Published online Jan 24, 2024. doi: 10.5306/wjco.v15.i1.115
Peer-review started: November 20, 2023
First decision: December 5, 2023
Revised: December 13, 2023
Accepted: January 2, 2024
Article in press: January 2, 2024
Published online: January 24, 2024
Processing time: 64 Days and 2 Hours
Multiple myeloma (MM) is a terminal differentiated B-cell tumor disease characterized by clonal proliferation of malignant plasma cells and excessive levels of monoclonal immunoglobulins in the bone marrow. The translocation, (t)(4;14), results in high-risk MM with limited treatment alternatives. Thus, there is an urgent need for identification and validation of potential treatments for this MM subtype. Microarray data and sequencing information from public databases could offer opportunities for the discovery of new diagnostic or therapeutic targets.
To elucidate the molecular basis and search for potential effective drugs of t(4;14) MM subtype by employing a comprehensive approach.
The transcriptional signature of t(4;14) MM was sourced from the Gene Expression Omnibus. Two datasets, GSE16558 and GSE116294, which included 17 and 15 t(4;14) MM bone marrow samples, and five and four normal bone marrow samples, respectively. After the differentially expressed genes were identified, the Cytohubba tool was used to screen for hub genes. Then, the hub genes were analyzed using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis. Using the STRING database and Cytoscape, protein–protein interaction networks and core targets were identified. Potential small-molecule drugs were identified and validated using the Connectivity Map database and molecular docking analysis, respectively.
In this study, a total of 258 differentially expressed genes with enriched functions in cancer pathways, namely cytokine receptor interactions, nuclear factor (NF)-κB signaling pathway, lipid metabolism, atherosclerosis, and Hippo signaling pathway, were identified. Ten hub genes (cd45, vcam1, ccl3, cd56, app, cd48, btk, ccr2, cybb, and cxcl12) were identified. Nine drugs, including ivermectin, deforolimus, and isoliquiritigenin, were predicted by the Connectivity Map database to have potential therapeutic effects on t (4;14) MM. In molecular docking, ivermectin showed strong binding affinity to all 10 identified targets, especially cd45 and cybb. Ivermectin inhibited t(4;14) MM cell growth via the NF-κB pathway and induced MM cell apoptosis in vitro. Furthermore, ivermectin increased reactive oxygen species accumulation and altered the mitochondrial membrane potential in t(4;14) MM cells.
Collectively, the findings offer valuable molecular insights for biomarker validation and potential drug deve
Core Tip: Multiple myeloma is a hematological malignancy with a significant impact on public health, and the t(4;14) subtype is particularly aggressive and resistant to existing treatments. Our study addresses the urgent need for new therapeutic approaches by employing a comprehensive approach that includes bioinformatics analysis, molecular docking, and experimental validation. We identified ten key genes associated with t(4;14) multiple myeloma (MM), shedding light on the molecular basis of this subtype. We explored the potential of ivermectin to assess whether it may be “repurposed” as a therapeutic agent for t(4;14) MM. Our findings indicate that ivermectin not only inhibits MM cell growth but also induces apoptosis via the nuclear factor-κB signaling pathway.
- Citation: Song Y, Zhang HJ, Song X, Geng J, Li HY, Zhang LZ, Yang B, Lu XC. Gene signatures to therapeutics: Assessing the potential of ivermectin against t(4;14) multiple myeloma. World J Clin Oncol 2024; 15(1): 115-129
- URL: https://www.wjgnet.com/2218-4333/full/v15/i1/115.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i1.115
Multiple myeloma (MM) represents a severe hematological malignancy, affecting 176404 individuals and resulting in117077 fatalities annually[1]. MM is characterized by uncontrolled plasma cell proliferation in the bone marrow, leading to severe complications, such as bone and kidney damage, anemia, and hypercalcemia[2]. Of particular concern is the t(4;14) subtype, comprising up to 15% of new MM cases, with notably low survival rates and strong resistance to existing therapies[3]. Thus, effectively addressing this subtype remains an important medical challenge. Although the seminal research of Foltz et al[4] and Ashby et al[5] have provided insights into the pivotal facets of t(4;14) MM (Supple
Translational medicine is increasingly considering drug repurposing as a potential strategy to develop efficient, safe, cost-effective, and readily available anticancer treatments[6]. Advances in high-throughput sequencing technology have expanded biomedical and computational resources, facilitating a deeper understanding of cancer etiology and drug-target interactions, thus enabling drug repurposing[7]. Notably, the Connectivity Map (CMap) tool, housing a comprehensive dataset of 7000 microarrays from various cancer cells treated with 1309 molecular compounds, has been instrumental in identifying potential treatments for various cancers[8]. Notably, Shi et al[9] used CMap to identify eugenol as a potential treatment for triple-negative breast cancer, and Qiu et al[10] computationally identified small-molecule drugs with the potential to treat cervical cancer.
There is an urgent need for identification and validation of potential treatments for this MM subtype, therefore, we aim to elucidate the molecular basis and search for potential effective drugs by employing a comprehensive approach. We hope to contribute to advancing early MM diagnosis and tailoring its treatment.
The Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds) is a key public repository housing high-throughput gene expression data, microarray data, and gene chip information. For this study, we sourced the t(4;14) MM-related expression dataset from GEO. Our search parameters were tailored to t(4;14) MM across all fields, combined with filters for "Homo sapiens" as the organism and "dataset." Two datasets, GSE16558 and GSE116294, which included 17 and 15 t(4;14) MM bone marrow samples, and five and four normal bone marrow samples, respectively (Supplementary Table 2), were further considered for our analysis. To identify DEGs between the t(4;14) MM samples and normal samples, we used the "limma" package in R software [version 4.2.2, (http://www.R-project.org/)]. Our selection criteria were a P value < 0.05 and absolute log2 fold change (FC) > 1.
To construct a functional PPI network for the identified DEGs, we used the STRING online database tool (https://string-db.org/)[11]. Our selected threshold required a credibility score of > 0.4 for inclusion within the network. The Cytoscape software [version 3.10.1, (https://cytoscape.org/)] was used to visually map the PPI, offering a clear understanding of protein interactions.
Using Cytoscape, the molecular complex detection (MCODE) plugin was activated to highlight prominent clusters, abiding by criteria, such as an MCODE score > 6 and a node count > 4. The CytoHubba plugin[12] within Cytoscape, paired with the Maximal Clique Centrality (MCC) method, was used to filter and identify the top-ranking genes. A combination of results drawn from the MCODE, MCC, and degree scores led to the identification of 10 key hub genes.
The enrichment analysis was performed using the “GSEApy” package within Python (v.3.11.4, [https://www.python.or
The CMap database (https://clue.io)[13], a key tool in systems biology, was used to identify potential therapeutic candidates. Our criteria were rigorous, with a cutoff score of < −80.
