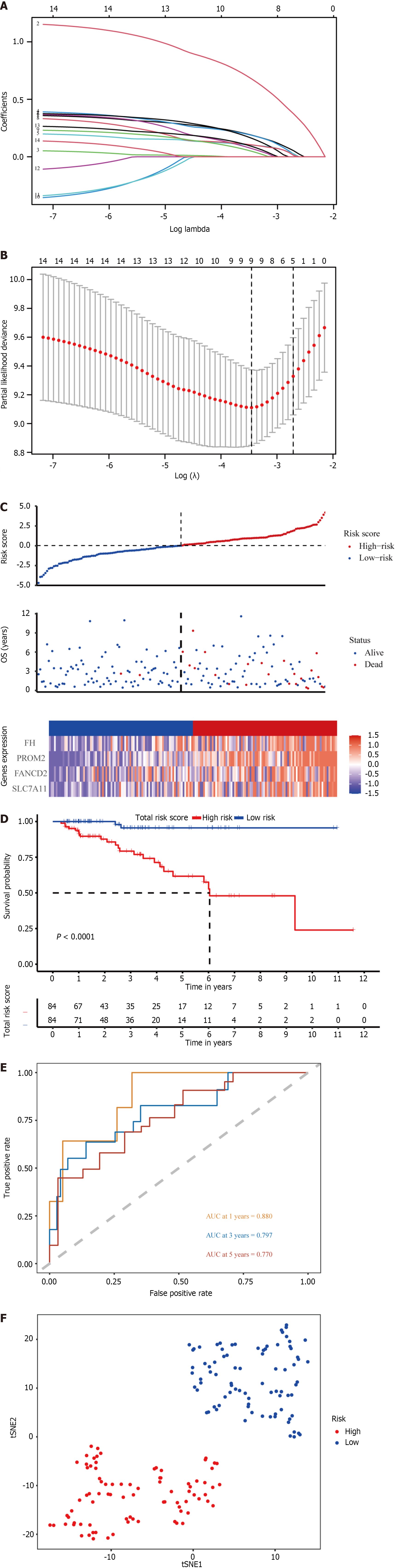
The ferroptosis-related gene expression profiles of the patients in the high- and low-risk groups are displayed in a heatmap (Figure 2A). In addition, the volcano plot showed 5481 upregulated genes and 3766 downregulated genes in tumor tissues (Figure 2B; P < 0.05). A total of 128 genes were differentially expressed in ferroptosis and tumor tissues (Figure 2C). Of the 128 ferroptosis-related genes, 18 were identified using the univariate Cox regression model as significantly associated with patient OS (P < 0.05). The results are shown as forest plots (Figure 2D). Lasso-penalized Cox regression analysis was further conducted to limit the scope of the gene screening (Figure 3A). The nine candidate gene markers had the best lambda values (Figure 3B). Finally, multivariate regression analysis revealed that four DEGs were significantly correlated with OS.
With respect to the TCGA cohort, a risk score was developed to determine the predictive power of the 4 genes associated with ferroptosis. The risk score was calculated using the formula below: Risk score = 1.05 × expression level of PROM2 + 0.532 × SLC7A11 + 0.447 × FANCD2 + 0.453 × FH. Patients were classified into high-risk (n = 84) and low-risk (n = 84) groups based on the median risk score cutoff (Figure 3C; Table 1).
| TCGA | Metabric | |||||
| High risk group, n = 84 | Low risk group, n = 84 | P value | High risk group, n = 54 | Low risk group, n = 72 | P value | |
| Age | ||||||
| < 60 | 38 (0.45) | 40 (0.48) | 0.003 | 34 (0.63) | 41 (0.57) | 0.606 |
| ≥ 60 | 46 (0.55) | 44 (0.52) | 20 (0.37) | 31 (0.43) | ||
| Stage | ||||||
| Ⅰ/Ⅱ | 59 (0.70) | 58 (0.69) | 0.033 | 40 (0.74) | 60 (0.83) | 0.101 |
| Ⅲ/Ⅳ | 25 (0.30) | 26 (0.31) | 14 (0.26) | 12 (0.17) | ||
| Radio therapy | ||||||
| Yes | 3 (0.04) | 5 (0.06) | 0.293 | 41 (0.76) | 49 (0.68) | 0.548 |
| No | 7 (0.08) | 6 (0.07) | 13 (0.24) | 23 (0.32) | ||
| Unknown | 74 (0.88) | 73 (0.87) | 0 | 0 | ||
According to K-M curves, patients in the TCGA cohort with lower risk scores had better prognoses (Figure 3D; P < 0.05). Using time-dependent ROC curve analysis, the area under the curve (AUC) was evaluated. The AUCs of the four ferroptosis-related genes at 3, 5, and 8 years were 0.797, 0.770, and 0.664, respectively (Figure 3E), with the third year having the most significant AUC. These four genes are anticipated to be associated with OS. Patients in different risk groups were dispersed in both directions according to the t-distributed stochastic neighbor embedding (t-SNE) analysis (Figure 3F).
We chose the independent database METABRIC for validation to confirm the ability of the four-gene signature to predict survival. The patients were divided into high-risk and low-risk groups using the same algorithm used for the TCGA cohort (Table 1, Supplementary Figure 1A). Those in the high-risk group demonstrated significantly worse OS than did those in the low-risk group (Supplementary Figure 1B, P < 0.05), consistent with the findings in the TCGA cohort. The AUC for 3-, 5-, and 8-year OS were 0.653, 0.648, and 0.560, respectively (Supplementary Figure 1C). Additionally, T-SNE analysis verified that the two patient subgroups spread in opposite directions (Supplementary Figure 1D). These findings showed that the four-gene signature could accurately predict OS in patients with the HER2+ subtype of BC.
