Published online Aug 24, 2021. doi: 10.5306/wjco.v12.i8.656
Peer-review started: February 25, 2021
First decision: May 7, 2021
Revised: May 11, 2021
Accepted: August 6, 2021
Article in press: August 6, 2021
Published online: August 24, 2021
Processing time: 178 Days and 17.1 Hours
Over the last twenty years, with the development of gene-driven therapies, numerous new drugs have entered clinical use. Very few of these new drugs are suitable for a large number of patients, and all require molecular genetic testing. In lung cancer, gene-targeted therapy has evolved rapidly and has placed demands on the development of diagnostics and tissue sample preparation and logistics. Rapid diagnosis and prevalence assessment are necessary to determine the prognosis of a lung cancer patient based on the latest research findings. Therefore, the molecular-genetic diagnostic pathway must also be accelerated and matured to do the necessary analyses on small samples. Because lung cancer rebiopsy can be difficult, liquid biopsy techniques should be developed to cover more of the treatable mutations. There are obstacles related to tissue sampling, new genomic techniques and access to gene-driven cancer drugs, including their affordability. With this review and case study, we go into the obstacles faced by our clinic and discuss how to tackle these obstacles in lung cancer. We use lung cancer as an example due to its complexity, though these same obstacles are found in different cancers on a minor scale.
Core Tip: Several gene-driven therapy drugs and molecular testing, together with immunohistochemistry, have evolved for lung cancer in recent years. Lung cancer is mutation dense and has more predictive genes that potentially influence treatment decisions than any other cancer type. In our case study, we ran into obstacles and delays in the diagnostic pathway both in tissue sampling and in gene testing. However, the major obstacle was the financial availability of new drugs. All elements need to be in place before a new drug can be given to patients, following the idea of the right drug at the right time.
- Citation: Saarenheimo J, Andersen H, Eigeliene N, Jekunen AP. Current challenges in applying gene-driven therapies in clinical lung cancer practice. World J Clin Oncol 2021; 12(8): 656-663
- URL: https://www.wjgnet.com/2218-4333/full/v12/i8/656.htm
- DOI: https://dx.doi.org/10.5306/wjco.v12.i8.656
Since the sequencing of the whole human genome was completed in 2003[1], new principles of genomic approaches have eagerly been used in cancer drug therapy[2]. Early enthusiasm about genes in cancer drug therapy was overwhelming, and overall the outlook was positive. Gene-targeted drug development took new steps, and the first major drug was imatinib (Glivec). Imatinib served as a breakthrough by changing attitudes from one-size-fits-all concept to an outlook that focused on small, restricted indications. Ph-positive chronic myelogenous leukemia was found to inhibit the BCR-ABL1 tyrosine kinase with increased activity because of a definite genetic alteration[3], but given its high effectiveness, it rapidly dominated the whole set of indications and became an example of the importance of highly targeted drugs in restricted indications, achieving commercial success. After Glivec’s success for limited indications, an increasing number of developers targeted limited indications with their therapeutic agents. The dominant thinking was that it would be better to have a highly active agent in limited indications than a less active one in a broader indication. It was important that the general attitude changed swiftly to allow specific approaches with gene-driven drugs.
Swift from cytotoxic chemotherapy agents towards targeted cancer drugs has been transpiring. By the end of 2014, the United States Food and Drug Administration (FDA) had approved 150 anticancer drugs, of which 89 were target-based and 61 were cytotoxic drugs[4]. Regarding solid cancers, drug agencies have become active in approving new agents based on solid early clinical data and scientific rationale. The FDA approved pembrolizumab in microsatellite-high tumors without any restrictions of cancer type[5]. Recently, both the FDA and the European Medicines Agency (EMA) approved larotrectinib, based on limited phase II data, for tumors with neurotrophic receptor tyrosine kinase (NTRK) gene fusion with all indications[6]. While these targeting agents add treatment possibilities, they also demand appropriate testing before their use.
