Published online Oct 24, 2022. doi: 10.5306/wjco.v13.i10.779
Peer-review started: July 26, 2022
First decision: August 18, 2022
Revised: August 25, 2022
Accepted: September 15, 2022
Article in press: September 15, 2022
Published online: October 24, 2022
Processing time: 85 Days and 7.8 Hours
The FAT cadherin family members (FAT1, FAT2, FAT3 and FAT4) are conserved tumor suppressors that are recurrently mutated in several types of human cancers, including colorectal carcinoma (CRC).
To characterize the clinicopathologic features of CRC patients with somatic mutations in FAT cadherin family members.
We analyzed 526 CRC cases from The Cancer Genome Atlas PanCancer Atlas dataset. CRC samples were subclassified into 2 groups based on the presence or absence of somatic mutations in FAT1, FAT2, FAT3 and FAT4. Individual clinic
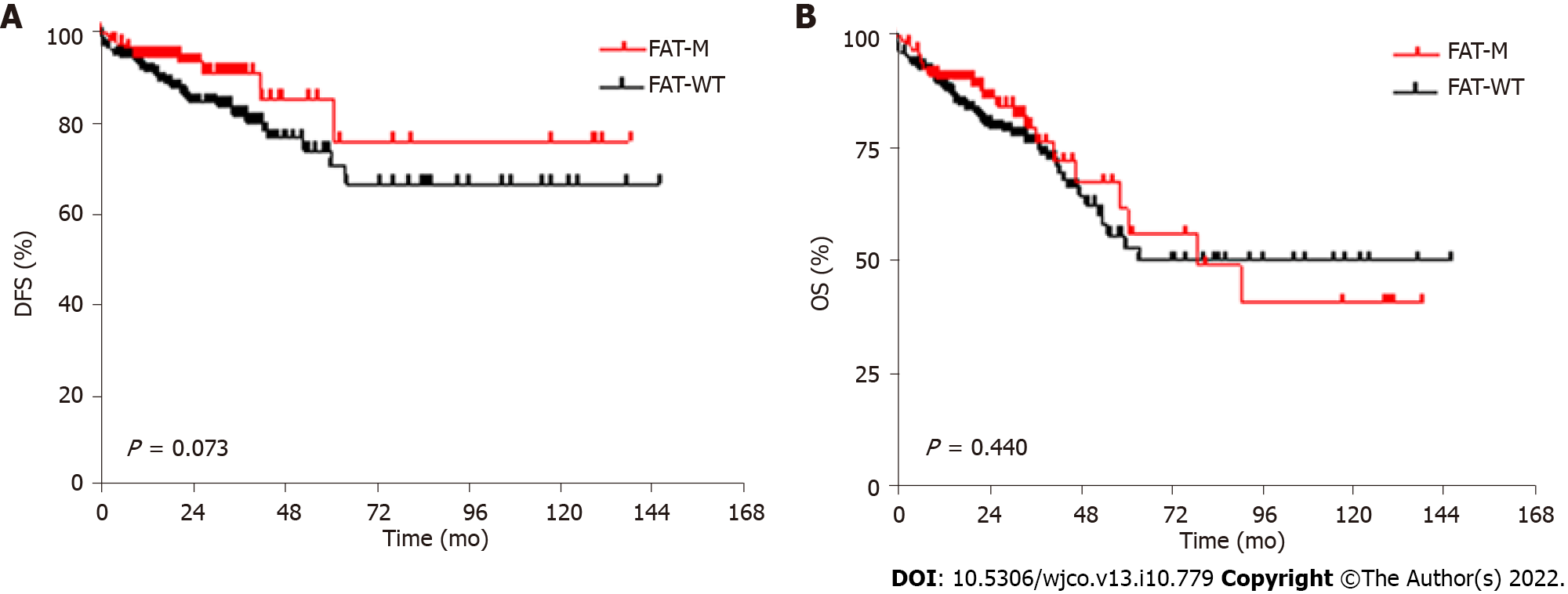
This CRC study cohort had frequent mutations in the FAT1 (10.5%), FAT2 (11.2%), FAT3 (15.4%) and FAT4 (23.4%) genes. Two hundred CRC patients (38.0%) harbored somatic mutations in one or more of the FAT family genes and were grouped into the FAT mutated CRC subtype. The FAT-mutated CRC subtype was more commonly located on the right side of the colon (51.0%) than in the rest of the cohort (30.1%, P < 0.001). It showed favorable clinicopathologic features, including a lower rate of positive lymph nodes (pN1-2: 33.5% vs 46.4%, P = 0.005), a lower rate of metastasis to another site or organ (pM1: 7.5% vs 16.3%, P = 0.006), and a trend toward an early tumor stage (pT1-2: 25.0% vs 18.7%, P = 0.093). FAT somatic mutations were significantly enriched in microsatellite instability CRC (28.0% vs 2.1%, P < 0.001). However, FAT somatic mutations in microsatellite stable CRC demonstrated similar clinicopathologic behaviors, as well as a trend of a better disease-free survival rate (hazard ratio = 0.539; 95% confidence interval: 0.301-0.967; log-rank P = 0.073).