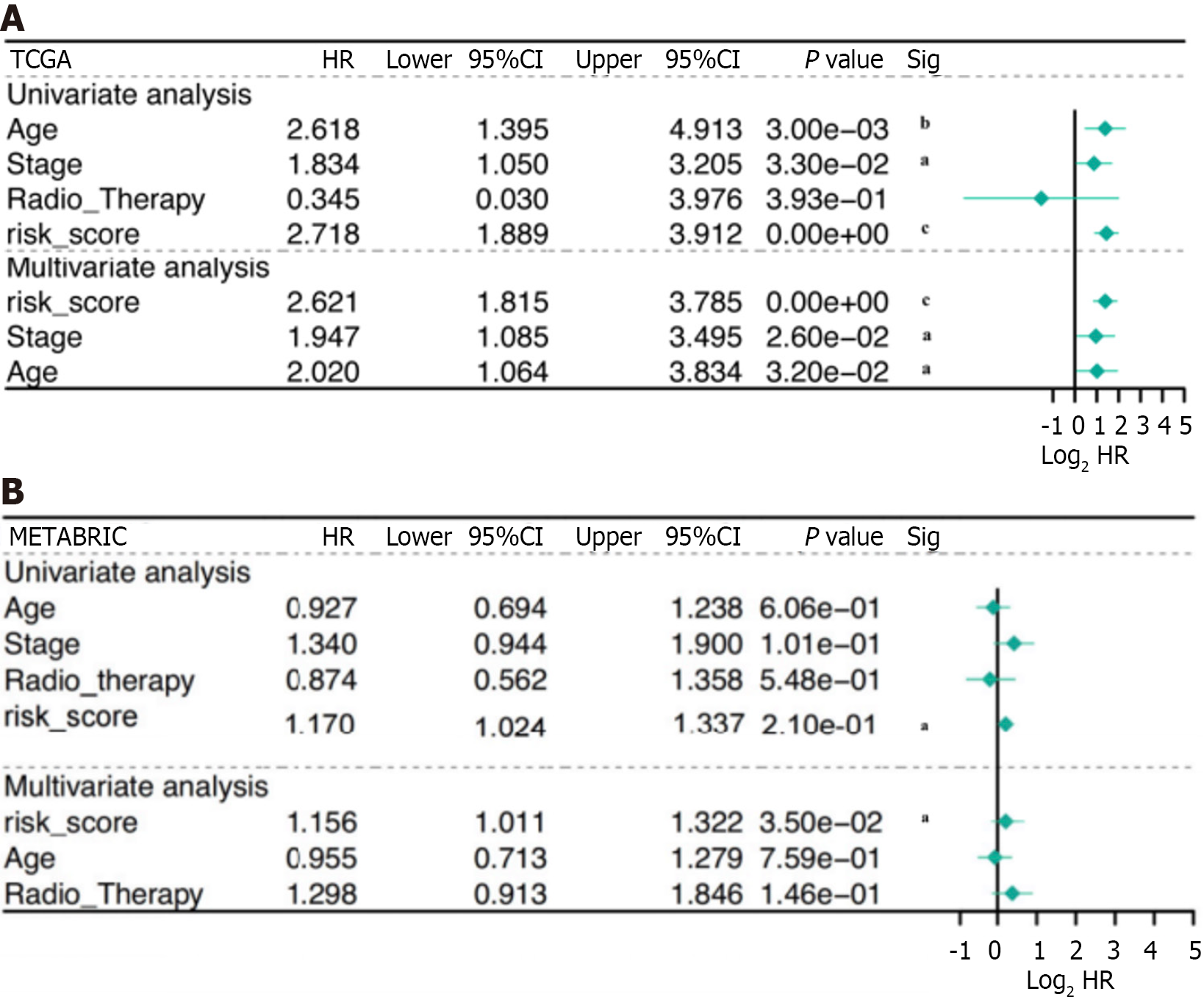
Patients with complete data, including age, stage, radiation therapy, and risk score, were enrolled for additional analysis. The risk score was identified as a significant prognostic risk factor in the TCGA cohort [P < 0.001, hazard ratio (HR) = 2.72, 95%CI = 1.889-3.912] and in the METABRIC cohort (P < 0.001, HR = 1.17, 95%CI = 1.024-1.337) by univariate Cox analysis (Figure 4A). The risk score was also found to be an independent predictive factor for OS in the TCGA cohort (P < 0.001, HR = 2.62, 95%CI = 1.815-3.785) and the METABRIC cohort (P < 0.001, HR = 1.16, 95%CI = 1.011-1.322) according to multivariate Cox regression analysis (Figure 4B). Consequently, the risk score derived from the four-gene profile was an independent prognostic factor.
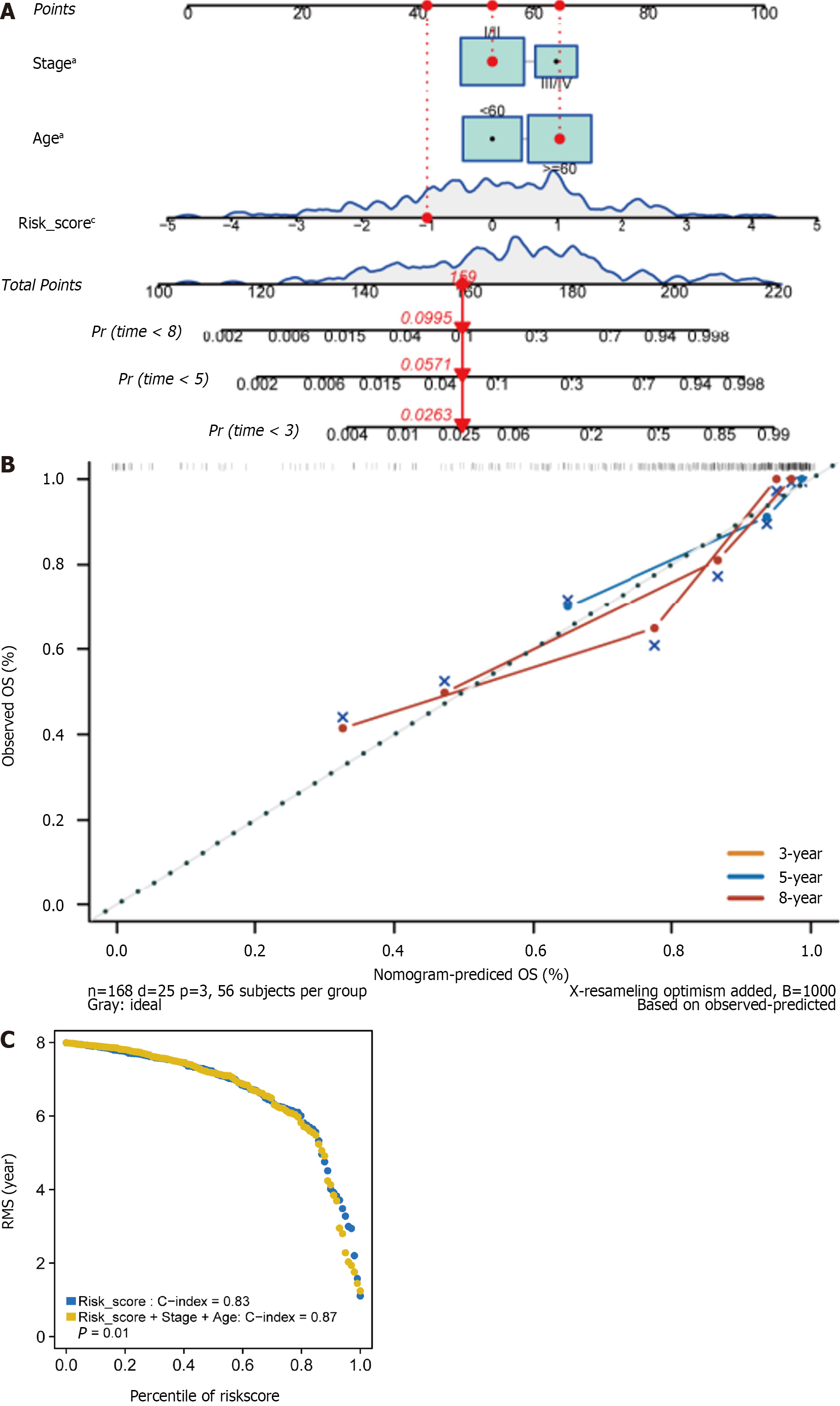
We subsequently developed a nomogram employing three independent prognostic parameters, cancer stage, age, and risk score, to predict 3-, 5-, and 8-year OS in 168 HER2+ BC patients (Figure 5A). The calibration plot showed that the nomogram might under- or overestimate mortality (Figure 5B). The C-index of the model, which considered risk score, age, and stage, was 0.87 (Figure 5C). In addition, we repeated these steps in the METABRIC cohort to validate the efficacy of the training cohort. It is important to note that there was significant agreement between the predicted and observed survival rates, suggesting that the nomogram has excellent predictive value (Supple
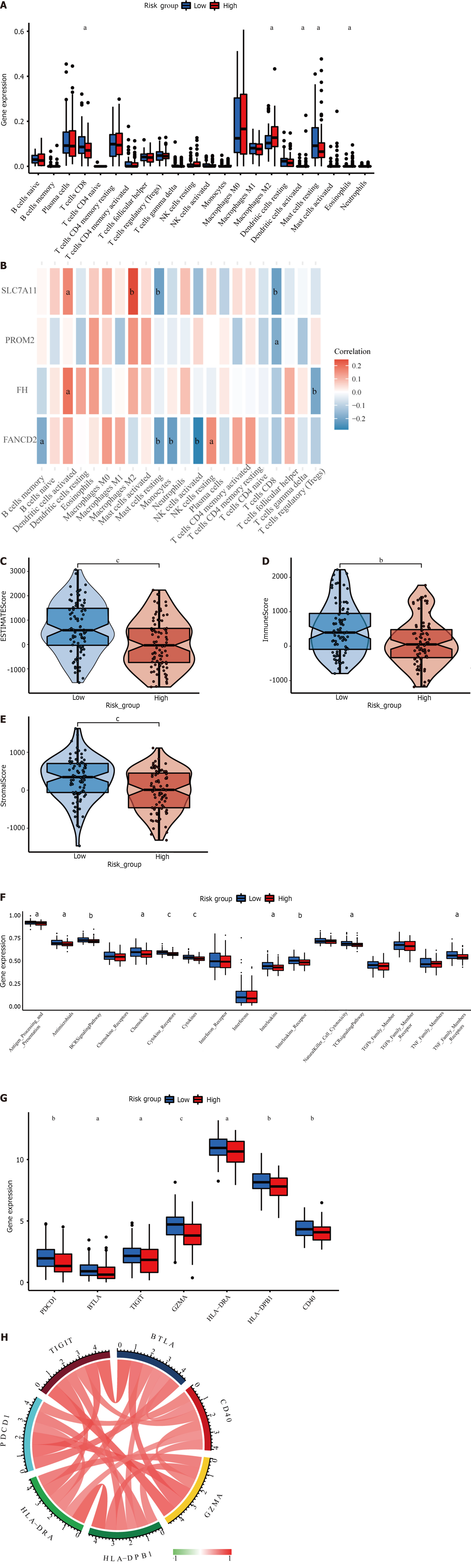