Many targetable genes have emerged. Most new cancer drugs are linked to one gene mutation. Fortunately, molecular techniques have also been further developed, and gene tests have become more affordable. Now we have knowledge of multiple gene-driven cancers with certain gene alterations and the drugs that target them. Lung cancer is a prime example of a cancer with a wide variety of actionable mutations, partially because of the aggressive nature of the disease but also because of the existence of many mutations, some of which are recognized druggable driver mutations. Several gene-driven drugs are included in lung cancer guidelines. Even more drugs are in development, which require extensive clinical research before approval and implementation in clinical practice.
This article will address the practical obstacles in access to gene-driven cancer drugs in non-small-cell lung cancer (NSCLC). Small-cell lung cancer was not included due to high molecular heterogeneity and lack of current target genes with available treatment[7]. More than an approved cancer drug at the pharmacy is required. First, there need to be proper techniques for analysis; second, there must be enough tumor samples available; third, treatment procedures will need to allow the use of the drug; and finally, there needs to be funding for new drugs. We present the challenges involved in using gene-driven therapies based on our clinical experience with lung cancer at a Finnish central hospital. Our practices are reviewed in the form of a case study.
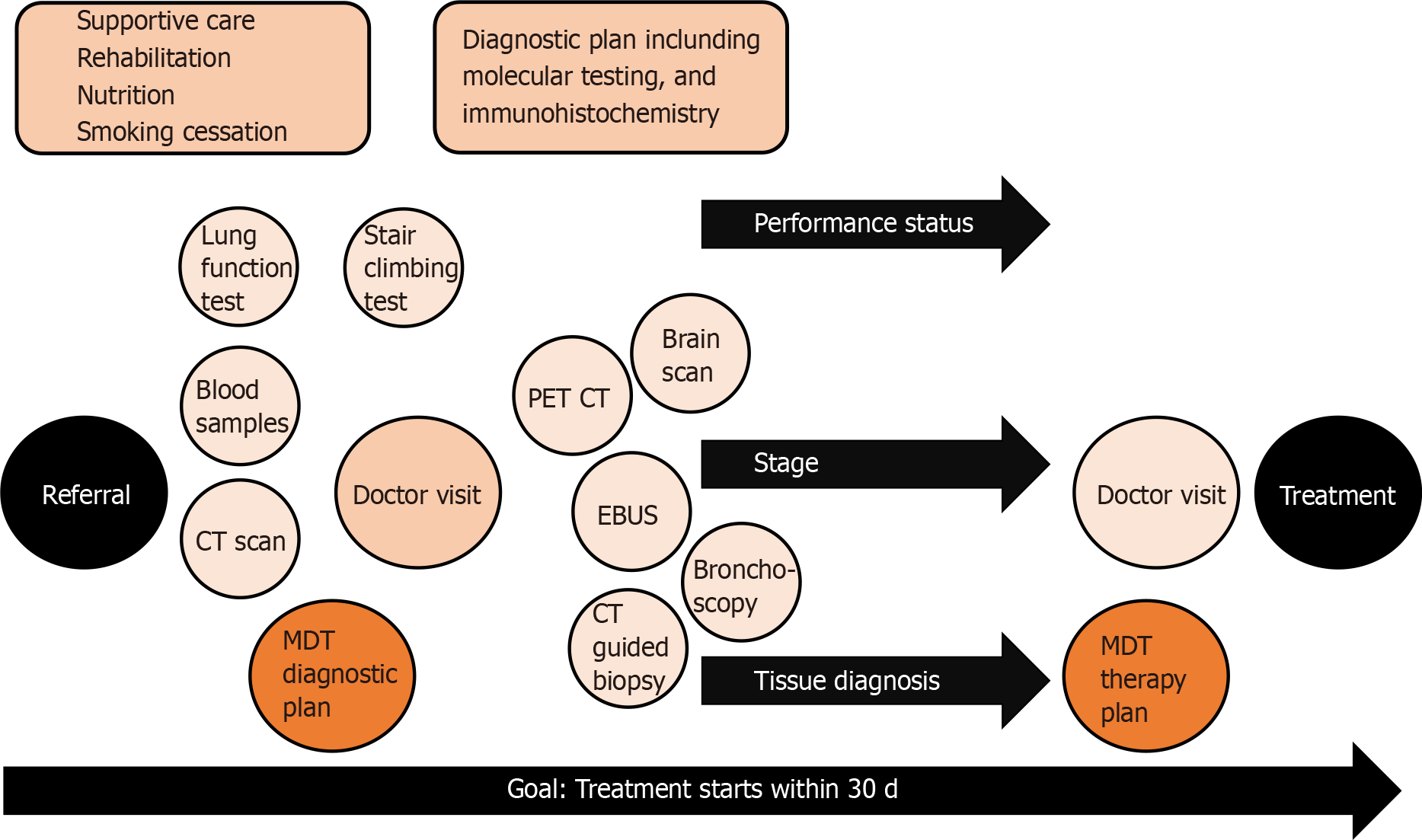
Process of lung cancer treatment decision is complex and requires many elements as illustrated in Figure 1. Poor performance status, lung diseases and other comorbidities are patient-related factors that often make lung cancer diagnostics challenging. The patient’s cooperation, sedation during tissue sampling and safe interruption of anticoagulation need to be considered during the planning phase of a safe procedure. Localized sampling of tissue consumes both time and resources, and both can be saved with good planning[8]. In lung cancer, sampling approaches vary based on the location and spread of disease. The most common sampling techniques are bronchoscopy, computed tomography (CT)-guided transthoracic lung biopsy and endosonography.