FAT cadherin family genes are frequently mutated in CRC, and their mutation profile defines a subtype of CRC with favorable clinicopathologic characteristics.
Core Tip: Colorectal carcinoma (CRC) is the third most common cancer and the second leading cause of cancer-related deaths worldwide. In this study, we aimed to characterize the clinicopathologic features of CRC patients with somatic mutations in FAT cadherin family members. CRC cases have frequent mutations in FAT family genes. The FAT-mutated CRC subtype is more commonly located on the right side of the colon and shows favorable clinicopathologic features, including a lower rate of positive lymph nodes and a lower rate of metastasis to another site or organ, suggesting that the FAT somatic mutation is a potentially independent prognostic factor in CRC.
- Citation: Wang LL, Zheng W, Liu XL, Yin F. Somatic mutations in FAT cadherin family members constitute an underrecognized subtype of colorectal adenocarcinoma with unique clinicopathologic features. World J Clin Oncol 2022; 13(10): 779-788
- URL: https://www.wjgnet.com/2218-4333/full/v13/i10/779.htm
- DOI: https://dx.doi.org/10.5306/wjco.v13.i10.779
Colorectal carcinoma (CRC) is the third most common cancer and the second leading cause of cancer-related deaths worldwide, with more than 1.9 million new cases and 935000 deaths in 2020[1]. Except for a few CRC cases (5%-10%) with inherited gene mutations, most CRC cases occur sporadically and exhibit chromosomal instability that leads to changes in chromosome numbers and structure, featuring aneuploidy, loss of heterozygosity, subkaryotypic amplification, and chromosomal rearrangement. Along with karyotypic abnormalities, mutations in specific tumor suppressor genes and oncogenes, such as the adenomatous polyposis (APC) gene, tumor protein p53 (TP53) and KRAS proto-oncogene GTPase, also contribute to CRC tumorigenesis. Notably, mutation of the APC gene, which leads to the activation of Wnt/β-catenin signaling, is an essential and early event in the development of CRC[2,3].
Despite the well-defined genetic and epigenetic alterations in CRC initiation and progression, recent studies have shown that the Hippo pathway may interact with Wnt/β-catenin signaling and play a crucial role in controlling intestinal stem cell proliferation and CRC development[4]. The Hippo pathway is an emerging tumor suppressor pathway. As a proposed upstream component of the Hippo pathway, the atypical cadherin FAT acts as a receptor to activate the Hippo pathway[5], and its mutation appears to be a recurrent event in human cancers in association with dysregulation of the Hippo pathway[6].
The human FAT cadherin gene family comprises the FAT1, FAT2, FAT3 and FAT4 genes[7-10]. The encoded proteins FAT1-4 are human homologs of Drosophila FAT, of which FAT1 and FAT4 have been reported to be involved in the regulation of planar cell polarity[11] and tumor suppression[12,13]. FAT1 also promotes actin-mediated cell migration[14,15] and plays a role in epithelial mesenchymal transition[16]. Somatic mutations of FAT family genes have been detected in different human cancers, including squamous cell carcinoma of the head and neck (FAT1, FAT2 and FAT4)[17-20], breast cancer (FAT1)[21], melanomas (FAT4)[22], leukemia (FAT1)[23,24], hepatocellular cancer (FAT1, FAT4)[25,26], esophageal squamous cell carcinoma (FAT1)[27-29], pancreatic cancer (FAT1, FAT3 and FAT4)[30,31], and gastric cancer (FAT4)[32,33]. Alterations in FAT family genes are associated with tumorigenesis and prognosis. For instance, upregulation of the FAT1 gene is associated with poor prognosis and early relapse in acute lymphoblastic leukemia patients[24] and invasive progression of ductal carcinoma in situ[21], while loss of FAT4 is associated with invasiveness in gastric cancer[34]. Until now, the role of FAT family genes in CRC tumorigenesis has not been well studied. In this study, we characterized the clinicopathologic features of FAT family gene mutations in CRC patients.
In total, 526 CRC cases were selected from The Cancer Genome Atlas (TCGA) PanCancer Atlas dataset. cBioPortal (https://www.cbioportal.org/) was used to download whole-exome somatic mutation data and clinical information. There are certain sample inclusion criteria for the TCGA PanCancer Atlas on colorectal adenocarcinoma. The biospecimens were collected from newly diagnosed colorectal adenocarcinoma patients undergoing surgical resection, regardless of histologic grade or tumor stage. The patients had not received prior chemoradiation therapy. The histological sections contained an average of 60% tumor cells with less than 20% necrosis[35].