The four-gene signature may be correlated with the immunological characteristics of cancer patients, providing future guidance for immunotherapy for HER2+ BC patients. A significant association between the risk score and essential immune cell infiltration or immunological aspects was assessed using the CIBERSORT algorithm. We discovered that the high- and low-risk groups had distinct immune cell infiltration rates. The infiltration of M2-type macrophages, activated dendritic cells, and eosinophils was greater than that of CD8+ T cells and resting mast cells in the high-risk group (Figure 6A). We also constructed heatmaps to evaluate the correlation between immune cells and prognostic genes (Figure 6B). In the present analysis, the ESTIMATE score also revealed higher immune, stromal, and ESTIMATE scores in the low-risk subgroup than in the high-risk subgroup (Figure 6C-E). In addition, the distribution of immune-related signal transduction pathways was significantly different between the two risk subgroups, with lower infiltration of cytokine receptors, cytokines, the BCR signaling pathway, interleukin receptors, antimicrobials, chemokines, interleukins, T cell receptor signal transduction pathways and tumor necrosis factor receptors in the high-risk subgroup(Figure 6F).
Our analysis also included genes related to immune checkpoints, PD-1 (PDCD1), BTLA, TIGIT, GZMA, HLA-DRA, HLA-DPB1, and CD40. The expression of these seven well-known immune checkpoint genes varied between the low- and high-risk groups. Figure 6G indicates that immune checkpoint mRNA expression was decreased in HER2+ BC patients with higher risk scores. In addition, there was a significant positive correlation between the mRNA levels of the seven immune checkpoint receptors (Figure 6H).