For our molecular docking investigations, 3D structural models of the proteins encoded by target genes were retrieved from UniProt[14] and Protein Data Bank databases and augmented with AlphaFold data. Focusing on structures determined via X-ray diffraction with a co-crystal resolution < 2.5 Å, we downloaded the 2D structural model of ivermectin from PubChem[15] and converted it into a 3D structure using the LigPrep module in Maestro. AutoDockTools were used to preprocess the ligands. Optimization included water removal, hydrogenation, charge determination, and torsion centers and bond selection, yielding a pdbqt file suitable for docking studies. Macromolecular docking calculations were performed using Vina software [version 1.1.2, (http://vina.scripps.edu/)]. To illustrate the post-docking ligand-receptor interactions, we used Ligplot+[16] for 2D mapping and PyMOL[17] for in-depth 3D analysis.
Ivermectin was purchased from Aladdin Biological Technology (Shanghai, China). The following primary antibodies were purchased from CST (Danvers, MA, United States): Anti- poly (ADP-ribose) polymerase (PARP) (9532; 1:1000), anti-cleaved PARP (5625; 1:1000), anti-cleaved caspase 3 (9661; 1:500), anti-cleaved caspase 9 (20750; 1:1000), and β-actin (3700; 1:1000). The following primary antibodies were purchased from Abcam (Cambridge, United Kingdom): Anti-B-cell lymphoma 2 (Bcl2; ab182858; 1:1000), anti-Bcl-2-associated X protein (Bax; ab32503; 1:1000), anti-NF-κB p65 (ab32536; 1:1000), anti-NF-κB p65 (phospho S536) (ab86299; 1:1000), anti-inhibitor of NF-κB (IκB) alpha (ab32518; 1:1000), and anti-IκB alpha (phospho-S36) (ab133462; 1:1000). The following secondary antibodies were purchased from Beyotime Biotechnology (Shanghai, China): anti-rabbit IgG (A0208; 1:5000) and anti-mouse IgG (A0216; 1:5000).
NCI-H929, a t(4;14) MM cell line, was obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 medium (Gibco, Billings, MO, United States) with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco) at 37 °C with 5% CO2. Preliminary tests confirmed that the cell lines were Mycoplasma-free.
To assess cytotoxic activity, 5000 cells were seeded per well into a 96-well plate and exposed to ivermectin concentrations ranging from 0–20 μmol/L for 24–48 h. After incubation, cell viability was evaluated using Cell Counting Kit-8 (CCK8) (DojinDo, Shanghai, China), which required an additional 2-h incubation with 10 μL of CCK8 solution at 37 °C. The absorbance of the reaction mixture was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, California, United States). The inhibitory concentration 50% (IC50) of ivermectin for each cell type was determined from the viability data using GraphPad Prism v.10 and the following equation:
[(As−Ab)/(Ac−Ab)] × 100%, where “As,” “Ac,” and “Ab” are the absorbance values in the experimental, control, and blank wells, respectively.
Cells were seeded into 6-well plates and treated with ivermectin for 24 h. After treatment, the cells were collected by centrifugation. They were then stained using the fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection Kit with 7-aminoactinomycin D (7-AAD; 640922; BioLegend, San Diego, CA, United States) by adding 5 µL of FITC conjugated with annexin V and 5 µL of 7-AAD solution. This was followed by 15 min incubation in the dark at 25 °C. Subsequently, the stained cells were analyzed using a flow cytometer (LSRFortessa; BD, Franklin Lakes, NJ, United States), and the data obtained were processed and interpreted using FlowJo software (v.10.4).
The cells were cultured in 6-well plates overnight. Fresh RPMI-1640 medium containing the indicated concentrations of ivermectin was added, and the cells were cultured for 24 h. A reactive oxygen species (ROS) assay kit (S0033S; Beyotime) was used to detect intracellular ROS levels using the following protocol: treated cells were incubated with 2’,7’-dichlorodihydrofluorescein diacetate for 20 min at 37 °C. Cells were washed thrice with serum-free medium, and images were acquired using a fluorescent inverted microscope (Olympus, Tokyo, Japan).
Cells were first allowed to settle in 6-well plates, after which they were treated with ivermectin for 24 h. A JC-1 assay kit (M34152; Thermo Fisher Scientific) was used to evaluate the mitochondrial membrane potential within these cells. The assessment was performed in strict accordance with the manufacturer’s guidelines. Readings from this assessment were captured using a flow cytometer (LSRFortessa; BD).
After 48 h of ivermectin treatment, the cells were harvested and lysed using Cell Lysis Buffer (P0013; Beyotime), which included a protease inhibitor. The subsequent lysate was subjected to a 30-min ice bath before centrifugation at 12000 rpm for 20 min at 4 °C. Proteins were quantified using a BCA assay kit (P0010; Beyotime), separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis, and transferred onto polyvinylidene fluoride membranes. Subsequently, these membranes were blocked with 5% skim milk for 2 h and incubated with primary antibodies overnight at 4 °C. After thorough washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at 25 °C. The proteins were visualized using an electrochemiluminescence kit (NCM Biotech, Suzhou, China).
All statistical evaluations were performed using both the R package and GraphPad Prism v.9.0 (GraphPad Software Inc., San Diego, CA, United States). Student’s t-test was used to compare the means between two groups. Comparisons across multiple groups were conducted using one- and two-way ANOVA. All results are presented as the mean ± SD. Statistical significance was set at P levels < 0.05.
We identified 1,100 DEGs (270 upregulated and 830 downregulated) and 1746 DEGs (808 upregulated and 938 downregulated) in the GSE16558 and GSE116294 datasets, respectively. These microarray findings have been presented using heat maps and volcano graphs, applying a threshold of P < 0.05, logFC<1 for downregulated genes, and logFC>1 for upregulated genes (Supplementary Figure 1).