In the TCGA PanCancer Atlas dataset, the somatic mutation profiles of FAT1, FAT2, FAT3 and FAT4 were analyzed for each tumor. Furthermore, the CRC cases were categorized into two groups based on their mutational status on FAT family genes: The cases with mutant FAT1-4 and the cases with wild-type FAT1-4. Standard demographic and clinicopathological data were retrieved for each patient, including age, sex, tumor location, pT stage, pN stage, pM stage, differentiation grade, tumor type, lymphovascular invasion, month of disease-free survival (DFS) and overall survival (OS).
Demographic and clinicopathological details were stratified according to FAT1-4 mutation. Quantitative and qualitative variables were expressed as the means ± SD and the frequencies. Comparisons between the groups were analyzed with t tests and chi-square tests. DFS and OS were analyzed using the Kaplan-Meier method, and the log-rank test was used to assess differences. The figure was prepared using GraphPad Prism 9 software (GraphPad Software, San Diego, California, United States). P values less than 0.05 were considered statistically significant.
The study included 526 patients with CRC from TCGA PanCancer Atlas Dataset. The mean age of the patients was 65.8 years (SD 13.0 years; range: 31-90 years). Based on the available clinicodemographic information, two hundred fifty-two patients were female, and two hundred seventy-two patients were male. Of them, 254 (48.3%) patients had left-sided colon cancer, and 197 (37.5%) patients had right-sided colon cancer. The majority (72.4%) of the CRCs were moderately differentiated adenocarcinomas. The detailed demographics, histopathologic stage and features are summarized in Table 1.
| Feature | Level | Number | MSS number |
| Age (yr), mean ± SD | 65.8 ± 13.0 | 65.4 ± 12.7 | |
| Gender | Female | 252 (47.9%) | 218 (47.1%) |
| Male | 272 (51.7%) | 243 (52.5%) | |
| Unknown | 2 (0.4%) | 2 (0.4%) | |
| Histopathologic differentiation | Well | 19 (3.6%) | 18 (3.9%) |
| Moderate | 381 (72.4%) | 351 (75.8%) | |
| Poor | 114 (21.7%) | 83 (17.9%) | |
| Unknown | 12 (2.3%) | 11 (2.4%) | |
| Tumor location | Left | 254 (48.3%) | 248 (53.6%) |
| Right | 197 (37.5%) | 149 (32.2%) | |
| Left and right | 3 (0.6%) | 3 (6.5%) | |
| Unknown | 72 (13.7%) | 63 (13.6%) | |
| Tumor staging (pT) | T1 | 18 (3.4%) | 17 (3.7%) |
| T2 | 94 (17.9%) | 83 (17.9%) | |
| T3 | 355 (67.5%) | 310 (67.0%) | |
| T4 | 57 (10.8%) | 52 (11.2%) | |
| TX | 2 (0.4%) | 2 (0.4%) | |
| Nodal staging (pN) | N0 | 305 (58.0%) | 255 (55.1%) |
| N1 | 128 (24.3%) | 120 (25.9%) | |
| N2 | 90 (17.1%) | 85 (18.4%) | |
| NX | 3 (0.6%) | 3 (6.5%) | |
| Metastasis (pM) | M0 | 388 (73.8%) | 338 (73.0%) |
| M1 | 68 (12.9%) | 66 (14.3%) | |
| MX | 70 (13.3%) | 59 (12.7%) | |
| Lymphovascular invasion | Present | 178 (33.8%) | 157 (33.9%) |
| Absent | 230 (43.7%) | 202 (43.6%) | |
| Unknown | 118 (22.4%) | 104 (22.5%) | |
| Ethnicity | Caucasian | 273 (51.9%) | 236 (51.0%) |
| African-American | 60 (11.4%) | 51 (11.0%) | |
| Asian | 12 (2.3%) | 11 (2.4%) | |
| Unknown | 181 (34.4%) | 165 (35.6%) | |
| Subtype | CIN | 328 (62.4%) | |
| MSI | 63 (12.0%) | ||
| GS | 58 (11.0%) | ||
| POLE | 10 (1.9%) | ||
| Unknown | 57 (10.8%) | ||
| Total | 526 | 463 |
Among the 526 CRC cases, 200 (38.0%) patients harbored one or more somatic mutations of the FAT cadherin family genes, including mutations in the FAT1 (10.5%), FAT2 (11.2%), FAT3 (15.4%), and FAT4 (23.4%) genes. The somatic mutation types of the FAT family genes include missense mutation, nonsense mutation, splicing mutation, frameshift deletion, frameshift insertion and in-frame deletion, with missense mutation being the most common somatic mutation type (Table 2). Interestingly, these somatic mutations were significantly enriched in the extracellular cadherin domain (FAT1, 49.0%; FAT2, 63.4%; FAT3, 40.1%; FAT4, 57.8%) (Table 2).
| Gene | Missense mutation | Nonsense mutation | Splicing mutation | Frame shift deletion | Frame shift insertion | Inflame deletion | Total mutation | Mutation in Cadherin domains |
| FAT1 | 85 | 5 | 2 | 3 | 2 | 1 | 98 | 48 (49.0%) |
| FAT2 | 90 | 2 | 3 | 5 | 1 | 0 | 101 | 64 (63.4%) |
| FAT3 | 124 | 6 | 0 | 5 | 2 | 0 | 137 | 55 (40.1%) |
| FAT4 | 198 | 19 | 0 | 10 | 4 | 0 | 230 | 133 (57.8%) |
Based on the presence or absence of somatic mutations in FAT1-4 genes, these cases were subclassified into 2 groups in our study. The clinicopathologic features of these 2 subtypes are summarized in Table 3. In the FAT-mutated CRC subtype, the median patient age was 66.5 years (range: 33-90 years), and 102 (51.0%) patients were male. Compared with the rest of the cohort, the FAT-mutated CRC subtype was more commonly located on the right side of the colon (51.0% vs 30.1%, P < 0.001) and more commonly associated with favorable histopathologic features, including lower pathological nodal stage (pN0: 66.5% vs 52.8%, P = 0.005), lower rate of metastasis to another site or organ (pM1: 7.5% vs 16.3%, P = 0.006), and a trend of lower pathological tumor stage (pT1-2: 25.0% vs 18.7%, P = 0.093).
| Clinicopathologic features | Mutated FAT genes | Wildtype FAT genes | P value | Mutated FAT genes (MSS) | Wildtype FAT genes (MSS) | P value |
| Mean age (mean ± SD) | 66.5 ± 12.9 | 65.3 ± 13.0 | 0.912 | 65.6 ± 12.1 | 65.3 ± 12.9 | |
| Sex | 0.689 | 0.825 | ||||
| Female | 98 (49.0%) | 154 (47.2%) | 67 (46.5%) | 151 (47.3%) | ||
| Male | 102 (51.0%) | 170 (52.1%) | 77 (53.5%) | 166 (52.0%) | ||
| Location | < 0.001a | 0.038a | ||||
| Left side | 65 (32.5%) | 181 (55.5%) | 70 (48.6%) | 178 (55.8%) | ||
| Right side | 102 (51.0) | 98 (30.1%) | 57 (39.6%) | 92 (28.8%) | ||
| pT stage | 0.093 | 0.083 | ||||
| pT1-2 | 50 (25.0%) | 61 (18.7%) | 38 (26.4%) | 61 (19.1%) | ||
| pT3-4 | 150 (75.0%) | 263 (80.7%) | 106 (73.6%) | 256 (80.3%) | ||
| pN stage | 0.005a | 0.079 | ||||
| pN0 | 133 (66.5%) | 172 (52.8%) | 87 (60.4%) | 168 (52.7%) | ||
| pN1 | 44 (22.0%) | 84 (25.8%) | 39 (27.1%) | 81 (25.4%) | ||
| pN2 | 23 (11.5%) | 67 (20.6%) | 18 (12.5%) | 67 (21.0%) | ||
| pM stage | 0.006a | 0.038a | ||||
| pM0 | 153 (76.5%) | 235 (72.1%) | 110 (76.4%) | 228 (71.5%) | ||
| pM1 | 15 (7.5%) | 53 (16.3%) | 13 (9.0%) | 53 (16.6%) | ||
| Differentiation grade | 0.332 | 0.172 | ||||
| G1-2 | 145 (72.5%) | 255 (78.2%) | 117 (81.3%) | 252 (79.0%) | ||
| G3 | 47 (23.5%) | 67 (20.6%) | 20 (13.9%) | 63 (19.7%) | ||
| Subtype | < 0.001a | |||||
| CIN | 92 (46.0%) | 236 (72.4%) | ||||
| MSI | 56 (28.0%) | 7 (2.1%) | ||||
| GS | 25 (12.5%) | 33 (10.1%) | ||||
| Lymphovascular invasion | 0.313 | 0.516 | ||||
| Positive | 61 (30.5%) | 117 (35.9%) | 44 (30.6%) | 113 (35.4%) | ||
| Negative | 90 (45.0%) | 140 (42.9%) | 63 (43.8%) | 139 (43.6%) | ||
| Total | 200 (38.0%) | 326 (62.0%) | 144 (31.1%) | 319 (68.9%) |
Human FAT family genes encode large atypical cadherin proteins with a large number of cadherin repeats. Given the overlapping features found in the FAT-mutated CRC subtype and microsatellite-instable (MSI) CRC (right sided with favorable clinicopathological features), we further explored the association between FAT mutations and MSI. Interestingly, FAT somatic mutations were significantly enriched in MSI CRC (28.0% vs 2.1%, P < 0.001) (Table 3).
To control for confounding in the analysis, we focused on cases of microsatellite-stable (MSS) CRC. As shown in Table 1, the MSS CRC cases showed similar clinicodemographic and histologic features as the entire cohort. We also categorized the MSS CRC cases into 2 groups based on the mutation status of FAT family genes. Similar to the entire cohort we described earlier, the FAT-mutated MSS CRC subtype was also more commonly located on the right side of the colon (39.6% vs 28.8%, P = 0.038) and more commonly associated with favorable histopathologic features, such as a lower rate of metastasis to another site or organ (pM1: 9.0% vs 16.6%, P = 0.038). It also showed a trend of lower pathological tumor stage (pT1-2: 26.4% vs 19.1%, P = 0.083) and lower pathological nodal stage (pN0: 60.4% vs 52.7%, P = 0.079) (Table 3). Therefore, even though it is enriched in MSI CRC, the FAT somatic mutation is a potentially independent prognostic factor in CRC.
The median DFS for CRC patients was 26.0 mo (0.5-148.0 mo), and the OS was 21.0 mo (0-148.0 mo). Consistent with the favorable pathologic features, the FAT-mutated MSS CRC subgroup showed a trend toward a better DFS rate [hazard ratio (HR) = 0.539; 95% confidence interval (CI): 0.301-0.967; log-rank P = 0.073]. However, FAT mutation status did not show a significant impact on the OS rate (HR = 1.198; 95%CI: 0.770-1.864; log-rank P = 0.440) (Figure 1).
To our knowledge, this is the first study to assess the impact of somatic mutations in FAT family genes on clinicopathologic features, with an emphasis on prognosis in CRC patients. Our study shows that somatic mutations in FAT family genes are associated with favorable clinicopathologic features, including a lower rate of lymph node and distal metastasis. It also showed a trend toward a lower tumor stage with a relatively favorable DFS.
In addition to the APC-β-catenin pathway, which represents the most prominent signaling pathway in CRC, components of the Hippo pathway have been reported to be involved in CRC tumorigenesis[36-40] and have been proposed as prognostic factors in CRC[41-44]. As an upstream organizer and activator of the Hippo pathway[6], FAT family genes have emerged as an important mechanism that orchestrates epithelial development as well as human cancer initiation and progression. The FAT family genes (FAT1-4) encode atypical cadherins that contain multiple extracellular cadherin repeats, laminin G motifs and EGF-like motifs[45]. Among these, FAT1 and FAT4 are relatively well studied. Loss of FAT4 expression has been reported in some primary breast cancers and breast cancer cell lines[46]. Low FAT4 expression was also observed in gastric cancers and was associated with a poor prognosis, including high pathologic T stage, an increase in perineural invasion, high lymph node metastasis and reduced DFS[47]. Similarly, a study reported recurrent FAT1 mutations in multiple human cancers, including glioblastoma, CRC, and head and neck cancer, and FAT1 mutations affected patient survival by promoting Wnt signaling and tumorigenesis[48]. Our study demonstrates that somatic mutations in FAT family genes are frequent recurrent events in CRC and that FAT mutations are associated with favorable clinicopathologic features. These somatic mutations are highly enriched in the extracellular cadherin domains (Table 2). FAT proteins are large single transmembrane receptors characterized by 32-34 extracellular cadherin repeats. These cadherin repeats contain highly conserved binding sites for proteins, such as beta-catenin and p120-catenin, which are important for the FAT protein to execute its role in migration, polarity and cell adhesion by linking it to the actin cytoskeleton.
Our study also revealed the significant enrichment of FAT-mutated CRC (28.0%) in the MSI subgroup. However, the clinicopathologic characteristics in FAT-mutated MSS CRC are quite compatible with the entire FAT-mutated CRC cohort in our study, suggesting that MSI only partially contributes to its pathologic features and clinical outcomes. Interestingly, FAT-mutated MSS CRC cases showed a trend of favorable DFS but not OS. The underlying mechanisms of this discrepancy are currently unclear. Notably, DFS does not always correlate with OS in CRC, such as in the case of liver-only metastatic CRC[49].
Similar to the findings in our study, Wang et al[33] reported a superior prognosis in gastric adenocarcinoma with FAT family gene mutations. In their study, FAT gene mutations were significantly associated with better progression-free survival and OS, which was likely attributed to the significantly higher tumor mutational burden and an inflamed tumor microenvironment[33]. Whether the tumor microenvironment plays a similar role in CRC still awaits further investigation.
Our study has several limitations. First, our findings were obtained from a bioinformatics study on somatic mutation profiles through the TCGA PanCancer Atlas dataset. The protein expression levels of individual FAT family members were not systemically examined in the study, and the underlying molecular mechanisms related to the prognostic role of the FAT family in colorectal cancer need further experimental validation. Second, all the patients in the study were untreated, with no therapy response data and a short follow-up. Therefore, the evaluation of advanced-stage CRC is relatively limited. Third, we tried to address the impact of MSI status, a confounding factor, by analyzing the MSS samples. However, there are still additional potential confounding factors, such as histopathological subtypes, TP53 mutation status, and intratumoral spatial and temporal heterogeneity. The ability of our study to address these potential confounding factors is hampered by intrinsic limitations of the TCGA database, the landmark cancer program heavily focused on cancer genomics datasets. A randomized, large-scale clinical cohort is necessary to validate our conclusion and to establish somatic mutations in FAT family genes as independent prognostic factors for CRC in future studies.
In summary, our study shows that somatic mutations in FAT family genes are recurrent genetic events detected in approximately 38% of CRC cases and therefore represent an underrecognized subtype of CRC. The FAT-mutated CRC subtype shows unique clinicopathologic features, including a right-side location, a lower rate of positive lymph nodes, a lower rate of metastasis to another site or organ, and a trend toward favorable DFS. Our study suggests that somatic mutations in FAT family genes are potential prognostic biomarkers for CRC.
The human FAT cadherin gene family comprises the FAT1, FAT2, FAT3 and FAT4 genes. Somatic mutations of FAT family genes have been detected in different human cancers.
Until now, the role of FAT family genes in colorectal carcinoma (CRC) tumorigenesis has not been well studied. In this study, we characterized the clinicopathologic features of FAT family gene mutations in CRC patients.
In total, 526 CRC cases were selected from The Cancer Genome Atlas PanCancer Atlas dataset.
CRC cases were categorized into two groups based on their mutational status on FAT family genes: The cases with mutant FAT1-4 and the cases with wild-type FAT1-4. Standard demographic and clinicopathological data were retrieved for each patient, including age, sex, tumor location, pT stage, pN stage, pM stage, differentiation grade, tumor type, lymphovascular invasion, month of disease-free survival and overall survival.
The FAT-mutated CRC subtype is more commonly located on the right side of the colon and shows favorable clinicopathologic features, including a lower rate of positive lymph nodes and a lower rate of metastasis to another site or organ.
FAT cadherin family genes are frequently mutated in CRC, and their mutation profile defines a subtype of CRC with favorable clinicopathologic characteristics.
FAT somatic mutation is a potentially independent prognostic factor in CRC.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Aktekin A, Turkey; Osera S, Japan; Yang Z, China S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64121] [Article Influence: 16030.3] [Reference Citation Analysis (174)] |
| 2. | Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol Lett. 2018;16:9-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (1)] |
| 3. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 2813] [Article Influence: 104.2] [Reference Citation Analysis (0)] |
| 4. | Wierzbicki PM, Rybarczyk A. The Hippo pathway in colorectal cancer. Folia Histochem Cytobiol. 2015;53:105-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 5. | Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090-2100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 270] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 6. | Martin D, Degese MS, Vitale-Cross L, Iglesias-Bartolome R, Valera JLC, Wang Z, Feng X, Yeerna H, Vadmal V, Moroishi T, Thorne RF, Zaida M, Siegele B, Cheong SC, Molinolo AA, Samuels Y, Tamayo P, Guan KL, Lippman SM, Lyons JG, Gutkind JS. Assembly and activation of the Hippo signalome by FAT1 tumor suppressor. Nat Commun. 2018;9:2372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 137] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 7. | Katoh Y, Katoh M. Comparative integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med. 2006;18:523-528. [PubMed] |
| 8. | Hoeng JC, Ivanov NV, Hodor P, Xia M, Wei N, Blevins R, Gerhold D, Borodovsky M, Liu Y. Identification of new human cadherin genes using a combination of protein motif search and gene finding methods. J Mol Biol. 2004;337:307-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Wu Q, Maniatis T. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc Natl Acad Sci U S A. 2000;97:3124-3129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Dunne J, Hanby AM, Poulsom R, Jones TA, Sheer D, Chin WG, Da SM, Zhao Q, Beverley PC, Owen MJ. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics. 1995;30:207-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 124] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Saburi S, Hester I, Goodrich L, McNeill H. Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development. 2012;139:1806-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 12. | Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101-2110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 261] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 13. | Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081-2089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 251] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 14. | Tanoue T, Takeichi M. New insights into Fat cadherins. J Cell Sci. 2005;118:2347-2353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 116] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 15. | Badouel C, Zander MA, Liscio N, Bagherie-Lachidan M, Sopko R, Coyaud E, Raught B, Miller FD, McNeill H. Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development. 2015;142:2781-2791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Pastushenko I, Mauri F, Song Y, de Cock F, Meeusen B, Swedlund B, Impens F, Van Haver D, Opitz M, Thery M, Bareche Y, Lapouge G, Vermeersch M, Van Eycke YR, Balsat C, Decaestecker C, Sokolow Y, Hassid S, Perez-Bustillo A, Agreda-Moreno B, Rios-Buceta L, Jaen P, Redondo P, Sieira-Gil R, Millan-Cayetano JF, Sanmatrtin O, D'Haene N, Moers V, Rozzi M, Blondeau J, Lemaire S, Scozzaro S, Janssens V, De Troya M, Dubois C, Pérez-Morga D, Salmon I, Sotiriou C, Helmbacher F, Blanpain C. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature. 2021;589:448-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 283] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 17. | Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Treviño L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154-1157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1453] [Cited by in RCA: 1368] [Article Influence: 97.7] [Reference Citation Analysis (1)] |
| 18. | Kim KT, Kim BS, Kim JH. Association between FAT1 mutation and overall survival in patients with human papillomavirus-negative head and neck squamous cell carcinoma. Head Neck. 2016;38 Suppl 1:E2021-E2029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Nishikawa Y, Miyazaki T, Nakashiro K, Yamagata H, Isokane M, Goda H, Tanaka H, Oka R, Hamakawa H. Human FAT1 cadherin controls cell migration and invasion of oral squamous cell carcinoma through the localization of β-catenin. Oncol Rep. 2011;26:587-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Hsu TN, Huang CM, Huang CS, Huang MS, Yeh CT, Chao TY, Bamodu OA. Targeting FAT1 Inhibits Carcinogenesis, Induces Oxidative Stress and Enhances Cisplatin Sensitivity through Deregulation of LRP5/WNT2/GSS Signaling Axis in Oral Squamous Cell Carcinoma. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 21. | Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, Medina D, Allred DC. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 2012;72:4574-4586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 126] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 22. | Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, Harshman K, Guipponi M, Bukach O, Zoete V, Michielin O, Muehlethaler K, Speiser D, Beckmann JS, Xenarios I, Halazonetis TD, Jongeneel CV, Stevenson BJ, Antonarakis SE. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2011;44:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 328] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 23. | Neumann M, Seehawer M, Schlee C, Vosberg S, Heesch S, von der Heide EK, Graf A, Krebs S, Blum H, Gökbuget N, Schwartz S, Hoelzer D, Greif PA, Baldus CD. FAT1 expression and mutations in adult acute lymphoblastic leukemia. Blood Cancer J. 2014;4:e224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | de Bock CE, Ardjmand A, Molloy TJ, Bone SM, Johnstone D, Campbell DM, Shipman KL, Yeadon TM, Holst J, Spanevello MD, Nelmes G, Catchpoole DR, Lincz LF, Boyd AW, Burns GF, Thorne RF. The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia. 2012;26:918-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 25. | Piao Z, Park C, Park JH, Kim H. Deletion mapping of chromosome 4q in hepatocellular carcinoma. Int J Cancer. 1998;79:356-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 26. | Zhu HY, Cao GY, Wang SP, Chen Y, Liu GD, Gao YJ, Hu JP. POU2F1 promotes growth and metastasis of hepatocellular carcinoma through the FAT1 signaling pathway. Am J Cancer Res. 2017;7:1665-1679. [PubMed] |
| 27. | Hu X, Zhai Y, Shi R, Qian Y, Cui H, Yang J, Bi Y, Yan T, Ma Y, Zhang L, Liu Y, Li G, Zhang M, Cui Y, Kong P, Cheng X. FAT1 inhibits cell migration and invasion by affecting cellular mechanical properties in esophageal squamous cell carcinoma. Oncol Rep. 2018;39:2136-2146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Wang Y, Wang G, Ma Y, Teng J, Wang Y, Cui Y, Dong Y, Shao S, Zhan Q, Liu X. FAT1, a direct transcriptional target of E2F1, suppresses cell proliferation, migration and invasion in esophageal squamous cell carcinoma. Chin J Cancer Res. 2019;31:609-619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Zhai Y, Shan C, Zhang H, Kong P, Zhang L, Wang Y, Hu X, Cheng X. FAT1 downregulation enhances stemness and cisplatin resistance in esophageal squamous cell carcinoma. Mol Cell Biochem. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 30. | Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801-1806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3216] [Cited by in RCA: 3022] [Article Influence: 177.8] [Reference Citation Analysis (0)] |
| 31. | Furukawa T, Sakamoto H, Takeuchi S, Ameri M, Kuboki Y, Yamamoto T, Hatori T, Yamamoto M, Sugiyama M, Ohike N, Yamaguchi H, Shimizu M, Shibata N, Shimizu K, Shiratori K. Whole exome sequencing reveals recurrent mutations in BRCA2 and FAT genes in acinar cell carcinomas of the pancreas. Sci Rep. 2015;5:8829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 32. | Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 511] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 33. | Wang Q, Cui L, Li P, Wang Y. Somatic Mutation of FAT Family Genes Implicated Superior Prognosis in Patients With Stomach Adenocarcinoma. Front Med (Lausanne). 2022;9:873836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 34. | Jung HY, Cho H, Oh MH, Lee JH, Lee HJ, Jang SH, Lee MS. Loss of FAT Atypical Cadherin 4 Expression Is Associated with High Pathologic T Stage in Radically Resected Gastric Cancer. J Gastric Cancer. 2015;15:39-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 35. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6641] [Article Influence: 510.8] [Reference Citation Analysis (0)] |
| 36. | Yang R, Cai TT, Wu XJ, Liu YN, He J, Zhang XS, Ma G, Li J. Tumour YAP1 and PTEN expression correlates with tumour-associated myeloid suppressor cell expansion and reduced survival in colorectal cancer. Immunology. 2018;155:263-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 37. | Bhat IP, Rather TB, Bhat GA, Maqbool I, Akhtar K, Rashid G, Parray FQ, Besina S, Mudassar S. TEAD4 nuclear localization and regulation by miR-4269 and miR-1343-3p in colorectal carcinoma. Pathol Res Pract. 2022;231:153791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 38. | Pan J, Liu F, Xiao X, Xu R, Dai L, Zhu M, Xu H, Xu Y, Zhao A, Zhou W, Dang Y, Ji G. METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp Clin Cancer Res. 2022;41:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 53] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 39. | Jiao S, Li C, Hao Q, Miao H, Zhang L, Li L, Zhou Z. VGLL4 targets a TCF4-TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat Commun. 2017;8:14058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 40. | Konsavage WM Jr, Kyler SL, Rennoll SA, Jin G, Yochum GS. Wnt/β-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J Biol Chem. 2012;287:11730-11739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 236] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 41. | Yuen HF, McCrudden CM, Huang YH, Tham JM, Zhang X, Zeng Q, Zhang SD, Hong W. TAZ expression as a prognostic indicator in colorectal cancer. PLoS One. 2013;8:e54211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 125] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 42. | Wang L, Shi S, Guo Z, Zhang X, Han S, Yang A, Wen W, Zhu Q. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS One. 2013;8:e65539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 211] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 43. | Xu Z, Wang H, Gao L, Zhang H, Wang X. YAP Levels Combined with Plasma CEA Levels Are Prognostic Biomarkers for Early-Clinical-Stage Patients of Colorectal Cancer. Biomed Res Int. 2019;2019:2170830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 44. | Yang C, Xu W, Meng X, Zhou S, Zhang M, Cui D. SCC-S2 Facilitates Tumor Proliferation and Invasion via Activating Wnt Signaling and Depressing Hippo Signaling in Colorectal Cancer Cells and Predicts Poor Prognosis of Patients. J Histochem Cytochem. 2019;67:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 45. | Zhang X, Liu J, Liang X, Chen J, Hong J, Li L, He Q, Cai X. History and progression of Fat cadherins in health and disease. Onco Targets Ther. 2016;9:7337-7343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 46. | Qi C, Zhu YT, Hu L, Zhu YJ. Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int J Cancer. 2009;124:793-798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 85] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 47. | Jiang X, Liu Z, Xia Y, Luo J, Xu J, He X, Tao H. Low FAT4 expression is associated with a poor prognosis in gastric cancer patients. Oncotarget. 2018;9:5137-5154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 48. | Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan Ş, Eng S, Kannan K, Zou Y, Peng L, Banuchi VE, Paty P, Zeng Z, Vakiani E, Solit D, Singh B, Ganly I, Liau L, Cloughesy TC, Mischel PS, Mellinghoff IK, Chan TA. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253-261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 277] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 49. | Kanemitsu Y, Shimizu Y, Mizusawa J, Inaba Y, Hamaguchi T, Shida D, Ohue M, Komori K, Shiomi A, Shiozawa M, Watanabe J, Suto T, Kinugasa Y, Takii Y, Bando H, Kobatake T, Inomata M, Shimada Y, Katayama H, Fukuda H; JCOG Colorectal Cancer Study Group. Hepatectomy Followed by mFOLFOX6 Versus Hepatectomy Alone for Liver-Only Metastatic Colorectal Cancer (JCOG0603): A Phase II or III Randomized Controlled Trial. J Clin Oncol. 2021;39:3789-3799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 143] [Article Influence: 35.8] [Reference Citation Analysis (0)] |