SNP analysis was performed on 155 samples comprising 80 samples in the high-risk group and 75 samples in the low-risk group, with significant mutation frequency genes screened out using the “MAFtools” package. Supplementary Figure 3A and B lists the top 10 mutated genes in samples from the high- and low-risk groups, with TTN and TP53 mutations being the most frequent in the two groups. Supplementary Figure 3C and D lists the top 20 genes in the sample, revealing that the most significant mutation types were missense, nonsense, missense, and multihit mutations. In patients, TP53, PIK3CA, and TTN were strongly associated with the development of HER2+ BC.
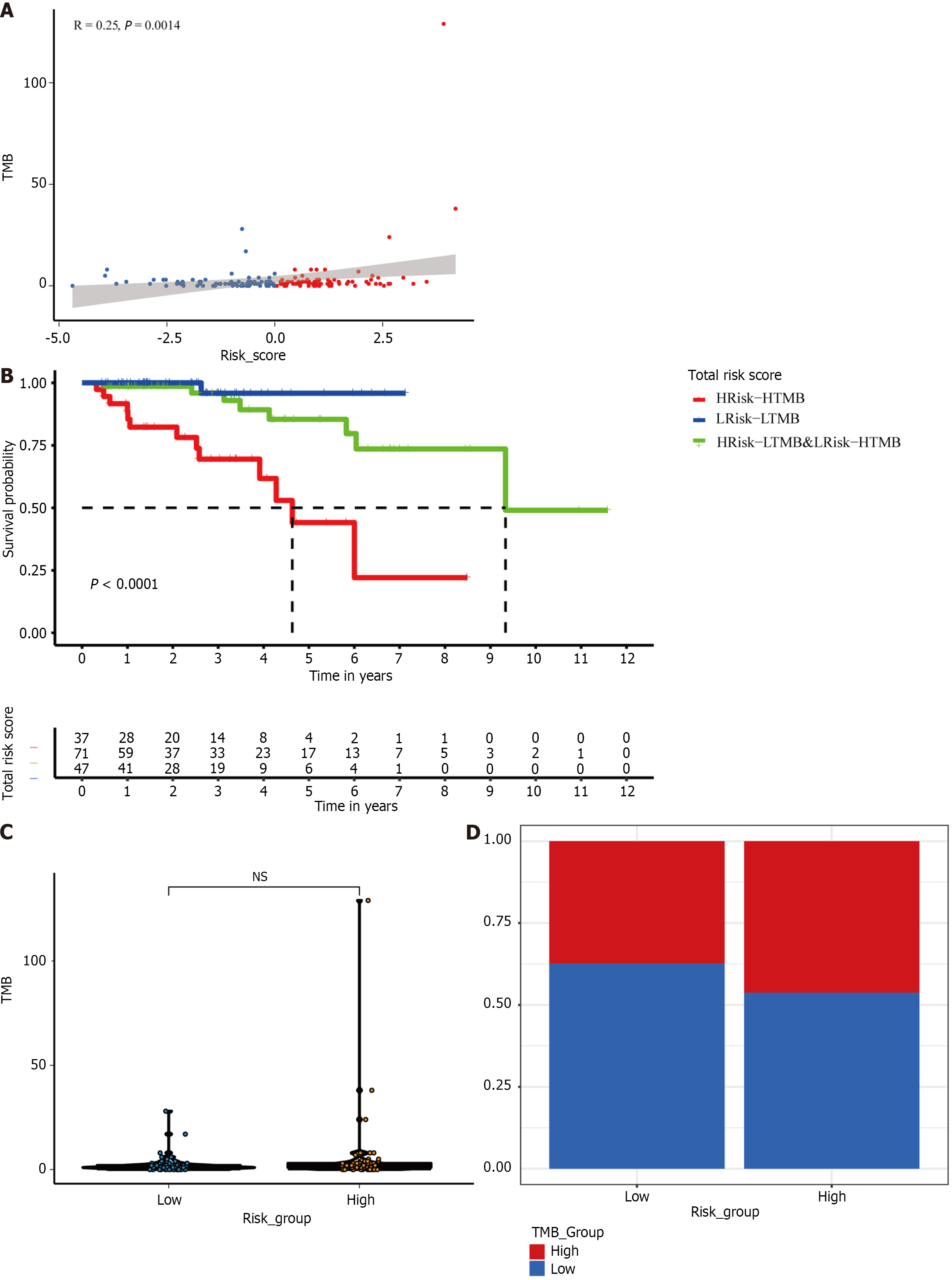
We also extracted the TMB subgroups in the high-risk and low-risk groups. We found a positive correlation between risk score and TMB (P < 0.01; Figure 7A and B). However, there was no significant difference in the TMB between the two risk groups (Figure 7C). Therefore, to explore whether combining the TMB and risk score provides better predictive ability, we combined the TMB and risk score to form a new subgroup. Kaplan-Meier survival curves for the new subset revealed significant differences in survival outcomes. The prognosis was worst for patients with HRisk-HTMB but best for those with LRisk-LTMB (P < 0.0001; Figure 7D).
We then extracted data on 138 drugs from the GDSC database and analyzed patient sensitivities to 138 medications between the high- and low-risk cancer groups. We found 13 drugs with significantly different sensitivities between the two risk groups (Supplementary Figure 4; P < 0.05).