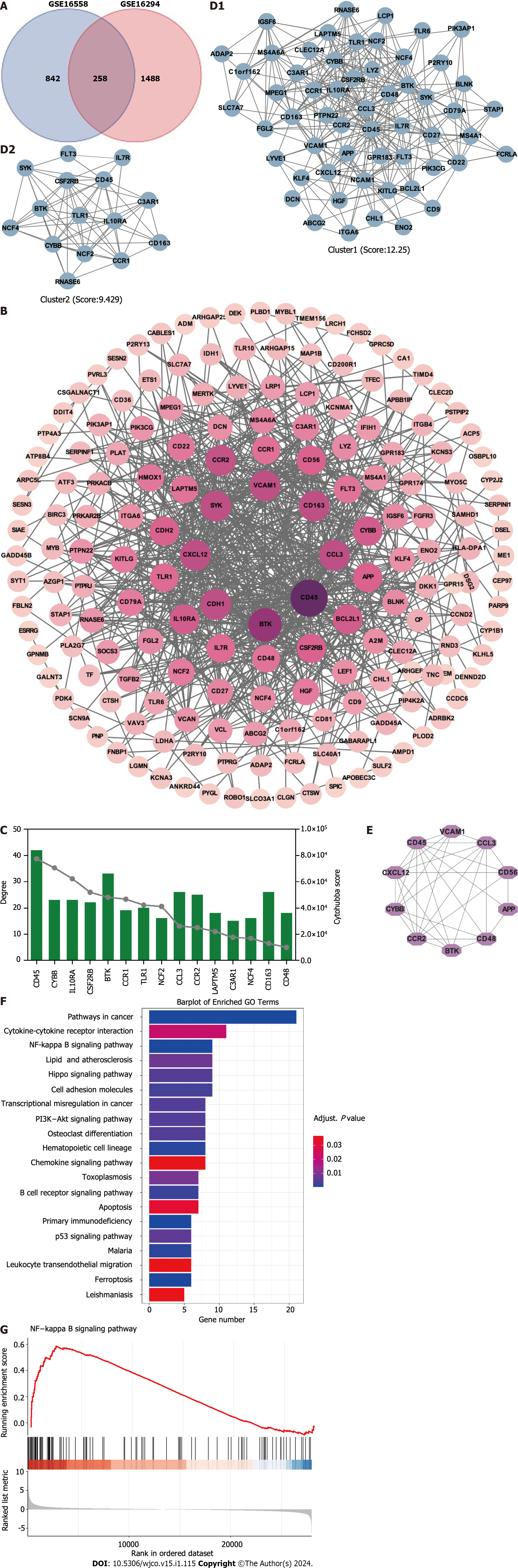
Potential targets involved in t(4;14) MM and PPI network: To better understand the therapeutic pathways involved in t(4;14) MM, we constructed a Venn diagram and identified 258 probable targets related to t(4;14) MM (Figure 1A). Subsequently, a PPI network containing these targets was constructed to elucidate the target interrelations. The PPI network comprised 175 nodes and 697 edges (Figure 1B). Three targets that lacked interactions with other targets were excluded from the assessment. The top five identified nodes were cd45, btk, ccl3, cdh1, and cxcl12, highlighting their pivotal roles in the network.
Using the Cytoscape MCODE plugin, we identified two clusters with scores > 6 (Figure 1C). The first group, with 57 nodes and 343 edges, was centered on the seed gene, CD45, and was predominantly related to altered transcriptional regulation in cancer. The second smaller cluster, which included 15 nodes and 66 edges, had TLR1 as its seed gene and was primarily related to the negative regulation of apoptosis and signal transduction. Subsequently, the CytoHubba score was used to identify the top 15 genes (Figure 1D). Each of these 15 genes had a degree score >15, indicating comprehensive interactions among them. Combining insights from MCODE, MCC, and degree scores, we identified 10 central hub genes: cd45, vcam1, ccl3, cd56, app, cd48, btk, ccr2, cybb, and cxcl12 (Figure 1E).
To better understand the potential therapeutic targets of t(4;14) MM, we performed functional enrichment analysis of the 255 shared targets. Using the KEGG database, we identified 232 enriched signal pathway terms through enrichment analysis. With an adjusted P value of < 0.05, the top 20 pathways, including those related to cancer, cytokine-cytokine receptor interaction, NF-κB signaling, and the Hippo signaling pathway, were identified (Figure 1F). The GSEA for the NF-κB signaling pathway is illustrated in Figure 1G. Based on these findings, we focused on the NF-κB signaling pathway to elucidate the potential mechanisms related to t(4;14) MM.
We used 258 DEGs as potential drug targets for t(4; 14) MM and assessed the CMap database to identify small compounds that could serve as prospective drugs. Table 1 lists the top nine small-molecule drugs believed to hold therapeutic potential in countering the gene expression pattern of t(4;14) MM (with a cut-off score of < −80). Notably, ivermectin, which was listed among the identified small molecules, was considered for further investigation, as it is a high-ranking approved non-chemotherapeutic drug. Thus, we evaluated the potential inhibitory effects of ivermectin on t(4;14) MM cells.