For mediastinal nodal staging in patients with suspected or proven NSCLC with abnormal mediastinal or hilar nodes on CT or positron emission tomography, endosonography is recommended over surgical staging as the initial procedure[9]. The three most common complications following endobronchial ultrasound with real-time guided transbronchial needle aspiration (EBUS-TBNA) are hemorrhage (0.6%), pneumothorax (0.4%) and infection (0.3%)[10]. Bronchoscopy and tissue sampling of intrabronchial cancer can be achieved in a combined session with EBUS-TBNA with proper sedation and cooperation. However, the tissue samples are small, most often cytological, materials, limiting the ability to extensively test them.
More material is available from core biopsies, but the risks are higher. The main complications in CT-guided transthoracic lung biopsy were pneumothorax (25% vs 19%), pulmonary hemorrhage (18% vs 6%) and hemoptysis (4% vs 2%) for core biopsy and fine needle aspiration (FNA), respectively. As shown by CT-guided core biopsy and FNA, the rate of pneumothorax requiring intervention was 6% and 4%, respectively[11]. Tissue sampling is an invasive procedure with risks, so the diagnostic pathway should be optimally planned to reduce the number of procedures needed to obtain enough material diagnosis and staging.
At the early stages, staging and tissue sampling require more advanced methods and time, whereas advanced metastatic cancer sampling is usually fast and easy. This might be one of the reasons why diagnosis time in lung cancer does not affect prognosis[12,13], while the stage of lung cancer at diagnosis does determine survival[14]. The faster pathway was shown to be beneficial in three studies[15-17]. In Denmark, improved diagnostic accuracy has led to significant improvements in survival rates and reduced mortality[18].
It has been estimated that in the United States, approximately 80% of cancer patients are treated at community hospitals without access to advanced genomic testing[19]. Clinical implementations of advanced cancer genomics have been limited to academic cancer centers[19]. Advanced testing includes a combination of next-generation sequencing and computational data analysis. In lung cancer, targetable gene alterations are looked for first, including sensitizing epidermal growth factor receptor (EGFR) mutations, anaplastic lymphoma kinase (ALK) and ROS-1 rearrangements, the BRAF V600E mutation, NTRK 1/2/3 gene fusions, the METex14 skipping mutation and RET rearrangement[20]. However, in practice, the list may be shorter if there is no access to the cancer drug related to a given gene alterations. For example, in Finland it is common practice to sequence only EGFR, ALK, ROS-1 and NTRK, because their corresponding drugs have reimbursements available, and to leave others untested.
Keeling et al[21] defined common barriers to the clinical implementations of companion diagnostic tests. Specifically, if physicians are not aware of biomarkers, tests are not requested, and the quality and appropriateness of samples are essential because accurate analysis requires enough sample of good quality. Tests are not always available in existing laboratories. The results should be reliable and consistent. If physicians lose confidence in using a certain cancer drug, test results might not be available in time. Cancer treatment decision making usually requires fast test turnaround times; if the results take more than 2 wk, this will decrease the willingness to use the therapy. If tests are insufficiently reimbursed, then financial issues become an obstacle. All technical obstacles need to be covered before patients can be indicated for the rights treatment at the right time.
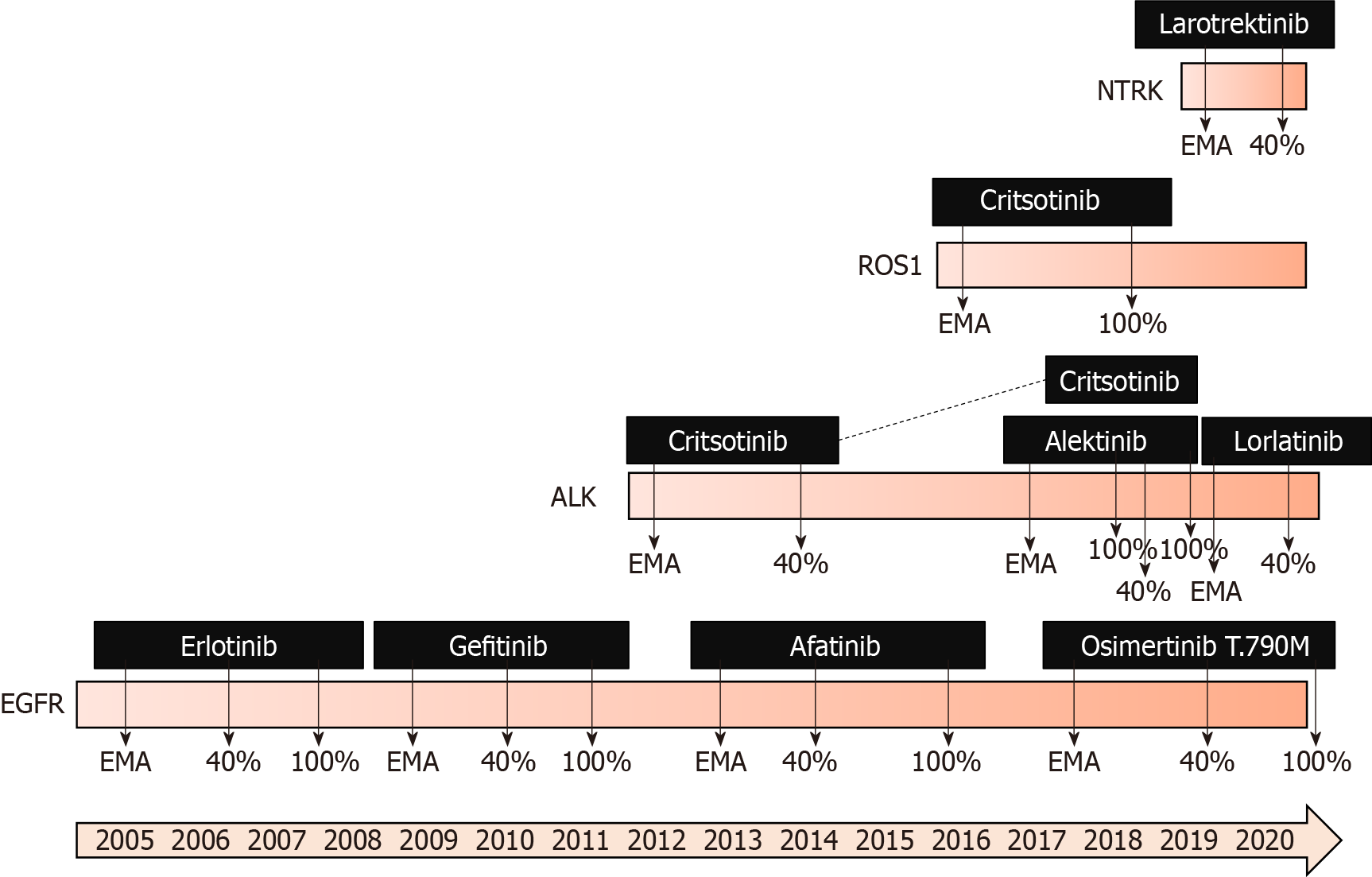
New drugs are entering clinical use via clinical trials, but not every country has access to the results of clinical trials. When a cancer drug is approved based on the results of clinical trials by drug registration agencies, it becomes available if the pharmaceutical company seeks authorization in a specific country. Usually, the FDA in the States is faster at approving than the EMA in Europe[22], but even faster approval might be done in other countries, such as Canada, Australia, Japan, or China. Although most applications rely on identical pivotal trials across regulatory agencies, only 57% of indications received either FDA or EMA approval and approval by an additional agency[23].
When a new cancer drug is approved, it becomes widely available, but there is an additional obstacle to its use, which is the price (Figure 2). All new cancer drugs, which are more expensive than old ones that are no longer covered by patents, should have an additional payer to at least partially support its high costs. Of course, the first payer is the patient, but support may come from insurance companies, government institutions or fraternal societies. Coverage varies widely. Every country has its own system based on available funds and evaluations that are performed mainly depen
The Vaasa Oncology Clinic covers 365000 people with approximately 50 annual first visit consultations for oncological treatment of lung cancer. Gene testing for all actionable mutations is implemented in our process. Altogether, 458 gene tests were analyzed during 2016-2019 (pyrosequencing 2016-2017 and then NGS 2017-2019), which covered 160 lung cancer patients (Table 1). Our focus was on EGFR driver gene mutations, as there has been a reimbursed cancer drug available for these patients. However, other actionable mutations, such as KRAS and BRAF mutations, were also observed and reported. As the reimbursement situation was improved during the study years, ALK and ROS-1 genes were included in the testing pattern (immunohistochemically analyzed and positive results verified by FISH) in 2018 and 2019, res
| Pyrosequencing (all patients) | NGS(all patients) | Lung cancer | cfDNA | ALK (IHC) | ROS-1 (IHC) | |
| 2016 | 128 | 1 | 39 | 5 | 8 | 0 |
| 2017 | 82 | 29 | 38 | 10 | 6 | 0 |
| 2018 | 0 | 125 | 49 | 28 | 51 | 0 |
| 2019 | 0 | 93 | 33 | 12 | 34 | 16 |
During 2016-2019, we found 16 EGFR-positive tumors from tissue biopsies (Table 2), corresponding to 5%-18% of tested patients/year (Table 2). The relative proportion of EGFR-positive patients was much higher during 2018-2019 (12% and 18%, respecti
| EGFR-pos | BRAF pos | KRAS pos | cfDNA EGFR-pos | ALK-pos | ROS-1-pos | |
| 2016 | 2 (5%) | NR | NR | 1 (20%) | 0 | 0 |
| 2017 | 2 (5%) | NR | NR | 2 (20%) | 0 | 0 |
| 2018 | 6 (12%) | 1 (2%) | 9 (18%) | 6 (21%) | 2 (4%) | 0 |
| 2019 | 6 (18%) | 2 (6%) | 10 (30%) | 3 (25%) | 0 | 0 |
We are confident that we can identify lung cancer patients who would benefit from targeted therapies based on gene morphology. Initial biopsy and rebiopsy in lung cancer are challenging, but with an adequate team, safe sampling is possible. Early planning of the diagnostic procedure together with radiologists is advisable, and this daily interaction needs to be based on actual data. For fast processing, pathologists and molecular pathologists need to stay informed about the current requirements. Extensive testing takes almost two weeks at the moment, and this process needs to be made faster. Liquid biopsy as a follow-up tool for T790M development is working well.