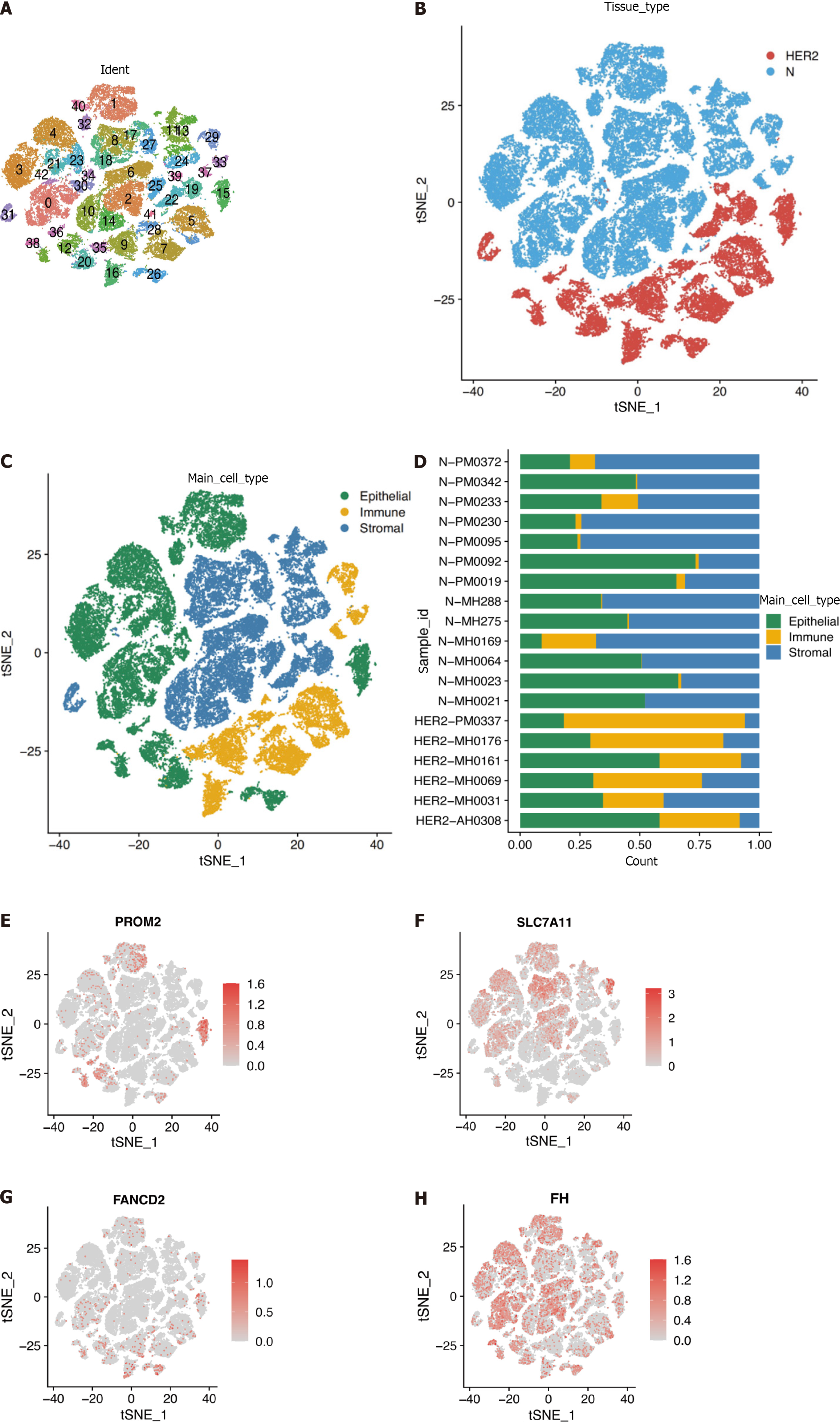
The scRNA-seq dataset (GSE161529) was used to characterize HER2+ BC heterogeneity from the GEO database. After gene filtering and normalization, the “Seurat” package of the FindCluster function was used to cluster cells into 42 clusters (Figure 8A and B). The identified clusters were labeled as cell types using marker genes (Supplementary Table 3). We ultimately annotated these clusters into three main clusters (Figure 8C, Supplementary Table 4), and Figure 8D shows the proportions of cell types in patients and healthy individuals. According to the scRNA-seq analysis, Figure 8E-H demonstrates that the identified prognostic genes were primarily expressed in epithelial cells. Differences in the expression of the four marker genes between healthy controls and patients were further verified in epithelial cells. Specifically, PROM2, SLC7A11, and FANCD2 but not FH were significantly differentially expressed, indicating that these genes were expressed in cancer epithelial cells (Table 2). However, the SLC7A11 results did not correspond to the trend observed via Bulk RNA-seq. This difference is most likely related to the somewhat small sample size.
| Gene | P value | Avg_log2FC | Pct.1 | Pct.2 | P value adjustment |
| PROM2 | 9.48E-134 | 0.115125977 | 0.096 | 0.017 | 1.98E-129 |
| SLC7A11 | 2.96E-77 | -0.101678852 | 0.016 | 0.08 | 6.18E-73 |
| FANCD2 | 4.01E-27 | 0.028728862 | 0.025 | 0.006 | 8.38E-23 |
Ferroptosis is a type of cell death distinguished by iron-dependent lipid peroxidation. It interferes with the progression of tumors, neurological diseases, and chronic inflammatory diseases. There has been much research recently on the role and mechanisms of ferroptosis under various conditions, particularly in the tumor research and treatment domains. Ferroptosis pathway activation increases the susceptibility of cancer cells to chemotherapy. One study demonstrated the importance of ferroptosis in tumor therapy by showing that combining the ferroptosis inducer erastin with cisplatin can significantly boost antitumor efficacy[14]. Using ferrostatin-1 (a ferroptosis inhibitor) knockdown, Chen et al[15] reported that cystine (Cys) starvation induces ferroptosis in TNBC cells[15]. Therefore, ferroptosis could advance our under
This study systematically investigated the potential mechanisms of action of 259 iron death-associated genes in HER2+ BC patients. Four ferroptosis-related genes were included in the new prognostic model, and the validity of the model was tested in an external cohort. The association of these genes with OS was also explored. In addition, the immune microenvironment and mutations were enriched in our study. Nearly half of the iron apoptosis-related genes (129/259) were differentially expressed between 91 normal cells and 168 HER2+ cells, 18 of which were associated with OS according to univariate Cox regression analysis. Finally, a four-gene signature was obtained from the model using LASSO regression analysis and multivariate Cox regression analysis. These results demonstrated that iron-induced cell death plays a significant role in HER2+ BC patients, and prognostic features based on iron-induced cell death-related genes could be constructed.