| ID | Name | Score | Mechanism of action |
| BRD-K29733039 | Deforolimus | −92.69 | mTOR inhibitor |
| BRD-A48570745 | Ivermectin | −91.74 | GABA receptor regulator |
| BRD-K33583600 | Isoliquiritigenin | −91.69 | Guanylate cyclase activator |
| BRD-K59456551 | Methotrexate | −90.69 | Dihydrofolate reductase inhibitor |
| BRD-A43150328 | Penicillic acid | −87.56 | Another antibiotic |
| BRD-A64479082 | Quinidine | −85.76 | Sodium channel blocker |
| BRD-K35377380 | I-OMe-AG-538 | −83.43 | IGF-1 inhibitor |
| BRD-K59570838 | Homoveratrylamine | −82.67 | Dopamine analog |
| BRD-A50675702 | Fipronil | −80.55 | GABA gated chloride channel blocker |
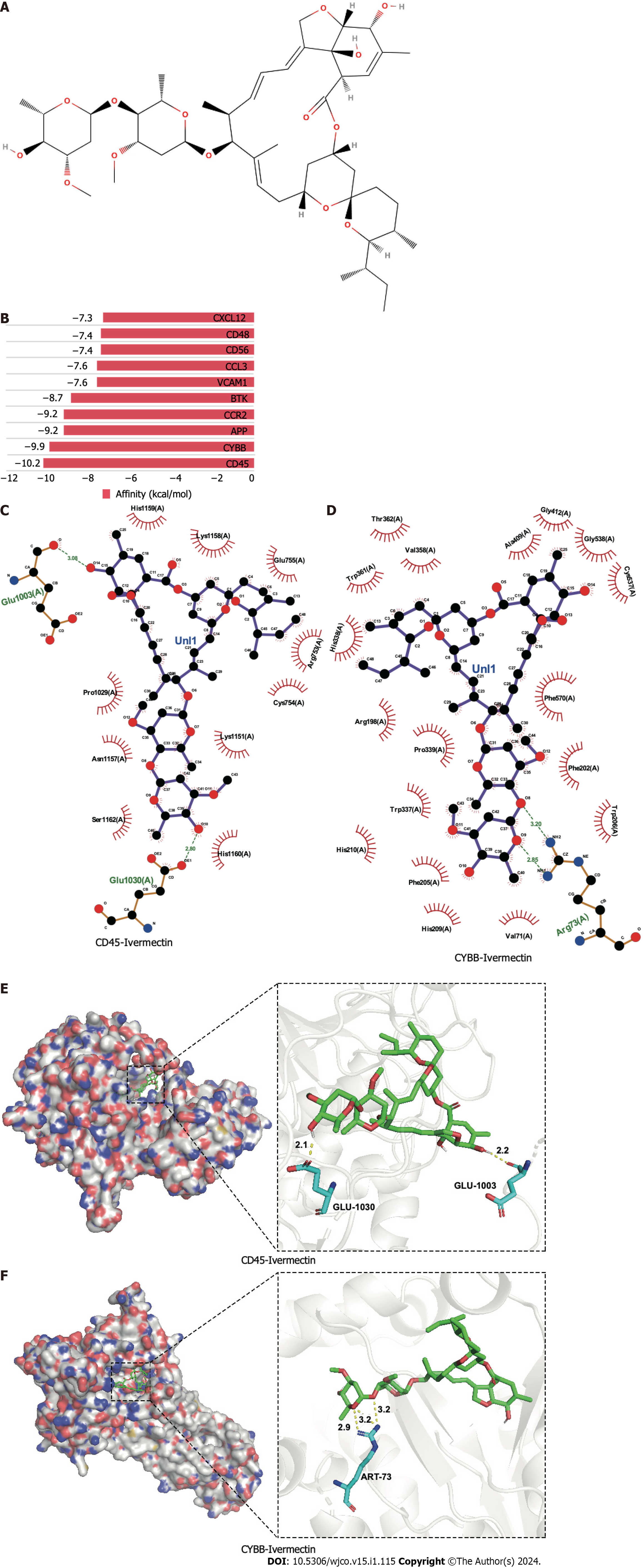
Molecular docking is a pivotal technique for the design of structure-based drugs. It facilitates the assessment of molecular interactions by determining the most favorable conformation between small-molecule targets and compounds. The 2D structure of ivermectin is shown in Figure 2A. In molecular docking simulations, ivermectin displayed strong binding affinity to all 10 identified targets, with docking energy scores below −7 kcal/mol (Figure 2B). Notably, CD45 and CYBB demonstrated the most potent binding, with binding energies of −10.2 kcal/mol and −9.9 kcal/mol, respectively. The 3D structural analysis revealedfavorable binding sites for ivermectin in both CD45 (Figure 2E) and CYBB (Figure 2F). Additionally, 2D interaction diagrams indicated hydrogen bond formation between ivermectin and specific amino acid residues in CD45 (Glu1030 and Glu1003) and CYBB (Art73), along with hydrophobic interactions in CD45 (Figure 2C) and water-mediated interactions in CYBB (Figure 2D). These findings shed light on the binding mechanisms underlying the strong affinity between ivermectin and its targets, providing valuable insights for future drug design.
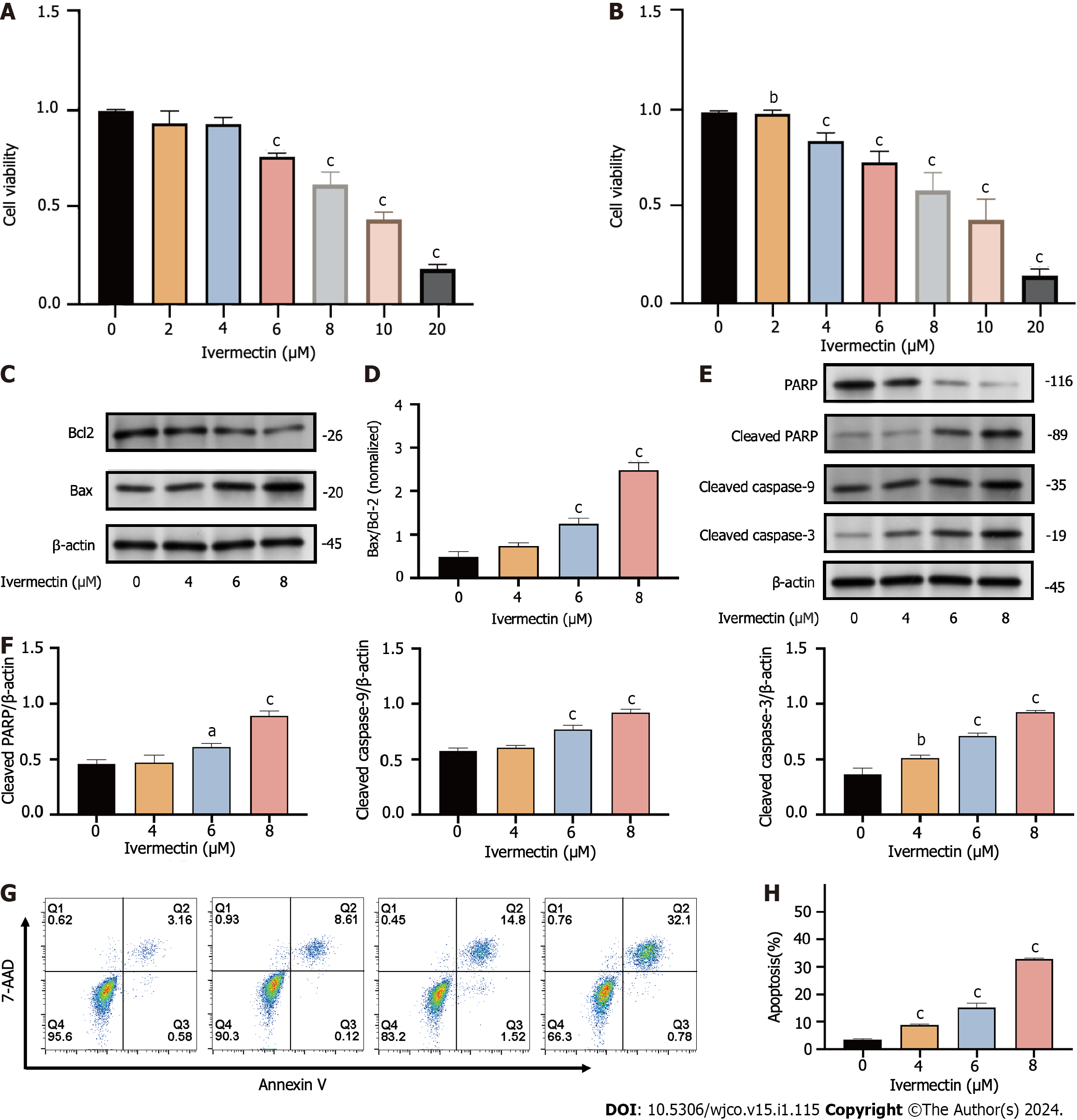
We evaluated the impact of ivermectin on t(4;14) MM cell proliferation by subjecting cells to varying concentrations (0, 4, 6, 8, 10, and 20 µmol/L) of ivermectin and time intervals (24 and 48 h) of exposure. Cell viability, determined using CCK8, demonstrated a significant, concentration- and time-dependent decrease in cancer cell viability (Figure 3A and B). The half-maximum inhibitory concentration (IC50) of ivermectin was approximately 9.4 µmol/L in NCI-H929 cells.