Rapid diagnosis and prevalence assessment influence the prognosis of a lung cancer patient based on the latest research findings. Therefore, the molecular genetic diagnostic pathway must also be accelerated and developed to run the necessary tests on small samples. Because lung cancer rebiopsy can be difficult, liquid biopsy techniques should be developed to cover more treatable mutations. Even though it was relatively easy to find a laboratory in Finland that could do the required analyses by the proper techniques, our challenges were sampling and handling the samples in time in the laboratory. Rebiopsy for tissue samples was difficult to organize, partially because only restricted personnel could take the biopsies but also because sampling was often deemed too difficult or to have excess risks. Blood samples of liquid biopsy were far easier to take, but there we ran into the problem that analyses were less comprehensive when done on liquid biopsies than on paraffin samples. In fact, the only recognized reliable tests in liquid biopsy were for EGFR and its resistance mutation T790M, so those were the only ones done routinely. Logistics was also challenging, as our limit for clinical answers was 2 wk, which was a demanding timeline that was often exceeded by a couple of days. However, if gene results are not obtained in 2 wk, the treatment decision is jeopardized, and delays in the initiation of treatment were frequent. Similarly, gene-driven therapies would have fit nicely into our treatment procedure, but the biggest limitation was the lack of reimbursements. New gene-driven drugs were so expensive that ordinary Finns could not afford them. There needed to be a copayment, and there was not, the drug was not prescribed at all, and no gene testing was needed. Therefore, the reimbursement system effectively dictated cancer therapy. Finally, based on our experience, it was not the availability of gene testing, samples or procedures but rather national reimbursement for particular drugs that had the greatest impact on the use of gene-driven therapies.
In a recent OECD report, challenges in access to oncological medicines were addressed from a financial perspective[18]. Patients expect rapid access to new cancer drugs, but that is not the reality, mainly because of both clinical and financial uncertainties and the position of the new drug in relation to the standard therapies. Societal willingness and capacity to pay vary across countries, and health technology assessments face the difficulties of estimating cost effectiveness, determining the position of new drugs, and, in the case of multiple indications, dealing with po
Similarly, we found in our case study that reimbursement was a major factor that actually influenced the use of gene-driven cancer drugs, as if there was no reim
The use of molecular testing and immunohistochemistry in lung cancer has evolved in recent years. We are still looking for the best combination of biomarkers, and negotiation among oncologists, invasive pulmonologists, invasive radiologists, lung pathologists and molecular pathologists continues. As the samples are small, both cytology and core biopsy, discussion what to prioritize is needed.
Lung cancer is mutation dense and has more predictive genes that potentially influence treatment decisions than any other cancer type. However, there are many obstacles to clear before a new drug becomes available for prescription by clinicians. The first requirement is the existence of a new cancer drug with efficacy relating to gene testing. Second, test methods and appropriate samples need to be available. Third, the new drug needs to be affordable to patients. All these elements need to be in place before the new drug is available to patients, following the concept of the right drug at the right time.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Finland
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Xu S, Zhao W S-Editor: Wu YXJ L-Editor: A P-Editor: Wang LYT
| 1. | National Human Genome Research Institute. International Human Genome Sequencing Consortium. [cited 25 February 2021]. Available from: https://www.genome.gov/12513430/2004-release-ihgsc-describes-finished-human-sequence. |
| 2. | International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3224] [Cited by in RCA: 2878] [Article Influence: 137.0] [Reference Citation Analysis (0)] |
| 3. | Cohen MH, Williams G, Johnson JR, Duan J, Gobburu J, Rahman A, Benson K, Leighton J, Kim SK, Wood R, Rothmann M, Chen G, U KM, Staten AM, Pazdur R. Approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia. Clin Cancer Res. 2002;8:935-942. [PubMed] |
| 4. | Niehues H. [Sleep and its pharmacologic treatment]. Landarzt. 1967;43:599-604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 213] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clin Cancer Res. 2019;25:3753-3758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 792] [Article Influence: 132.0] [Reference Citation Analysis (0)] |
| 6. | Scott LJ. Larotrectinib: First Global Approval. Drugs. 2019;79:201-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 7. | Friedman FB. So you always wanted to write about that patient who. J Pract Nurs. 1981;31:10-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 108] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Wanandi WB, Fluss SS. Technical cooperation in the pharmaceuticals sector: a new approach among Asian countries. WHO Chron. 1980;34:215-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 9. | Vilmann P, Clementsen PF, Colella S, Siemsen M, De Leyn P, Dumonceau JM, Herth FJ, Larghi A, Vazquez-Sequeiros E, Hassan C, Crombag L, Korevaar DA, Konge L, Annema JT. Combined endobronchial and esophageal endosonography for the diagnosis and staging of lung cancer: European society of gastrointestinal endoscopy (esge) guideline, in cooperation with the european respiratory society (ers) and the european society of thoracic surgeons (ests). Endoscopy. 2015;47:545-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 164] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 10. | Noble HB, Porter M. The role of the team physician in school athletics. IMJ Ill Med J. 1982;161:112-113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 11. | Arvela P, Kärki NT. Effect of cerium on drug metabolizing activity in rat liver. Experientia. 1971;27:1189-1190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 484] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 12. | Birch LL, Fisher JA. Appetite and eating behavior in children. Pediatr Clin North Am. 1995;42:931-953. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 383] [Cited by in RCA: 764] [Article Influence: 152.8] [Reference Citation Analysis (0)] |
| 13. | Alanen V, Koivunen JP. Association of diagnostic delays to survival in lung cancer: single center experience. Acta Oncol. 2019;58:1056-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P, Mitchell A, Bolejack V; International Association for the Study of Lung Cancer Staging and Prognostic Factors Committee; Advisory Boards, and Participating Institutions; International Association for the Study of Lung Cancer Staging and Prognostic Factors Committee Advisory Boards and Participating Institutions. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol. 2016;11:39-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3190] [Cited by in RCA: 3092] [Article Influence: 343.6] [Reference Citation Analysis (0)] |
| 15. | Lupi Herrera E, Horwitz S, Quintana F, Testelli M, Bialostozky D. [Radiology of Ebstein's anomaly. Report of new x-ray signs and review of the literature]. Arch Inst Cardiol Mex. 1972;42:861-874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 185] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 16. | Yang CJ, Wang H, Kumar A, Wang X, Hartwig MG, D'Amico TA, Berry MF. Impact of Timing of Lobectomy on Survival for Clinical Stage IA Lung Squamous Cell Carcinoma. Chest. 2017;152:1239-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 17. | Gomez DR, Liao KP, Swisher SG, Blumenschein GR, Erasmus JJ Jr, Buchholz TA, Giordano SH, Smith BD. Time to treatment as a quality metric in lung cancer: Staging studies, time to treatment, and patient survival. Radiother Oncol. 2015;115:257-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 18. | The Nordcan. The cancer statistics for the Nordic countries. [cited 25 February 2021]. Available from: https://www-dep.iarc.fr/NORDCAN/English/frame.asp. |
| 19. | Perales Palacios I, García Campos F, Michaus Oquiñena L, Blanco Guzmán S, Lantero Benedito M. [Isolation of Plesiomonas shigelloides in a case of gastroenteritis]. Rev Clin Esp. 1983;171:115-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 376] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 20. | NCCN Guidelines. [cited 25 February 2021]. Available from: https://www.nccn.org/professionals/physician_gls. |
| 21. | Keeling P, Clark J, Finucane S. Challenges in the clinical implementation of precision medicine companion diagnostics. Expert Rev Mol Diagn. 2020;20:593-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 22. | Lim SD, Woo CS, Youn JI, Kim YW, Kim DI, Fusaro RM. Leprosy. XII. T-cell subsets in lepromatous leprosy. Int J Dermatol. 1982;21:458-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 23. | Theissen JL, Zahn P, Theissen U, Brehler R. [Allergic and pseudo-allergic reactions in anesthesia. II: Symptoms, diagnosis, therapy, prevention]. Anasthesiol Intensivmed Notfallmed Schmerzther. 1995;30:71-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 44] [Article Influence: 1.5] [Reference Citation Analysis (0)] |