Four ferroptosis-related genes were included in the prognostic model in this investigation. PROM2, SLC7A11, FANCD2, and FH were highly expressed in HER2+ BC patients. The expression levels of these genes were positively correlated with patient survival risk. Previous studies have shown that iron, lipid, and antioxidative metabolism are the three key pathways regulating iron-related apoptosis[17]. In addition, energy metabolism is associated with iron-related apoptosis. Consistent with our four-gene prognostic model, PROM2, SLC7A11, and FANCD2 were previously reported to be involved in iron metabolism[18,19]. The PROM2 gene is significantly upregulated in tumor tissues. PROM2 contributes to iron transport and the inhibition of iron death by forming iron-containing multivesicular bodies and exosomes in BC cells[11,20]. SLC7A11 is an essential component of the glutamate/Cys antiporter (also known as xCT). SLC7A11 increases glutathione production and Cys absorption, reducing oxidative stress and iron cell death[21]. Depletion of SLC7A11 significantly reduces glutathione concentrations and triggers iron-related apoptosis. In addition, SLC7A11 is a central target of iron death regulation, and at high concentrations, it downregulates sensitivity to iron death in cancer cells[22,23]. Both PROM2 and SLC7A11 are regulated by GPX4, which increases the amount of peroxyl radicals required for lipid peroxidation, causing iron death. The regulation of FANCD2 gene expression helps maintain normal DNA replication, which prevents cancer progression by influencing the iron-related death process[24,25]. FANCD2 expression correlated with the characteristics of aggressive cancer: HER2 amplification, hormone receptor negativity, elevated p53 expression, proliferation, and high grade[26]. Several studies have demonstrated the positive association between FANCD2 and Ki-67 expression in BC cells[27]. Furthermore, high FANCD2 expression could independently predict a poor prognosis in patients in the sporadic BC cohort[28]. The fumarate complex enzyme FH belongs to three TCA cycle enzyme families. Some studies have indicated that the FH double allele is inactivated in BC patients and that mutations in the FH gene may affect the progression of BC[29,30]. The role of these genes in inducing iron-related death in HER2+ BC patients needs further investigation, as few relevant studies have reported the regulatory function of these genes. Moreover, the patterns of the relationships among TMB, risk score, and the combination of TMB grouping and risk grouping indicated the synergistic effect of TMB and the risk score in prognostic stratification. These findings may provide new insight into cancer prognosis.
The treatment landscape for HER2-positive BC has undergone significant advancements in recent years. A comprehensive understanding of tumor biology and the intricate signaling pathways associated with HER2 has played a pivotal role in developing novel therapeutic strategies to improve patient outcomes. Prominent among these emerging approaches is dual-HER2 inhibition utilizing monoclonal antibodies, exemplified by the combination of trastuzumab and pertuzumab. Additionally, antibody–drug conjugates, including T-DM1 and trastuzumab–deruxtecan[31], and TKIs, such as tucatinib and neratinib, have emerged as promising therapeutic options[32,33]. In this study, we selected 13 drugs from a list of 138 drugs by analyzing and visualizing their IC50s in high-risk and low-risk groups; these drugs included bibw2992 (afatinib) and ABT.263 (navitoclax), which have been previously reported to be effective treatments for HER2 + BC[34,35]. Bibw2992 (afatinib) is a tyrosine kinase and an irreversible blocker of the ErbB family[36]. It has been reported to be necessary for treating HER2+ BC[34,37]. Abt.263 (navitoclax) is a small molecule Bcl-2 inhibitor that induces apoptosis and treats HER2+ BC by blocking the interaction of Bcl-2 and Bcl XL with apoptotic precursor proteins[35]. Vinorelbine is an antimitotic semisynthetic drug that acts primarily by binding to tubulin, causing cells to become disorganized within microtubules during mitosis. It has been reported in the relevant literature that this approach is an effective treatment for metastatic BC. Vinorelbine is commonly used in combination therapy with trastuzumab and pyrotinib in HER2+ BC[38,39]. A-443654 is a potent pan-Akt inhibitor that has been reported to prolong survival in patients with HER2+ BC when combined with other medicines[40]. A c-Jun N-terminal kinase inhibitor called JNK.Inhibitor. VIII (TCS JNK 6o) inhibits invasive BC by reducing JNK activity. It can be used in combination with lapatinib[41].
In conclusion, our study developed a novel, previously unreported four-gene signature-associated prognostic model, which may be a useful prognostic classification tool for HER2+ BC patients. These genes were correlated with OS in the training cohort, and the association was confirmed in the validation cohort. A nomogram that combined our predictive signature with traditional clinical factors such as age and clinical stage performed noticeably better. As a result, the nomogram we developed can successfully direct clinical practice and help build a more individualized clinical follow-up approach.
In addition, some limitations relevant to our study should be noted. First, only four prognostic genes from the TCGA database were used to calculate the prognostic risk score; their somatic mutations or methylation status should have been considered. Second, the sample size of the single-cell expression data was relatively small; future analyses with larger sample sizes are needed to validate and explore the present results. Third, the database lacks targeted therapy information within its clinical data, limiting its contents to radiotherapy and chemotherapy information exclusively. However, the mechanism underlying the four-gene signature and its therapeutic implications for treating HER2+ BC require further study.
Our study identified a 4-gene model that, when combined with the tumor mutation burden (TMB) score, may have critical implications for clinical medical decisions and personalized treatment of patients with HER2-positive breast cancer.
This study aimed to identify and evaluate fresh ferroptosis-related biomarkers for HER2+ breast cancer (BC).
Identifying reliable prognostic biomarkers can direct clinical practice and help develop a more individualized clinical follow-up approach.
The prediction model was constructed using data from the TCGA and METABRIC databases. Subsequently, patients were categorized into high-risk and low-risk groups according to their median risk scores, independent predictors for overall survival (OS). We investigated immune infiltration, mutations, and drug sensitivity across risk groups. Moreover, we integrated tumor mutational burden (TMB) with risk scores to assess patient prognosis. Finally, we analyzed vital gene expression through single-cell RNA sequencing (scRNA-seq) in cancerous and normal epithelial cells.
Our model helps guide the prognosis of HER2+ breast cancer patients, and its combination with the TMB can aid in more accurate assessment of patient prognosis and provide new ideas for further diagnosis and treatment.