Drug-induced apoptosis is the primary mechanism underlying cancer cell death[18]. To assess this, we analyzed the expression of the pro- and anti-apoptotic proteins BAX and BCL2, respectively, after treatment and further inspected the BAX/BCL2 ratio. We observed that ivermectin induced apoptosis in t(4;14) MM cells, as evidenced by the upregulation of pro-apoptotic protein BAX and a decrease in anti-apoptotic protein BCL2 levels (Figure 3C). The BAX/BCL2 ratio significantly increased (Figure 3D), and the intrinsic mitochondrial apoptotic pathway was activated, as indicated by elevated caspase-9, caspase-3, downstream effector caspase-3, and PARP expression levels (Figure 3E and F). Annexin V-FITC/propidium iodide staining revealed a substantial increase in the proportion of apoptotic t(4;14) MM cells after ivermectin treatment compared with that of the control (Figure 3G and H), highlighting that the suppressive effect of ivermectin on t(4; 14) MM cells was associated with apoptosis.
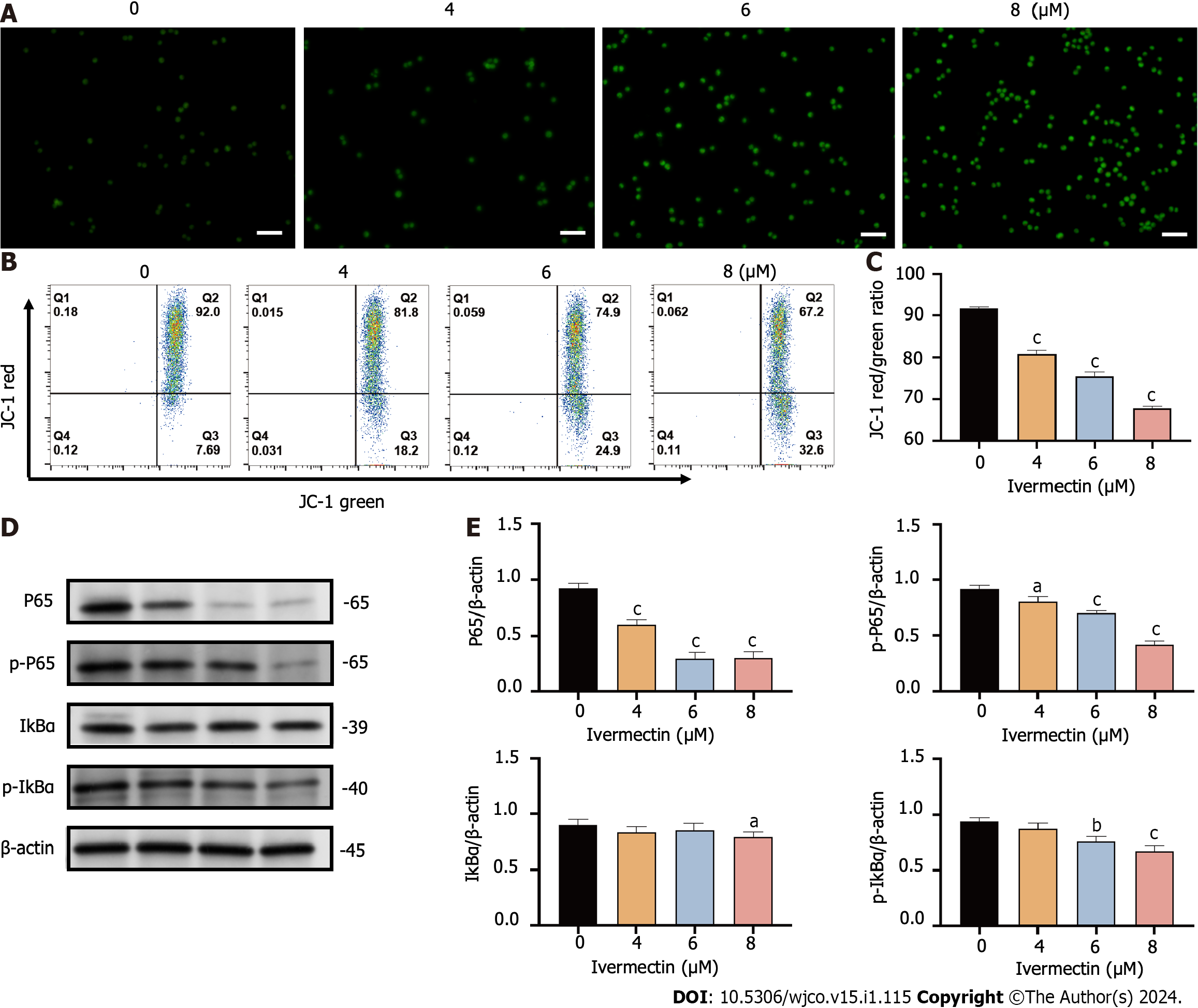
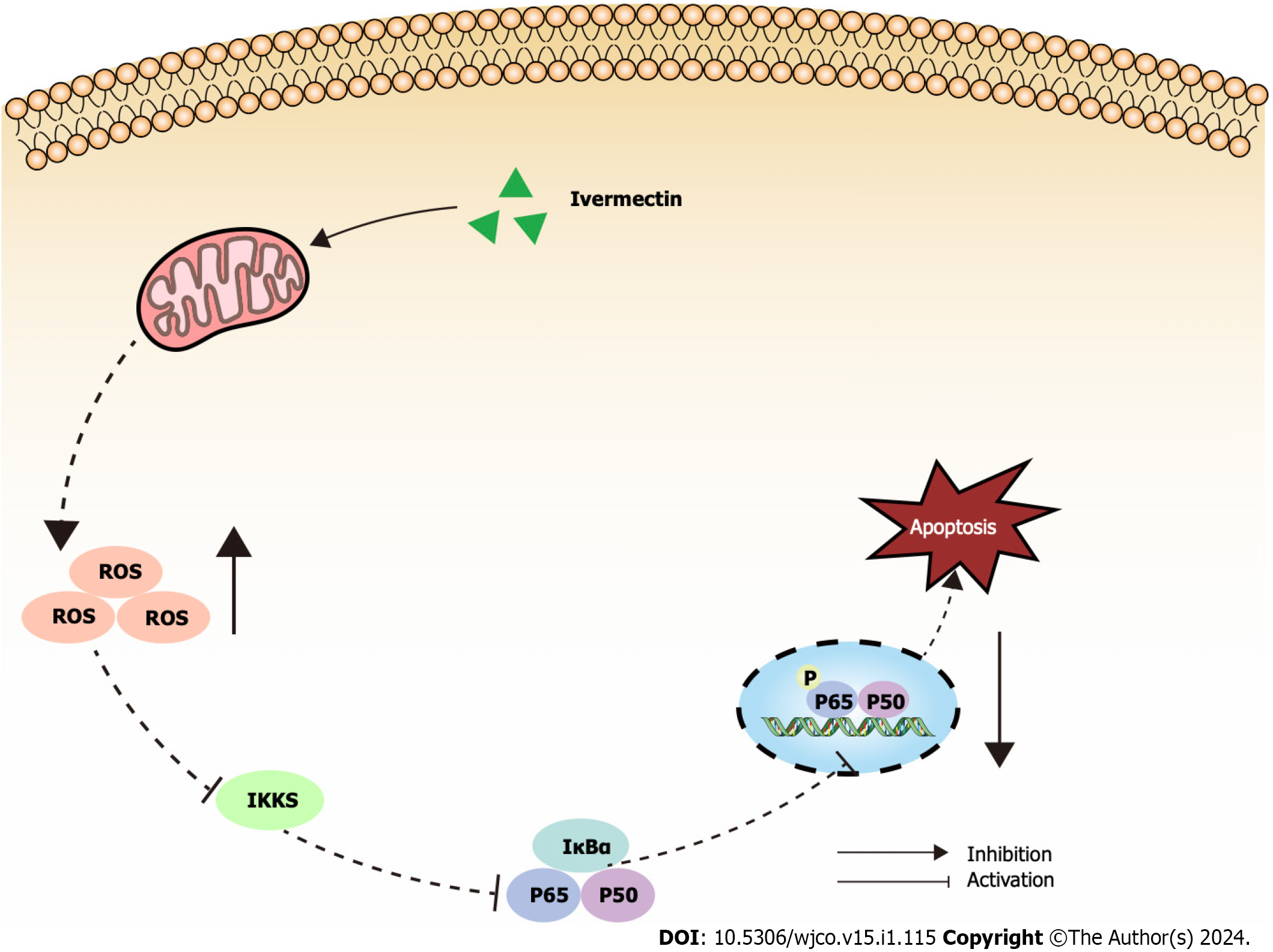
Mitochondria are widely known to be the intracellular source of ROS. These species can trigger oxidative damage, leading to a series of mitochondria-related events, including apoptosis[19]. We detected a significant accumulation of ROS in t(4;14) MM cells treated with ivermectin compared with that in untreated controls (Figure 4A). We also observed a notable reduction in the mitochondrial membrane potential (Figure 4B and C), suggesting that ivermectin-induced apoptosis in t(4;14) MM cells was related to mitochondrial function. Collectively, these findings suggest a relationship between ivermectin-induced apoptosis in t(4;14) MM cells and altered mitochondrial dynamics.
The NF-κB signaling pathway plays a crucial role in the onset and progression of the disease in t(4;14) MM, often exhibiting overactivation in these cells and promoting their survival[20]. Our results revealed that ivermectin treatment substantially reduced both the protein expression and phosphorylation levels of NF-κB p65 in a dose-dependent manner (Figure 4D and E). Furthermore, the protein expression and phosphorylation levels of IκBα, a regulator upstream of NF-κB, were also suppressed in t(4;14) MM cells post-ivermectin treatment.
MM is a multifaceted and incurable disease that displays vast heterogeneity in its clinical manifestations, genetic changes, therapeutic responses, and overall prognosis[2]. Guidelines from bodies, such as the International Myeloma Working Group, recognize t(4;14) MM as a high-risk cytogenetic abnormality[21]. The t(4;14) MM subtype continues to present a challenging prognosis despite advances in drug therapies[22]. Therefore, there is an urgent need to develop new therapeutic drugs to treat t(4;14) MM. Recent studies have highlighted the potential of identifying molecular targets and therapeutic drugs for MM through expression profiling and functional enrichment analyses. Di Meo et al[23] identified ILT3 as an immunotherapeutic target for MM, whereas Mereu et al[24] revealed that UNC0642 increasedcarfilzomib sensitivity and counteracted drug resistance in MM cell lines. In this study, we identified the key genes and prospective therapeutic agents for t(4;14) MM using gene expression profile analysis.
The regulation of the immune response is fundamental for the development and progression of MM[25]. We identified CD45, CD48, and CD56 as the key genes in t(4;14) MM. Compared with healthy bone marrow controls, patients with t(4;14) MM exhibited decreased CD45 expression levels, whereas CD48 and CD56 levels were notably increased. CD45, also known as protein tyrosine phosphatase receptor type C, was previously called the common leukocyte antigen. This protein is essential for modulating antigen receptor signaling, which is crucial for lymphocyte development, survival, and function[26]. However, the role of CD45 in MM remains elusive. Evidence suggests thatCD45 expression decreases during MM progression[27]. As mature MM cells are predominantly CD45-negative and have inactive SRC family kinases, they remain unaffected by elotuzumab[28]. Moreover, MM cells in high-risk genetic categories tend to express reduced CD45 levels[29]. CD48[30], a member of the signaling lymphocytic activation molecule family, plays a role in immune cell adhesion and activation. Although it is present in almost all MM plasma cells, it is absent from non-hematopoietic tissues. Van Acker et al[31] identified CD48 as a promising molecular target for the therapy of MM antibody therapy. CD56, a neural cell adhesion molecule, is a glycoprotein present in neural and muscle tissues, as well as in myeloma cells. Cottini et al[32] observed its high expression levels in the t(4;14) MM cell line NCI-H929. CD56 boosts MM cell growth and influences adhesion to stromal cells. Collectively, these findings highlight the potential for a deeper evaluation of immune phenotypes for both diagnosis and treatment of t(4;14) MM.
In this study, we also utilized the CMap database to identify potential small-molecule drugs targeting t(4;14) MM. Our primary focus was on molecular docking and subsequent experimental validation of the predicted drug, ivermectin. Traditionally, ivermectin is a macrocyclic lactone antibiotic used to treat parasitic diseases[33]. Recent studies have revealed its potential antitumor capabilities in various cancers, including breast[34] and pancreatic[35] cancer. Emerging evidence also suggests its synergistic lethal effects on MM cells when combined with proteasome inhibitors[36]. However, a comprehensive understanding of the specific action mechanisms of ivermectin against MM, especially the t(4;14) subtype, remains elusive. Our findings demonstrate that ivermectin suppresses the growth of t(4;14) MM cells, as well as triggers apoptosis.
The NF-κB signaling pathway is crucial for driving cancer progression, angiogenesis, and shaping the tumor microenvironment[37]. It orchestrates the production of pro-inflammatory cytokines, inflammatory mediators, and cell adhesion molecules, creating a favorable microenvironment for MM initiation and progression[38]. Consequently, many leading anti-MM drugs indirectly target the NF-κB signaling pathway[39]. Bortezomib hinders the proteasomal degradation of NF-κB and IκB proteins, inhibiting gene transcription activation[40]. Our findings revealed a pronounced suppression of the NF-κB signaling pathway in ivermectin-treated t(4;14) MM cells (Figure 5). This suggests that ivermectin, by acting as an external signaling agent, inhibits the activation of this pathway, reducing anti-apoptotic effects and thereby enhancing cell apoptosis.
Mitochondria are essential for cell survival, functioning as the primary source of ROS and producing adenosine triphosphate (ATP) through oxidative phosphorylation[41]. In a pathological state, mitochondrial dysfunction results inATP depletion[42]. In the current study, treatment of t(4;14) MM with ivermectin led to elevated ROS levels and decreased cell membrane potential. These findings, along with previous reports, suggest that ivermectin exposure can induce oxidative stress and disrupt the mitochondrial balance, facilitating the apoptosis of t(4;14) MM cells.
There are also some limitations of the present study. Firstly, the sample size may be insufficient, potentially leading to selection bias. Secondly, regarding the experiments on ivermectin against t(4;14) MM, this study has only completed a portion of the in vitro experiments, and further validation of its effectiveness and safety is still required in in vivo experiments. Furthermore, additional genetic and experimental studies are needed to elucidate the mechanisms and functions of these key genes in the occurrence and development of t(4;14) translocated multiple myeloma.
In conclusion, we utilized bioinformatics tools, molecular docking, and experimental validation to identify key genes and potential treatments for t(4;14) MM. Notably, we confirmed that ivermectin induced apoptosis in t(4;14) MM cells via the NF-κB signaling pathway. However, these insights require additional exploration and robust validation in further studies.
Multiple myeloma (MM) is a terminally differentiated B-cell tumor disease with a challenging prognosis. Specifically, the t(4;14) MM is categorized as a high-risk subtype within MM.
The t(4;14) MM tends to relapse, and currently, there is a lack of effective clinical treatment strategies.
This study aimed to elucidate the molecular basis of the t(4;14) MM and search for potential effective drugs through a comprehensive approach.
The transcriptional characteristics of t(4;14) multiple myeloma were obtained from the Gene Expression Omnibus and subjected to gene ontology and pathway enrichment analysis. Utilizing the STRING database and Cytoscape, a protein-protein interaction network was constructed, and core targets were identified. Connectivity Map identified potential small-molecule drugs, and these findings were validated through molecular docking analysis. One of these drugs, ivermectin, was further tested for its effects on t(4;14) multiple myeloma cells.
We identified 258 differentially expressed genes with enriched functions in cancer pathways, cytokine receptor interactions, the nuclear factor (NF)-kappa B signaling pathway. Ten key genes were pinpointed. Ivermectin emerged as a potential treatment. In vitro, ivermectin inhibited t(4;14) MM cell growth via the NF-kappa B pathway and induced t(4;14) MM cell apoptosis.
Ivermectin induced apoptosis in t(4;14) MM cells via the NF-κB signaling pathway.
Our study offers valuable molecular insights for biomarker validation and potential drug development in t(4;14) MM diagnosis and treatment.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Blanc R, France S-Editor: Liu JH L-Editor: A P-Editor: Zhang XD
| 1. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 9823] [Article Influence: 4911.5] [Reference Citation Analysis (1)] |
| 2. | Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA, Tuazon S, Gopal AK, Libby EN. Diagnosis and Management of Multiple Myeloma: A Review. JAMA. 2022;327:464-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 541] [Article Influence: 180.3] [Reference Citation Analysis (0)] |
| 3. | Abdallah N, Rajkumar SV, Greipp P, Kapoor P, Gertz MA, Dispenzieri A, Baughn LB, Lacy MQ, Hayman SR, Buadi FK, Dingli D, Go RS, Hwa YL, Fonder A, Hobbs M, Lin Y, Leung N, Kourelis T, Warsame R, Siddiqui M, Lust J, Kyle RA, Bergsagel L, Ketterling R, Kumar SK. Cytogenetic abnormalities in multiple myeloma: association with disease characteristics and treatment response. Blood Cancer J. 2020;10:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 4. | Foltz SM, Gao Q, Yoon CJ, Sun H, Yao L, Li Y, Jayasinghe RG, Cao S, King J, Kohnen DR, Fiala MA, Ding L, Vij R. Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nat Commun. 2020;11:2666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 5. | Ashby C, Boyle EM, Bauer MA, Mikulasova A, Wardell CP, Williams L, Siegel A, Blaney P, Braunstein M, Kaminetsky D, Keats J, Maura F, Landgren O, Walker BA, Davies FE, Morgan GJ. Structural variants shape the genomic landscape and clinical outcome of multiple myeloma. Blood Cancer J. 2022;12:85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 6. | Zhang Z, Zhou L, Xie N, Nice EC, Zhang T, Cui Y, Huang C. Overcoming cancer therapeutic bottleneck by drug repurposing. Signal Transduct Target Ther. 2020;5:113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 285] [Cited by in RCA: 314] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 7. | Hernández-Lemus E, Martínez-García M. Pathway-Based Drug-Repurposing Schemes in Cancer: The Role of Translational Bioinformatics. Front Oncol. 2020;10:605680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 8. | Martin KH, Slack JK, Boerner SA, Martin CC, Parsons JT. Integrin connections map: to infinity and beyond. Science. 2002;296:1652-1653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 173] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Shi X, Zhang W, Bao X, Liu X, Yang M, Yin C. Eugenol modulates the NOD1-NF-κB signaling pathway via targeting NF-κB protein in triple-negative breast cancer cells. Front Endocrinol (Lausanne). 2023;14:1136067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 10. | Qiu HZ, Huang J, Xiang CC, Li R, Zuo ED, Zhang Y, Shan L, Cheng X. Screening and Discovery of New Potential Biomarkers and Small Molecule Drugs for Cervical Cancer: A Bioinformatics Analysis. Technol Cancer Res Treat. 2020;19:1533033820980112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 11. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11669] [Article Influence: 1944.8] [Reference Citation Analysis (1)] |
| 12. | Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1658] [Cited by in RCA: 3720] [Article Influence: 338.2] [Reference Citation Analysis (0)] |
| 13. | Yang K, Dinasarapu AR, Reis ES, Deangelis RA, Ricklin D, Subramaniam S, Lambris JD. CMAP: Complement Map Database. Bioinformatics. 2013;29:1832-1833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204-D212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3457] [Cited by in RCA: 3658] [Article Influence: 332.5] [Reference Citation Analysis (0)] |
| 15. | Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102-D1109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1569] [Cited by in RCA: 1909] [Article Influence: 381.8] [Reference Citation Analysis (0)] |
| 16. | Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778-2786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2939] [Cited by in RCA: 4111] [Article Influence: 293.6] [Reference Citation Analysis (0)] |
| 17. | Delano WL. Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr. 2002;40:82-92. [DOI] [Full Text] |
| 18. | Singh P, Lim B. Targeting Apoptosis in Cancer. Curr Oncol Rep. 2022;24:273-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 115] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 19. | Nakamura H, Takada K. Reactive oxygen species in cancer: Current findings and future directions. Cancer Sci. 2021;112:3945-3952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 461] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 20. | Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Signal Transduct Target Ther. 2020;5:209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 821] [Cited by in RCA: 1242] [Article Influence: 248.4] [Reference Citation Analysis (0)] |
| 21. | Hagen P, Zhang J, Barton K. High-risk disease in newly diagnosed multiple myeloma: beyond the R-ISS and IMWG definitions. Blood Cancer J. 2022;12:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 47] [Reference Citation Analysis (0)] |
| 22. | Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC, Chng WJ, Moreau P, Attal M, Kyle RA, Caers J, Hillengass J, San Miguel J, van de Donk NW, Einsele H, Bladé J, Durie BG, Goldschmidt H, Mateos MV, Palumbo A, Orlowski R. Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood. 2016;127:2955-2962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 693] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 23. | Di Meo F, Iyer A, Akama K, Cheng R, Yu C, Cesarano A, Kurihara N, Tenshin H, Aljoufi A, Marino S, Soni RK, Roda J, Sissons J, Vu LP, Guzman M, Huang K, Laskowski T, Broxmeyer HE, Roodman DG, Perna F. A target discovery pipeline identified ILT3 as a target for immunotherapy of multiple myeloma. Cell Rep Med. 2023;4:101110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 24. | Mereu E, Abbo D, Paradzik T, Cumerlato M, Bandini C, Labrador M, Maccagno M, Ronchetti D, Manicardi V, Neri A, Piva R. Euchromatic Histone Lysine Methyltransferase 2 Inhibition Enhances Carfilzomib Sensitivity and Overcomes Drug Resistance in Multiple Myeloma Cell Lines. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 25. | Swamydas M, Murphy EV, Ignatz-Hoover JJ, Malek E, Driscoll JJ. Deciphering mechanisms of immune escape to inform immunotherapeutic strategies in multiple myeloma. J Hematol Oncol. 2022;15:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 68] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 26. | Al Barashdi MA, Ali A, McMullin MF, Mills K. Protein tyrosine phosphatase receptor type C (PTPRC or CD45). J Clin Pathol. 2021;74:548-552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 142] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 27. | Robillard N, Pellat-Deceunynck C, Bataille R. Phenotypic characterization of the human myeloma cell growth fraction. Blood. 2005;105:4845-4848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 28. | Guo H, Cruz-Munoz ME, Wu N, Robbins M, Veillette A. Immune cell inhibition by SLAMF7 is mediated by a mechanism requiring src kinases, CD45, and SHIP-1 that is defective in multiple myeloma cells. Mol Cell Biol. 2015;35:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 29. | Radzevičius M, Dirsė V, Klimienė I, Matuzevičienė R, Kučinskienė ZA, Pečeliūnas V. Multiple Myeloma Immunophenotype Related to Chromosomal Abnormalities Used in Risk Assessment. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 30. | McArdel SL, Terhorst C, Sharpe AH. Roles of CD48 in regulating immunity and tolerance. Clin Immunol. 2016;164:10-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 153] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 31. | Van Acker HH, Capsomidis A, Smits EL, Van Tendeloo VF. CD56 in the Immune System: More Than a Marker for Cytotoxicity? Front Immunol. 2017;8:892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 249] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 32. | Cottini F, Rodriguez J, Hughes T, Sharma N, Guo L, Lozanski G, Liu B, Cocucci E, Yang Y, Benson D. Redefining CD56 as a Biomarker and Therapeutic Target in Multiple Myeloma. Mol Cancer Res. 2022;20:1083-1095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | Laing R, Gillan V, Devaney E. Ivermectin - Old Drug, New Tricks? Trends Parasitol. 2017;33:463-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 244] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 34. | Draganov D, Han Z, Rana A, Bennett N, Irvine DJ, Lee PP. Ivermectin converts cold tumors hot and synergizes with immune checkpoint blockade for treatment of breast cancer. NPJ Breast Cancer. 2021;7:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 35. | Lee DE, Kang HW, Kim SY, Kim MJ, Jeong JW, Hong WC, Fang S, Kim HS, Lee YS, Kim HJ, Park JS. Ivermectin and gemcitabine combination treatment induces apoptosis of pancreatic cancer cells via mitochondrial dysfunction. Front Pharmacol. 2022;13:934746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 36. | Luo H, Feng Y, Wang F, Lin Z, Huang J, Li Q, Wang X, Liu X, Zhai X, Gao Q, Li L, Zhang Y, Wen J, Zhang L, Niu T, Zheng Y. Combinations of ivermectin with proteasome inhibitors induce synergistic lethality in multiple myeloma. Cancer Lett. 2023;565:216218. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 37. | Napetschnig J, Wu H. Molecular basis of NF-κB signaling. Annu Rev Biophys. 2013;42:443-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 594] [Cited by in RCA: 759] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 38. | Wong AH, Shin EM, Tergaonkar V, Chng WJ. Targeting NF-κB Signaling for Multiple Myeloma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 39. | Vrábel D, Pour L, Ševčíková S. The impact of NF-κB signaling on pathogenesis and current treatment strategies in multiple myeloma. Blood Rev. 2019;34:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 40. | Paramore A, Frantz S. Bortezomib. Nat Rev Drug Discov. 2003;2:611-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 182] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 41. | Zhang B, Pan C, Feng C, Yan C, Yu Y, Chen Z, Guo C, Wang X. Role of mitochondrial reactive oxygen species in homeostasis regulation. Redox Rep. 2022;27:45-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 206] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 42. | Ma Z, Han H, Zhao Y. Mitochondrial dysfunction-targeted nanosystems for precise tumor therapeutics. Biomaterials. 2023;293:121947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 42] [Reference Citation Analysis (0)] |