By analyzing the RNA expression data of HER2-positive breast cancer patients, we constructed a risk score model (PROM2, SLC7A11, FANCD2, and FH) for ferroptosis and evaluated the relationship between the high-risk score and patient prognosis. We verified that the high-risk group was associated with poorer immune infiltration and a greater tumor mutation load. By combining the risk score with the TMB, we found that patients with a high TMB-score had the worst prognosis, while patients with a low TMB-score had the best prognosis.
The prediction model was constructed using data from the TCGA and METABRIC cohorts. Patients were subsequently categorized into high-risk and low-risk groups according to their median risk score, an independent predictor of overall survival. We investigated immune infiltration, mutations, and drug sensitivity across risk groups. Moreover, we integrated the TMB with risk scores to assess patient prognosis. Finally, we analyzed vital gene expression through single-cell RNA sequencing in cancerous and normal epithelial cells.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bansal C, India S-Editor: Li L L-Editor: A P-Editor: Zhang XD
| 1. | Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, Bray F. Cancer statistics for the year 2020: An overview. Int J Cancer. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2411] [Cited by in RCA: 2969] [Article Influence: 742.3] [Reference Citation Analysis (7)] |
| 2. | Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9574] [Cited by in RCA: 9415] [Article Influence: 724.2] [Reference Citation Analysis (0)] |
| 3. | Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF; American Society of Clinical Oncology; College of American Pathologists. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997-4013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2910] [Cited by in RCA: 3055] [Article Influence: 254.6] [Reference Citation Analysis (0)] |
| 4. | Dowsett M, Procter M, McCaskill-Stevens W, de Azambuja E, Dafni U, Rueschoff J, Jordan B, Dolci S, Abramovitz M, Stoss O, Viale G, Gelber RD, Piccart-Gebhart M, Leyland-Jones B. Disease-free survival according to degree of HER2 amplification for patients treated with adjuvant chemotherapy with or without 1 year of trastuzumab: the HERA Trial. J Clin Oncol. 2009;27:2962-2969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 146] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 5. | Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, Safari E, Farahmand L. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol. 2020;84:106535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 478] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 6. | Blumenthal GM, Scher NS, Cortazar P, Chattopadhyay S, Tang S, Song P, Liu Q, Ringgold K, Pilaro AM, Tilley A, King KE, Graham L, Rellahan BL, Weinberg WC, Chi B, Thomas C, Hughes P, Ibrahim A, Justice R, Pazdur R. First FDA approval of dual anti-HER2 regimen: pertuzumab in combination with trastuzumab and docetaxel for HER2-positive metastatic breast cancer. Clin Cancer Res. 2013;19:4911-4916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 7. | Shao Z, Pang D, Yang H, Li W, Wang S, Cui S, Liao N, Wang Y, Wang C, Chang YC, Wang H, Kang SY, Seo JH, Shen K, Laohawiriyakamol S, Jiang Z, Li J, Zhou J, Althaus B, Mao Y, Eng-Wong J. Efficacy, Safety, and Tolerability of Pertuzumab, Trastuzumab, and Docetaxel for Patients With Early or Locally Advanced ERBB2-Positive Breast Cancer in Asia: The PEONY Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020;6:e193692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 134] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 8. | Wang L, Chen Y, Zhao J, Luo D, Tian W. Analysis and prediction model of ferroptosis related genes in breast cancer. Transl Cancer Res. 2022;11:1970-1976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 9. | Wang D, Wei G, Ma J, Cheng S, Jia L, Song X, Zhang M, Ju M, Wang L, Zhao L, Xin S. Identification of the prognostic value of ferroptosis-related gene signature in breast cancer patients. BMC Cancer. 2021;21:645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 10. | Jin LY, Gu YL, Zhu Q, Li XH, Jiang GQ. The role of ferroptosis-related genes for overall survival prediction in breast cancer. J Clin Lab Anal. 2021;35:e24094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Brown CW, Amante JJ, Chhoy P, Elaimy AL, Liu H, Zhu LJ, Baer CE, Dixon SJ, Mercurio AM. Prominin2 Drives Ferroptosis Resistance by Stimulating Iron Export. Dev Cell. 2019;51:575-586.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 431] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 12. | Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4608] [Cited by in RCA: 4983] [Article Influence: 622.9] [Reference Citation Analysis (0)] |
| 13. | Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4711] [Cited by in RCA: 11761] [Article Influence: 904.7] [Reference Citation Analysis (1)] |
| 14. | Sato M, Kusumi R, Hamashima S, Kobayashi S, Sasaki S, Komiyama Y, Izumikawa T, Conrad M, Bannai S, Sato H. The ferroptosis inducer erastin irreversibly inhibits system x(c)- and synergizes with cisplatin to increase cisplatin's cytotoxicity in cancer cells. Sci Rep. 2018;8:968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 265] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 15. | Chen MS, Wang SF, Hsu CY, Yin PH, Yeh TS, Lee HC, Tseng LM. CHAC1 degradation of glutathione enhances cystine-starvation-induced necroptosis and ferroptosis in human triple negative breast cancer cells via the GCN2-eIF2α-ATF4 pathway. Oncotarget. 2017;8:114588-114602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 220] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 16. | Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, Jemal A, Siegel RL. Breast Cancer Statistics, 2022. CA Cancer J Clin. 2022;72:524-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 1182] [Article Influence: 394.0] [Reference Citation Analysis (0)] |
| 17. | Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao N, Sun B, Wang G. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 639] [Cited by in RCA: 2536] [Article Influence: 507.2] [Reference Citation Analysis (0)] |
| 18. | Wei W, Hu Q, Li W, Li M, Dong S, Peng Y, Yin J, Lu Y, Liu L, Zhao Q. The Role of Ferroptosis Signature in Overall Survival and Chemotherapy of Pancreatic Adenocarcinoma. DNA Cell Biol. 2022;41:116-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Chen GH, Song CC, Pantopoulos K, Wei XL, Zheng H, Luo Z. Mitochondrial oxidative stress mediated Fe-induced ferroptosis via the NRF2-ARE pathway. Free Radic Biol Med. 2022;180:95-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 190] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 20. | Luo W, Wang J, Xu W, Ma C, Wan F, Huang Y, Yao M, Zhang H, Qu Y, Ye D, Zhu Y. LncRNA RP11-89 facilitates tumorigenesis and ferroptosis resistance through PROM2-activated iron export by sponging miR-129-5p in bladder cancer. Cell Death Dis. 2021;12:1043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 122] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 21. | Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12:599-620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 1464] [Article Influence: 292.8] [Reference Citation Analysis (0)] |
| 22. | He J, Ding H, Li H, Pan Z, Chen Q. Intra-Tumoral Expression of SLC7A11 Is Associated with Immune Microenvironment, Drug Resistance, and Prognosis in Cancers: A Pan-Cancer Analysis. Front Genet. 2021;12:770857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 23. | Sun X, Niu X, Chen R, He W, Chen D, Kang R, Tang D. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology. 2016;64:488-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 529] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 24. | Han B, Shen Y, Zhang P, Jayabal P, Che R, Zhang J, Yu H, Fei P. Overlooked FANCD2 variant encodes a promising, portent tumor suppressor, and alternative polyadenylation contributes to its expression. Oncotarget. 2017;8:22490-22500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 25. | Song X, Xie Y, Kang R, Hou W, Sun X, Epperly MW, Greenberger JS, Tang D. FANCD2 protects against bone marrow injury from ferroptosis. Biochem Biophys Res Commun. 2016;480:443-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 156] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 26. | Fagerholm R, Sprott K, Heikkinen T, Bartkova J, Heikkilä P, Aittomäki K, Bartek J, Weaver D, Blomqvist C, Nevanlinna H. Overabundant FANCD2, alone and combined with NQO1, is a sensitive marker of adverse prognosis in breast cancer. Ann Oncol. 2013;24:2780-2785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Rudland PS, Platt-Higgins AM, Davies LM, de Silva Rudland S, Wilson JB, Aladwani A, Winstanley JH, Barraclough DL, Barraclough R, West CR, Jones NJ. Significance of the Fanconi anemia FANCD2 protein in sporadic and metastatic human breast cancer. Am J Pathol. 2010;176:2935-2947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Feng L, Jin F. Expression and prognostic significance of Fanconi anemia group D2 protein and breast cancer type 1 susceptibility protein in familial and sporadic breast cancer. Oncol Lett. 2019;17:3687-3700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Zhang Q, Liang Z, Gao Y, Teng M, Niu L. Differentially expressed mitochondrial genes in breast cancer cells: Potential new targets for anti-cancer therapies. Gene. 2017;596:45-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Schmidt C, Sciacovelli M, Frezza C. Fumarate hydratase in cancer: A multifaceted tumour suppressor. Semin Cell Dev Biol. 2020;98:15-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 31. | Fu Z, Li S, Han S, Shi C, Zhang Y. Antibody drug conjugate: the "biological missile" for targeted cancer therapy. Signal Transduct Target Ther. 2022;7:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 535] [Cited by in RCA: 798] [Article Influence: 266.0] [Reference Citation Analysis (0)] |
| 32. | Jacobs AT, Martinez Castaneda-Cruz D, Rose MM, Connelly L. Targeted therapy for breast cancer: An overview of drug classes and outcomes. Biochem Pharmacol. 2022;204:115209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 72] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 33. | Xu B, Yan M, Ma F, Hu X, Feng J, Ouyang Q, Tong Z, Li H, Zhang Q, Sun T, Wang X, Yin Y, Cheng Y, Li W, Gu Y, Chen Q, Liu J, Cheng J, Geng C, Qin S, Wang S, Lu J, Shen K, Liu Q, Wang H, Luo T, Yang J, Wu Y, Yu Z, Zhu X, Chen C, Zou J; PHOEBE Investigators. Pyrotinib plus capecitabine vs lapatinib plus capecitabine for the treatment of HER2-positive metastatic breast cancer (PHOEBE): a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22:351-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 252] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 34. | Collins DM, Madden SF, Gaynor N, AlSultan D, Le Gal M, Eustace AJ, Gately KA, Hughes C, Davies AM, Mahgoub T, Ballot J, Toomey S, O'Connor DP, Gallagher WM, Holmes FA, Espina V, Liotta L, Hennessy BT, O'Byrne KJ, Hasmann M, Bossenmaier B, O'Donovan N, Crown J. Effects of HER Family-targeting Tyrosine Kinase Inhibitors on Antibody-dependent Cell-mediated Cytotoxicity in HER2-expressing Breast Cancer. Clin Cancer Res. 2021;27:807-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 35. | Zoeller JJ, Vagodny A, Taneja K, Tan BY, O'Brien N, Slamon DJ, Sampath D, Leverson JD, Bronson RT, Dillon DA, Brugge JS. Neutralization of BCL-2/X(L) Enhances the Cytotoxicity of T-DM1 In Vivo. Mol Cancer Ther. 2019;18:1115-1126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 36. | Wecker H, Waller CF. Afatinib. Recent Results Cancer Res. 2018;211:199-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 37. | Duchnowska R, Loibl S, Jassem J. Tyrosine kinase inhibitors for brain metastases in HER2-positive breast cancer. Cancer Treat Rev. 2018;67:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 38. | Li Y, Qiu Y, Li H, Luo T, Li W, Wang H, Shao B, Wang B, Ge R. Pyrotinib Combined With Vinorelbine in HER2-Positive Metastatic Breast Cancer: A Multicenter Retrospective Study. Front Oncol. 2021;11:664429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 39. | Aitelhaj M, Lkhoyaali S, Rais G, Boutayeb S, Errihani H. First line chemotherapy plus trastuzumab in metastatic breast cancer HER2 positive - Observational institutional study. Pan Afr Med J. 2016;24:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 40. | Yndestad S, Austreid E, Svanberg IR, Knappskog S, Lønning PE, Eikesdal HP. Activation of Akt characterizes estrogen receptor positive human breast cancers which respond to anthracyclines. Oncotarget. 2017;8:41227-41241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Ebelt ND, Kaoud TS, Edupuganti R, Van Ravenstein S, Dalby KN, Van Den Berg CL. A c-Jun N-terminal kinase inhibitor, JNK-IN-8, sensitizes triple negative breast cancer cells to lapatinib. Oncotarget. 2017;8:104894-104912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |