Published online Aug 24, 2020. doi: 10.5306/wjco.v11.i8.614
Peer-review started: January 20, 2020
First decision: April 21, 2020
Revised: May 21, 2020
Accepted: July 1, 2020
Article in press: July 1, 2020
Published online: August 24, 2020
Processing time: 213 Days and 12.9 Hours
Mutational activation of Ras genes is established as a prognostic factor for the genesis of a constitutively active RAS-mitogen activated protein kinase pathway that leads to cancer. Heterogeneity among the distribution of the most frequent mutations in Ras isoforms is reported in different patient populations with urothelial carcinoma of the bladder (UCB).
To determine the presence/absence of mutations in Ras isoforms in patients with UCB in order to predict disease outcome.
This study was performed to determine the mutational spectrum at the hotspot regions of H-Ras, K-Ras and N-Ras genes by polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) and DNA sequencing followed by their clinical impact (if any) by examining the relationship of mutational spectrum with clinical histopathological variables in 87 UCB patients.
None of the 87 UCB patients showed point mutations in codon 12 of H-Ras gene; codon 61 of N-Ras gene and codons 12, 13 of K-Ras gene by PCR-RFLP. Direct DNA sequencing of tumor and normal control bladder mucosal specimens followed by Blastn alignment with the reference wild-type sequences failed to identify even one nucleotide difference in the coding exons 1 and 2 of H-Ras, N-Ras and K-Ras genes in the tumor and control bladder mucosal specimens.
Our findings on the lack of mutations in H-Ras, K-Ras and N-Ras genes could be explained on the basis of different etiological mechanisms involved in tumor development/progression, inherent genetic susceptibility, tissue specificity or alternative Ras dysfunction such as gene amplification and/or overexpression in a given cohort of patients.
Core tip: Mutant Ras has been shown to be associated with drug resistance, enhanced metastasis and shorter survival of patients. Due to reported heterogeneity among the distribution of the most frequent mutations in Ras isoforms in different patient populations with urothelial carcinoma of the bladder, it is necessary to examine these patients for Ras mutations in order to predict disease outcome. Our findings on the lack of Ras mutations could be explained on the basis of different etiological mechanisms involved in tumor development, inherent genetic susceptibility, tissue specificity or alternative Ras dysfunction including gene amplification or overexpression in a given cohort of patients.
- Citation: Tripathi K, Goel A, Singhai A, Garg M. Mutational analysis of Ras hotspots in patients with urothelial carcinoma of the bladder. World J Clin Oncol 2020; 11(8): 614-628
- URL: https://www.wjgnet.com/2218-4333/full/v11/i8/614.htm
- DOI: https://dx.doi.org/10.5306/wjco.v11.i8.614
Urinary bladder cancer is the second most common genitourinary cancer globally and its occurrence has very high gender variability (http://cancerindia.org.in/globocan-2018-india-factsheet/). It is the sixth most common cancer in men and the seventeenth most common cancer in women. The etiology of bladder cancer is very complex. Among many factors, tobacco chewing/smoking and environmental or occupational exposure to a number of carcinogens have been identified as the most important risk factors for bladder cancer[1-3].
Urothelial carcinoma of the bladder (UCB) originates in the cells of the innermost layer of bladder urothelium and accounts for approximately 90% of all bladder cancers. Clinically, two distinct forms of UCB namely, non-muscle invasive bladder cancer (NMIBC in 75%-80% of patients) and muscle invasive bladder cancer (MIBC in 20%-25% of patients) develop along papillary and non-papillary pathways[4]. Patients diagnosed with NMIBC can be successfully treated. Nevertheless, these tumors have a higher tendency to recur (50% to 90%) and 15% progress to invasive and metastatic tumors. Morbidity and mortality are associated with the high grade, non-papillary, muscle invasive form of the disease. Molecular studies to characterize the genotypic differences in the pathogenesis of NMIBC and MIBC may improve the diagnostic/prognostic outcome of the disease.
Rat sarcoma viral oncogene homolog (Ras) belongs to the family of small G proteins with intrinsic GTPase activity that governs various cellular signal transduction pathways. Alterations in the expression or functions of (Ras) genes caused by various point mutations within the gene have been established as prognostic factors in the genesis of a constitutively active RAS-mitogen activated protein kinase pathway that leads to cancer. Point mutations within the hotspot regions of Ras gene lead to reduced intrinsic GTPase activity, the protein is locked into a constitutively active state and results in aberrant cell signaling even in the absence of external signals[5]. In vitro and in vivo studies on tumor regression upon withdrawal of Ras expression indicate that mutant Ras is a therapeutically useful drug target even in advanced metastasis[6]. Mutant Ras gene has been shown to be associated with drug resistance, enhanced metastasis, poor prognosis and shorter survival of patients[7].
Approximately 30% of human cancers are known to harbor genomic mutations in the three functional isoforms of Ras genes (H-Ras; located at 11p15.5; K-Ras; located at 12p12.1; and N-Ras; located at 1p13.2). The most common mutational hotspots in the codons for amino acid residues 12, 13 or 61 are confined to exon 1 or 2 of H-Ras (G12V, G12S, G12A, G12D, G13D, Q61R); K-Ras (G12D, G12S, G12R, G12A, G12V, G12C, G13D); and N-Ras (G12D, Q61N, Q61L, Q61K). Tissue and organ specificities of Ras gene activation have been reported to vary with mutated codon and type of Ras gene isoform. K-Ras mutations occur frequently in non-small cell lung, colorectal, and pancreatic carcinomas; H-Ras mutations are common in bladder, kidney, and thyroid carcinomas; while N-Ras mutations have been identified in melanoma, hepatocellular carcinoma, and hematologic malignancies[8,9].
Published studies provide conflicting results regarding the frequency distribution of Ras mutational spectrum in UCB patients[10-14]. Out of a total of 11.67% mutations in exon 1 of K-Ras, maximum mutations were reported at codon 12 in bladder cancer patients[15]. Iranian patients with bladder cancer did not exhibit any mutation in the hotspot codons (12, 13, and 61)[16]. Various studies have examined 45%, 46.7% and 39% of H-Ras mutations in codon 12 in bladder cancer patients[17-19]. Due to the reported heterogeneity in the distribution of the most frequent mutations in Ras isoforms in bladder cancer specimens, it is necessary to examine the presence/absence of mutations in order to predict disease outcome[10,11].
Speculating the role of mutant Ras in bladder tumorigenesis, the present study has been conducted to determine its clinical impact by examining the relationship between clinical histopathological variables in UCB patients and the mutational spectrum. Frequency distribution and prevalence of mutations in the hotspot regions of H-Ras codon 12 (glycine to valine/serine/alanine/aspartic acid), K-Ras codon 12 (glycine to valine/aspartic acid/serine/arginine/alanine/cysteine), K-Ras codon 13 (glycine to aspartic acid) and N-Ras codon 61 (glutamine to lysine/arginine) were examined by polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) in a cohort of 87 North Indian UCB patients and 23 controls with normal bladder mucosa. The results were confirmed by direct DNA sequencing of coding exons 1 and 2 of H-Ras, K-Ras and N-Ras genes.
Patients were enrolled in the Urology OPD at King George’s Medical University (KGMU), Lucknow during 2018-2019. All 87 patients examined had symptoms of hematuria as a major sign followed by urinary frequency or irritative symptoms and were assessed for primary tumor. Patients underwent bimanual examination under anesthesia before and after endoscopic surgery (biopsy or transurethral resection) or histological verification of the absence or presence of tumor. Imaging of the chest, abdominal ultrasound and computed tomography of the abdomen (whenever required) were performed to detect common metastatic sites as well as lymph node involvement. Tumor tissues from 42 NMIBC (stage pTa-pT1) and 45 MIBC (stage pT2-pT4) were obtained after transurethral resection of the bladder tumor. Tissues were collected in RNAlater, snap frozen and stored at -80°C for future use. Clinical data on the UCB patients and pathological classification/records based on pathological TNM staging were provided by the Department of Urology and Department of Pathology, KGMU, Lucknow. After informed consent, normal bladder mucosal tissues were collected from 23 benign prostate hyperplasia (BPH) patients during cold cup biopsy. These patients underwent transurethral resection of the prostate for BPH and had known bladder lesions. Pathologists independently diagnosed and classified bladder tumors according to World Health Organization and International Society of Urologic Pathology 2004 classification system[20]. Ethical clearance was obtained from Bioethics Cell, Institutional Ethics Committee, KGMU (reference no. 89th ECM II A/P8).
Genomic DNA was extracted from 87 UCB and 23 control bladder mucosal tissues using proteinase K and phenol-chloroform extraction, followed by ethanol precipitation, and was quantified and then stored at -20°C.
PCR was performed to amplify DNA segments which span (1) codon 12 of H-Ras gene; (2) codon 12 and codon 13 of K-Ras gene; and (3) codon 61 of N-Ras gene in 87 UCB and 23 bladder mucosal tissues. The primer sequences used are listed in Table 1.
| Gene | Target codon | Strand | Primer sequences |
| H-Ras | 12 | + | 5’GACGGAATATAAGCTGGTGG 3’ |
| - | 5’AGGCACGTCTCCCCATCAAT 3’ | ||
| K-Ras | 12 and 13 | + | 5’ACTGAATATAAACTTGTGGTAGTTGGACCT 3’ |
| - | 5’TTCTCCATCAATTACTACTTGCTTCCTGTA 3’ | ||
| N-Ras | 61 | + | 5’GACATACTGGATACAGCTGGC 3’ |
| - | 5’CCTGTCCTGATGTATTGGTC 3’ |
PCR was carried out with 200 ng of DNA, 10 pmol of primer(s), and Emerald Amp max PCR master mix (TaKaRa, Clontech) using a thermal cycler (T100TM, BioRad, United States). Cycling conditions included initial denaturation at 98°C for 20 s, followed by 30 cycles of [denaturation: 98°C for 10 s, annealing: 60°C (for H-Ras and N-Ras) and 58°C for K-Ras for 30 s, and extension: 72°C for 30 s] followed by a final extension at 72°C for 5 min.
Restriction endonucleases MspI (Thermo Scientific), BstNI (Thermo Scientific), HphI (Thermo Scientific), and MScI (Thermo Scientific) were used to digest amplified PCR fragments containing codon 12 of H-Ras, codon 12 of K-Ras, codon 13 of K-Ras, and codon 61 of N-Ras, respectively. Buffers and incubation conditions (37°C for 1-16 h) were used according to the manufacturers’ recommendations. The digested and undigested fragments were subjected to electrophoresis on 3% agarose gel. A summary of the Ras gene assays is described in Table 2.
| Gene | Target codon | Restriction enzyme/site | Fragment size | ||
| Undigested | Mutant after digestion | Wild-type/normal after digestion | |||
| K-Ras | 12 Glycine (GGT) to Valine (GTT)/Aspartic acid (GAT)/Serine (AGT)/Arginine (CGT)/Alanine (GCT)/Cysteine (TGT) | MvaI (BstNI) | 144 bp | 144 bp | 115 bp and 29 bp |
| CC↓ WGG | |||||
| GGW↑ CC | |||||
| K-Ras | 13 Glycine (GGC) to Aspartic acid (GAC) | HphI | 144 bp | 101 bp and 43 bp | 144 bp |
| GGTGAN8↓ | |||||
| CCACTN7↑ | |||||
| H-Ras | 12 Glycine (GGC) to Valine (GTC)/Aspartic acid (GAC)/Serine (AGC)/Alanine (GCC) | Mspl (HpaII) | 420 bp | 420 bp | 390 bp and 30 bp |
| (C↓ CGG) | |||||
| (GGC↑ C) | |||||
| N-Ras | 61 Glutamine (CAA) to Arginine (CGA)/Lysine (AAA)/Leucine (CTA) | Mlsl (MscI) | 65 bp | 65bp | 44 bp and 21 bp |
| (TGG↓ CCA) | |||||
| (ACC↑ GGT) | |||||
Coding exons 1 and 2 each of H-Ras, N-Ras and K-Ras were amplified by the laboratory developed primer pairs (Table 3). Primers were designed for the GenBank reference sequence of H-Ras, N-Ras and K-Ras (accession numbers: NM_001130442.1.1, NM_004985.4.1, NM_002524.4.1, respectively) by Primer plus software. The 200 ng of DNA was amplified with 10 pmol primer using the Phusion high-fidelity PCR kit (Thermo Scientific). The thermal profile included initial denaturation at 98°C for 40 s, followed by 35 cycles of (1) H-Ras: [denaturation: 98°C for 5 s, annealing: 63.2°C (for exon 1) and 64.8°C (for exon 2) for 10 s, and extension: 72°C for 15 s]; (2) N-Ras: [denaturation: 98°C for 5 s, annealing: 62.1°C (for exon 1) and 61.4°C (for exon 2) for 10 s, and extension: 72°C for 15 s]; (3) K-Ras: [denaturation: 98°C for 5 s, annealing: 61.8°C (for exon 1) and 61.3°C (for exon 2) for 10 s, and extension: 72°C for 15 s]; followed by a final extension at 72°C for 7 min. Amplified PCR products were electrophoresed on 2% agarose gel, eluted and purified with a QIAquick® PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Sequencing reactions were performed for both the DNA strands by the BigDye™ Terminator v1.1 Cycle Sequencing Ready Reaction Kit version 3.1 (Applied Biosystems, Monza, Italy) on a total of 10 ng of purified PCR products. Sequence analysis was performed using a 3500 Genetic Analyzer. The files/electropherogram obtained were analyzed by seq scap_v5.2 software. Sequence results of bladder mucosa were aligned with the reference sequences (mentioned above) using Blastn. Furthermore, DNA sequences of the respective regions in bladder tumor specimens were compared with that of wild-type sequences to examine the presence/absence of mutations in the coding exons 1 and 2 of the H-Ras, N-Ras and K-Ras genes.
| Gene | Strand | Coding exon | Primer sequences | Length of amplified fragment |
| K-Ras | + | 1 | F 5'-TTAACCTTATGTGTGACATGTTCTAA-3' | 378 bp |
| K-Ras | - | 1 | R 5'-CCCTGACATACTCCCAAGGA-3' | |
| K-Ras | + | 2 | F 5'- TCAAGTCCTTTGCCCATTTT-3' | 375 bp |
| K-Ras | - | 2 | R 5'- TGCATGGCATTAGCAAAGAC-3' | |
| N-Ras | + | 1 | F 5'-GCCCAAGGACTGTTGAAAAA-3' | 477 bp |
| N-Ras | - | 1 | R 5'-TGCATAACTGAATGTATACCCAAAA-3' | |
| N-Ras | + | 2 | F 5'-GGCAGAAATGGGCTTGAATA-3' | 424 bp |
| N-Ras | - | 2 | R 5'-CCTAAAACCAACTCTTCCCATAA-3' | |
| H-Ras | + | 1 | F 5'-GTGGGTTTGCCCTTCAGAT-3' | 386 bp |
| H-Ras | - | 1 | R 5'-TCTAGAGGAAGCAGGAGACAGG-3' | |
| H-Ras | + | 2 | F 5'-CAGGACACAGCCAGGATAGG-3' | 492 bp |
| H-Ras | - | 2 | R 5'-ACATGCGCAGAGAGGACAG-3' |
The mean age of the patients included in the study was 58.3 years (range: 25-83 years) and 44 (50.17%) patients were older than 60 years. The male to female ratio was 11:1. Of 87 patients, 66 (75.86%) had a positive history of either smoking or a tobacco chewing habit. Twenty of 87 tumors (22.98%) were more than 3 cm in size, whereas 67 tumors (77.01%) were less than 3 cm. Pathologically, 42/87 (48.27%) tumors were classified as NMIBC and 45/87 (51.72%) as MIBC. According to the histopathological classification, 80.95% (34/42) non-muscle invasive tumors were of low grade and 19.04% (8/42) were of high grade. All MIBC patients had a high grade tumor. Of 87 tumors, 26/87 (29.88%) were recurrent type, while the remaining 61/87 (70.11%) were identified as primary tumors (Table 4).
| Clinicohistopathological variables | n (%) |
| Total no. of patients | 87 (100) |
| Age (yr) mean, range | 58.3, 25-83 |
| n < 60 | 44 (50.17) |
| n ≥ 60 | 43 (49.42) |
| Gender | |
| Male | 81 (93.10) |
| Female | 8 (9.19) |
| Hematuria | |
| Present | 87(100) |
| Absent | Nil |
| No information | Nil |
| Smoking/Tobacco chewing status | |
| Smokers | 66 (75.86) |
| Non-smokers | 21 (24.1) |
| Tumor grade | |
| Low | 34 (39.04) |
| High | 53 (60.91) |
| Tumor stage | |
| Ta-T1 (Low/NMIBC) | 42 (48.27) |
| T2-T4 (High/MIBC) | 45 (51.72) |
| Tumor type | |
| Primary | 61 (70.11) |
| Recurrent | 26 (29.88) |
| Tumor Size | |
| > 3 cm | 20 (22.98) |
| < 3 cm | 67 (77.01) |
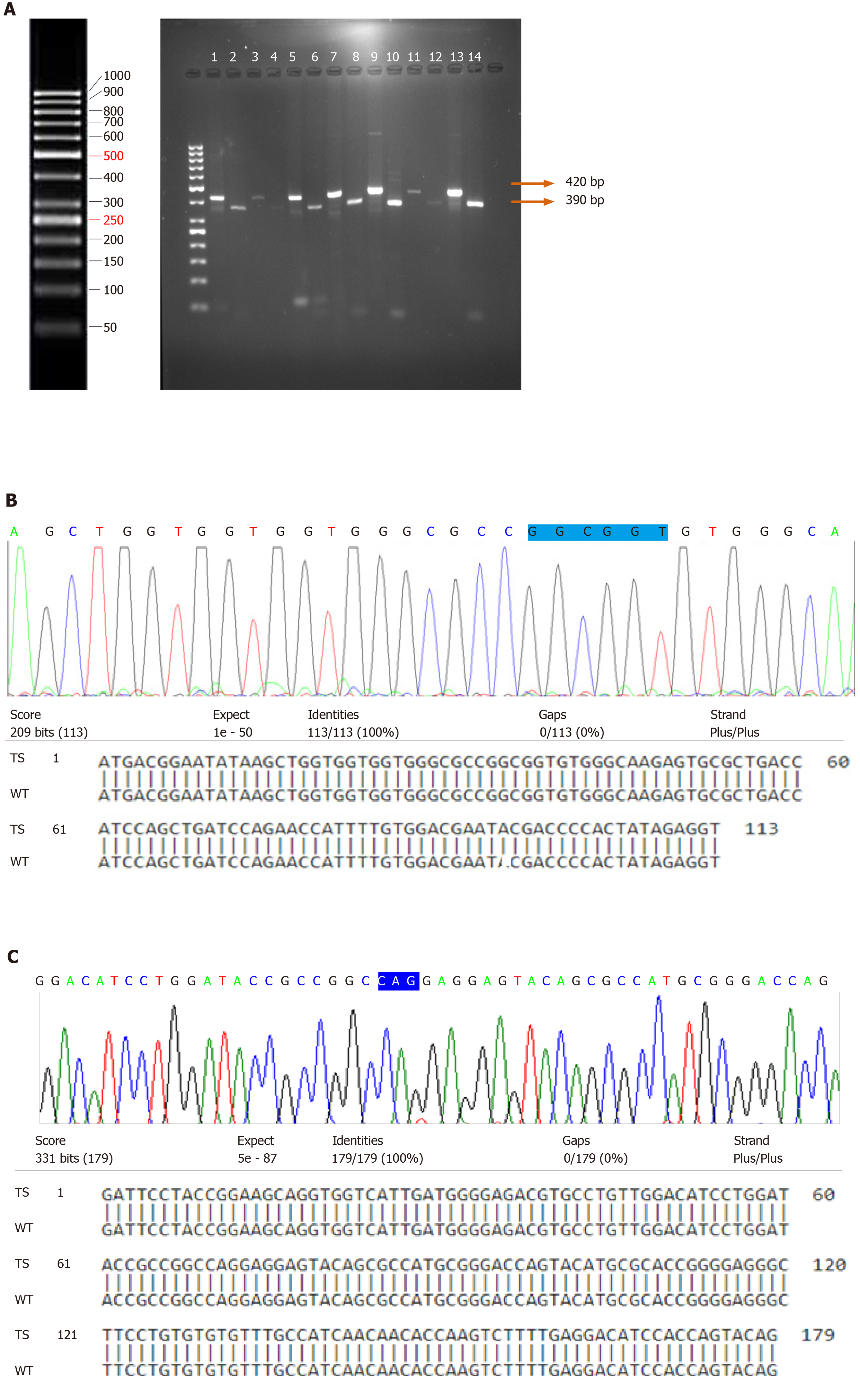
PCR-RFLP was carried out to examine the point mutation in codon 12 of H-Ras gene in 87 bladder tumor tissues and 23 normal bladder mucosal tissues. Digestion of the wild-type amplicon of 420 bp by MspI gave rise to two bands of 390 bp and 30 bp. The presence of a point mutation at codon 12 results in loss or modification of the endonuclease recognition site which is indicative of the translational change of glycine (GGC) to serine (AGC)/valine (GTC)/alanine (GCC)/aspartic acid (GAC). In our study, none of the tumors were examined for the presence of point mutation at codon 12 of H-Ras gene (Figure 1A).
Direct DNA sequencing of the coding exonic region 1 spanning the codons 12, 13 and exon 2 containing hotspot codon 61 of H-Ras gene was performed. Blastn results of DNA sequences in all the tumor specimens showed 100% alignment with that of the wild-type. Electropherogram analysis did not identify the presence of any point mutations in exons 1 and 2 of H-Ras genes in the tumor specimens (Figure 1B and C).
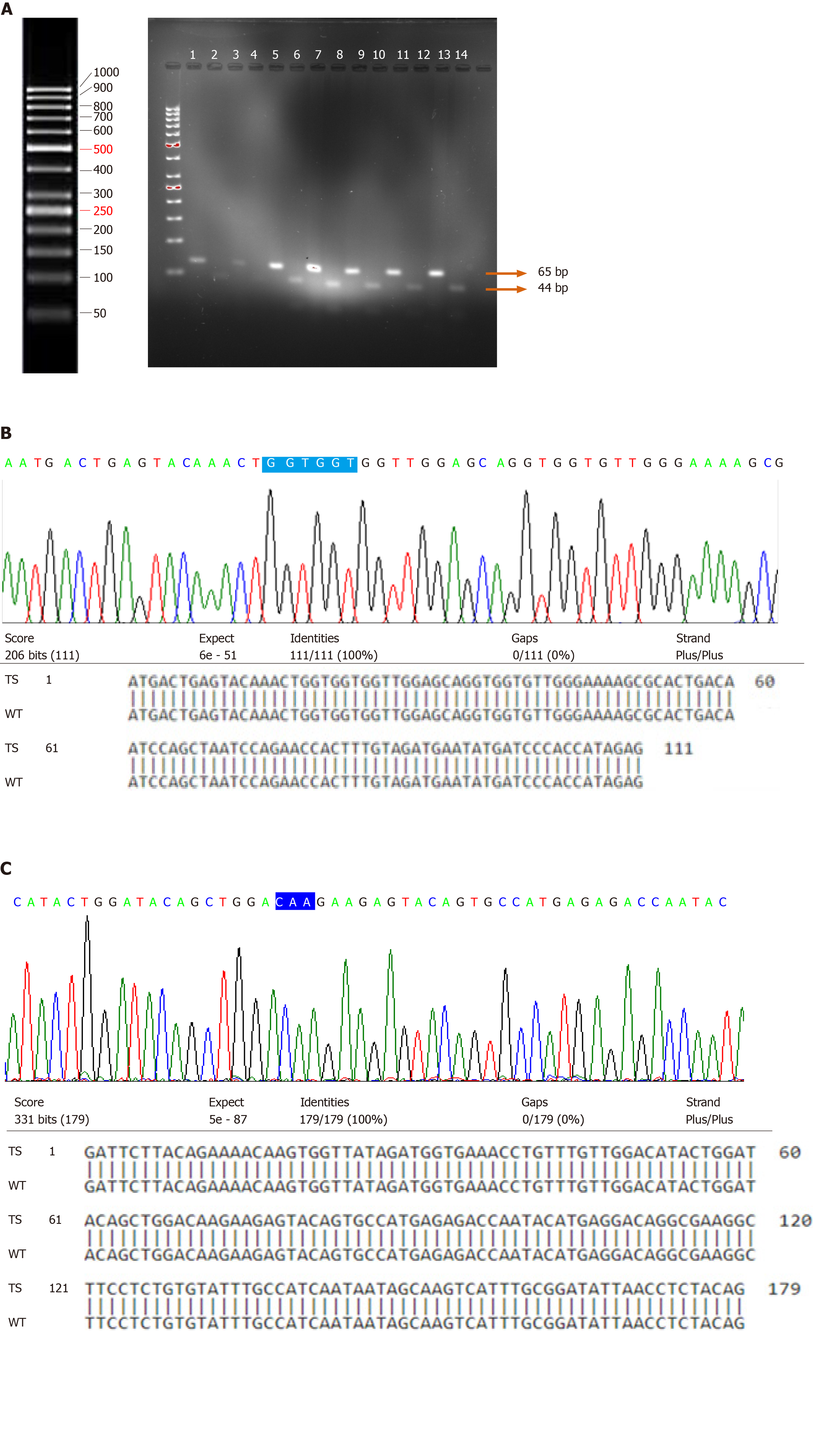
Tumor specimens from 87 UCB patients and 23 normal bladder mucosal tissues were examined by PCR-RFLP for the presence or absence of specific point mutations at codon 61. The presence of a point mutation at codon 61 may result in the conversion of glutamine (CAA) to lysine (AAA)/arginine (CGA)/leucine (CTA). The proper restriction site (TGG↓CCA) was created by changing only one nucleotide in a forward primer just before the start of codon 61. Restriction digestion of the wild-type amplicon of 65 bp by enzyme MscI resulted in its cleavage into 21 bp and 44 bp (Figure 2A). The present study failed to detect the presence of point mutations in 87 UCB and 23 normal mucosal specimens.
Direct DNA sequencing was performed to detect the point mutations in N-Ras coding exons 1 and 2 spanning codons 12, 13; and 61, respectively. Sequencing results in the wild-type and tumor specimens were analyzed and compared. The presence of point mutations in the hotspots of codon 12 and 13 of exon 1 and codon 61 of exon 2 of N-Ras gene was not detected in any of the bladder specimens (Figure 2B and C).
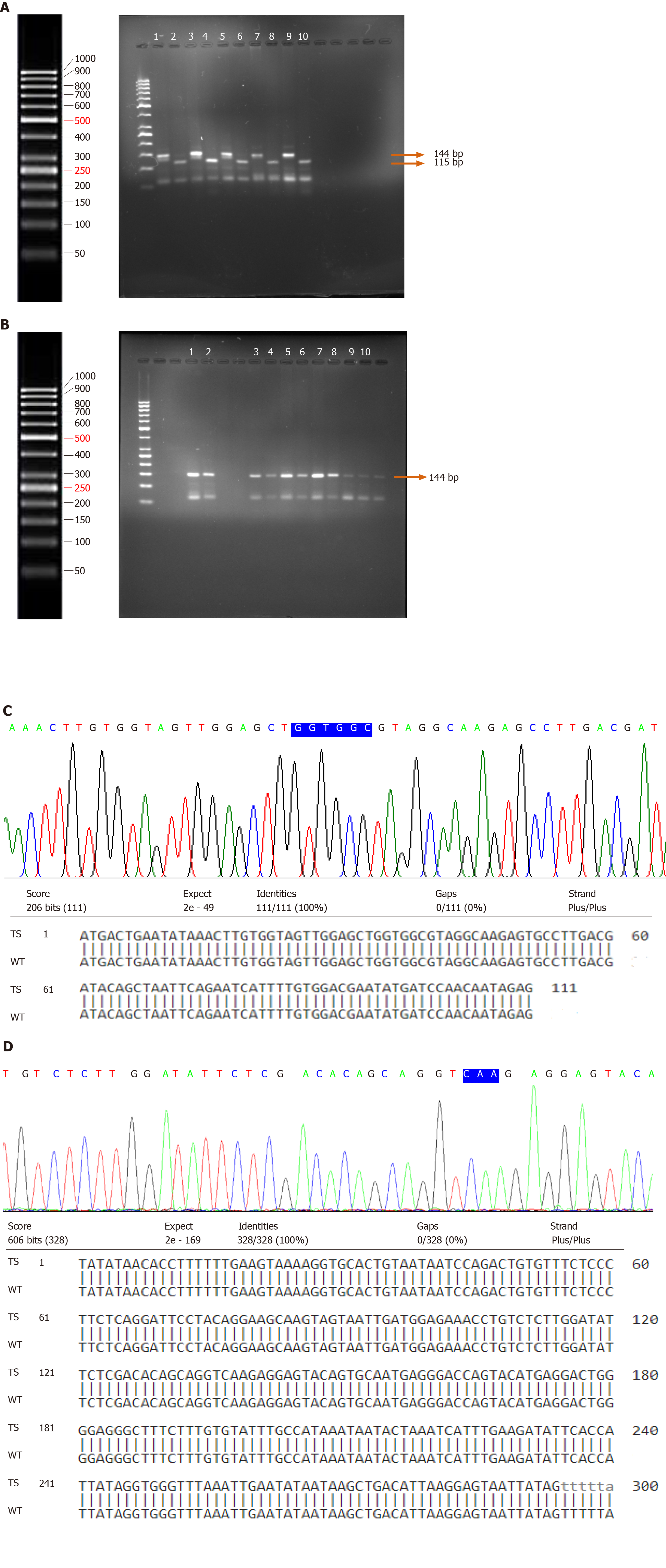
PCR amplification followed by RFLP was carried out to determine the presence of point mutations in codons 12 and 13 in K-Ras gene in 87 UCB and 23 normal bladder mucosal tissues. A primer was designed to create a restriction site just before the start of codon 12. Restriction digestion of the wild-type amplicon of 144 bp by enzyme BstNI resulted in its cleavage into 115 bp and 29 bp. The presence of a point mutation at codon 12 results in loss of the recognition site which is indicative of the translational change of glycine (GGT) to valine (GTT)/aspartic acid (GAT)/serine (AGT)/arginine (CGT)/alanine (GCT)/cysteine (TGT). The presence of a point mutation at codon 12 in K-Ras gene was not observed (Figure 3A).
Enzyme HphI was used to cleave the restriction site (GGTGA7/8↓) at codon 13 which is indicative of the conversion of glycine (GGC) to aspartic acid (GAC) in K-Ras gene. This site does not exist in the wild-type but tends to appear in mutants. The wild-type amplicon yielded a fragment of 144 bp when cut by HphI. Nevertheless, the presence of a mutation at codon 13 would yield two fragments of 101 bp and 43 bp oligonucleotides on restriction digestion (Figure 3B). PCR-RFLP failed to identify any mutational change in codon 13 of K-Ras gene in tumor and normal bladder mucosal tissues.
The results of direct DNA sequencing and Blastn of coding exons 1 (spanning codons 12 and 13) and 2 (spanning hotspot codon 61) of K-Ras genes in tumor and normal bladder mucosal tissues exhibited 100% alignment. DNA sequencing analysis verified the results of PCR-RFLP. No point mutations in the hotspots of exonic regions 1 and 2 of the K-Ras gene were observed (Figure 3C and D).
Considerable experimental evidence has demonstrated the significance of continual expression of mutant Ras in tumor maintenance. Withdrawal or suppression of Ras expression impairs the in vitro growth of Ras-mutant human cancer cell lines and tumor regression in mouse models driven by inducible mutant Ras. These findings indicate that mutant Ras is a therapeutically useful drug target even in advanced metastatic tumors[6].
Studies of a variety of tumors have demonstrated the prevalence of specific point mutations in the hotspots of Ras isoforms. These point mutations are known to transform Ras proto-oncogene into an oncogene and prevent normal deactivation of Ras proteins. Activated Ras proteins are associated with drug resistance, enhanced metastasis, poor prognosis and shorter survival of patients[7]. The present study examined the mutational spectrum at the hotspot regions of H-Ras codon 12, K-Ras codons 12, 13 and N-Ras codon 61 by PCR-RFLP followed by direct DNA sequencing of the coding exons 1 and 2 of the three Ras isoforms in 87 UCB patients and their clinical impact if any.
The incidence of Ras mutations varies, and greatly depends on the tissue or cell type from which the cancer cells are derived. Although Ras mutations occur in 75% to 95% of pancreatic carcinomas and in 50% of colon carcinomas, they are rare in several other neoplasms[15]. The H-Ras mutation was first detected in the human bladder cancer cell line T24. Subsequent studies demonstrated the frequent occurrence of H-Ras mutations in urinary tract tumors compared to mutations in K-Ras or N-Ras genes[21]. A number of studies has reported H-Ras mutations with variable frequencies in urinary bladder cancer specimens. Fitzgerald et al[22] reported mutations in the H-Ras gene in 44% of urine sediments from bladder cancer patients. Czerniak et al[17] observed H-Ras mutation specifically at codon G12 in 45% of bladder cancers. Zhu et al[19] and Buyru et al[18] showed 46.7% and 39% point mutations in H-Ras at codon 12, respectively. Cattan et al[23] detected only 1% of such alterations in bladder cancer patients[23]. In constrast, Przybojewska et al[24] observed H-Ras mutations in 84% of patients with bladder cancer using PCR-RFLP. In contradiction to many earlier published studies, we did not find mutations at H-Ras codon G12 (glycine to valine/serine/ alanine/aspartic acid) or in the coding exons 1 and 2 in a cohort of North Indian urothelial bladder cancer patients.
N-Ras gene mutations have mainly been associated with hematopoietic malignancies and melanoma[25,26]. Results of the study by Przybojewska et al[24] revealed the frequent prevalence (80%) of N-Ras gene mutations at codon Q61 (glutamine to lysine) in bladder tumor tissues. These tumor tissues were obtained following infiltration of urinary bladder walls as well as peripheral blood specimens from confirmed bladder cancer patients[24]. Of the total mutations detected in N-Ras gene, 60% of mutations were observed in codon 61 in a cohort of North Indian patients[26]. A strong association between the percentage of mutations in Ras genes and smoking status of patients (total of 78% mutations) and the age of patients (more than 60 years with total of 80% mutations) was observed. However, no association between the percentage mutation distribution and tumor stage/grade was reported[27]. Jebar et al[10] examined one mutation in codon 12 and three mutations in codon 61 in 98 urinary bladder tumors and 31 bladder cell lines. Our findings on the lack of N-Ras gene mutations in bladder cancer patients are not in accordance with published studies. Heterogeneity in the results/genomic alterations could be attributed to the differences in ethnicity, exposure to different environmental carcinogens, genetic susceptibility to carcinogens and or tissue specificity.
High frequency of K-Ras gene mutations has been detected in many forms of cancer, including pancreatic cancer (80%-90%) and adenocarcinoma of the lung (60%)[17,22]. Cancers of the lung, large intestine (including colon, rectal and anal), pancreas and biliary tract exhibited higher frequency of mutations in K-Ras gene[28]. Observed similarities in the percentage distribution of mutations in K-Ras codons in lung cancer (58%) and bladder cancer (47%) could be due to the effects of tobacco consumption. Tobacco is considered an important risk factor for both of these cancers and can induce local somatic mutations in genes[13]. These studies did not report an association between K-Ras mutations and tumor stage/grade[13]. Unlike the majority of tumors that harbor an activated K-Ras gene, changes in K-Ras gene have been observed as a rare event in urinary bladder tumors[24]. Studies examined the percentage mutation prevalence at codon G12 (glycine to valine/aspartic acid/serine/ arginine/ alanine/cysteine) and codon G13 (glycine to aspartic acid) in K-Ras gene as an infrequent event in bladder cancer[29,30]. A study by Nanda et al[15] identified 11.67% tumors which harbored K-Ras mutations as well as a significant correlation of the K-Ras mutant status with the smoking history of patients, high tumor grade, lymph node involvement and tumor recurrence. Yan et al[31] reported the ability of mutant K-Ras, but not H-Ras, to confer metastatic phenotype in cells by interfering with the maturation of cell surface integrins and disrupting cell-cell adhesion. A recently published study reported a higher prevalence of point mutations in all the Ras isoforms in NMIBC (27%) compared to MIBC (9.4%) patients.
In contrast to earlier published studies, our findings on the lack of H-Ras, K-Ras and N-Ras gene mutations in urothelial bladder cancer patients provide evidence for the tissue specific activation of Ras isoforms.
Discrepancies/heterogeneity in the frequency distribution of mutations at hotspot regions/codons in different isoforms of Ras gene among different cohorts of UCB patients belonging to different ethnic groups are reported in many published studies. Observed heterogeneity among different studies could be explained on the basis of different etiological mechanisms involved in disease development/progression, inherent genetic susceptibility or alternative Ras dysfunction such as gene amplification and/or overexpression.
In conclusion, Ras mutations are the most common genetic alterations known in human cancers. Single base changes/point mutations in codon 12, 13 and 61 of the three closely related isoforms of the Ras gene family namely, H-Ras, K-Ras and N-Ras cause loss of intrinsic GTPase activity and thereby confer oncogenic functions. Oncogenic activation of Ras genes is involved in urothelial malignancies.
The present study was conducted to determine the clinical impact of mutant Ras by examining the relationship of clinical histopathological variables in 87 UCB patients with the mutational spectrum at the hotspot regions of H-Ras, K-Ras and N-Ras genes by PCR-RFLP and direct DNA sequencing.
The current observations rule out the possible role of the mutations examined in the above-mentioned hotspot regions in Ras gene activation. Our findings on the lack of mutations in H-Ras, K-Ras and N-Ras genes could be explained on the basis of different etiological mechanisms involved in disease development/progression, inherent genetic susceptibility, tissue specificity or alternative Ras dysfunction such as gene amplification and/or overexpression.
Mutational activation of Ras genes has been established as a prognostic factor for the genesis of a constitutively active RAS-mitogen activated protein kinase pathway that leads to cancer.
Due to the reported heterogeneity among the distribution of the most frequent mutations in Ras isoforms in different patient populations with urothelial carcinoma of the bladder (UCB), it is necessary to determine the presence/absence of mutations in order to predict disease outcome.
The present study was conducted to determine the mutational spectrum at the hotspot regions of H-Ras, K-Ras and N-Ras genes.
PCR-RFLP and direct DNA sequencing were employed to determine the presence or absence of mutations in the Ras isoforms and their clinical impact, if any, in 87 UCB patients.
None of the 87 UCB patients showed point mutations in codon 12 of H-Ras gene; codon 61 of N-Ras gene and codons 12, 13 of K-Ras gene by PCR-RFLP. Direct DNA sequencing of tumor and control bladder mucosal specimens followed by Blastn alignment with the reference wild-type sequences failed to identify even a single nucleotide difference in the coding exons 1 and 2 of H-Ras, N-Ras and K-Ras genes in the tumor and normal bladder mucosal specimens.
Our findings on the lack of mutations in H-Ras, K-Ras and N-Ras genes could be explained on the basis of different etiological mechanisms involved in tumor development/progression, inherent genetic susceptibility, and or tissue specificity in a given cohort of patients.
Gene amplification and/or overexpression of Ras could further explain an alternative mechanism of its dysfunction in Ras driven cancers.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kai K S-Editor: Dou Y L-Editor: Webster JR P-Editor: Wang LL
| 1. | Janković S, Radosavljević V. Risk factors for bladder cancer. Tumori. 2007;93:4-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 2. | Letašiová S, Medve'ová A, Šovčíková A, Dušinská M, Volkovová K, Mosoiu C, Bartonová A. Bladder cancer, a review of the environmental risk factors. Environ Health. 2012;11 Suppl 1:S11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 179] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 3. | Garg M. Urothelial cancer stem cells and epithelial plasticity: current concepts and therapeutic implications in bladder cancer. Cancer Metastasis Rev. 2015;34:691-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 4. | Dalbagni G, Presti J, Reuter V, Fair WR, Cordon-Cardo C. Genetic alterations in bladder cancer. Lancet. 1993;342:469-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 150] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 5. | Boulalas I, Zaravinos A, Karyotis I, Delakas D, Spandidos DA. Activation of RAS family genes in urothelial carcinoma. J Urol. 2009;181:2312-2319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1154] [Cited by in RCA: 1476] [Article Influence: 134.2] [Reference Citation Analysis (0)] |
| 7. | Shrestha G, MacNeil SM, McQuerry JA, Jenkins DF, Sharma S, Bild AH. The value of genomics in dissecting the RAS-network and in guiding therapeutics for RAS-driven cancers. Semin Cell Dev Biol. 2016;58:108-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Haliassos A, Liloglou T, Likourinas M, Doumas C, Ricci N, Spandidos D. H-ras oncogene mutations in the urine of patients with bladder-tumors - description of a noninvasive method for the detection of neoplasia. Int J Oncol. 1992;1:731-734. [PubMed] |
| 9. | Tripathi K, Garg M. Mechanistic regulation of epithelial-to-mesenchymal transition through RAS signaling pathway and therapeutic implications in human cancer. J Cell Commun Signal. 2018;12:513-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 10. | Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218-5225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 251] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 11. | Kompier LC, Lurkin I, van der Aa MN, van Rhijn BW, van der Kwast TH, Zwarthoff EC. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One. 2010;5:e13821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 12. | Sjödahl G, Lauss M, Gudjonsson S, Liedberg F, Halldén C, Chebil G, Månsson W, Höglund M, Lindgren D. A systematic study of gene mutations in urothelial carcinoma; inactivating mutations in TSC2 and PIK3R1. PLoS One. 2011;6:e18583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 13. | Ouerhani S, Elgaaied AB. The mutational spectrum of HRAS, KRAS, NRAS and FGFR3 genes in bladder cancer. Cancer Biomark 2011-. 2012;10:259-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Kim J, Kim WT, Kim WJ. Advances in urinary biomarker discovery in urological research. Investig Clin Urol. 2020;61:S8-S22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Nanda MS, Sameer AS, Syeed N, Shah ZA, Murtaza I, Siddiqi MA, Ali A. Genetic aberrations of the K-Ras proto-oncogene in bladder cancer in Kashmiri population. Urol J. 2010;7:168-173. [PubMed] |
| 16. | Karimianpour N, Mousavi-Shafaei P, Ziaee AA, Akbari MT, Pourmand G, Abedi A, Ahmadi A, Afshin Alavi H. Mutations of RAS gene family in specimens of bladder cancer. Urol J. 2008;5:237-242. [PubMed] |
| 17. | Czerniak B, Cohen GL, Etkind P, Deitch D, Simmons H, Herz F, Koss LG. Concurrent mutations of coding and regulatory sequences of the Ha-Ras gene in urinary bladder carcinomas. Hum Pathol. 1992;23:1199-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 68] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Buyru N, Tigli H, Ozcan F, Dalay N. Ras oncogene mutations in urine sediments of patients with bladder cancer. J Biochem Mol Biol. 2003;36:399-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Zhu D, Xing D, Shen X, Liu J. A method to quantitatively detect H-Ras point mutation based on electrochemiluminescence. Biochem Biophys Res Commun. 2004;324:964-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Eble JN, Sauter G, Epstein JI, Sesterhenn IA. Tumors of the urinary system. In: World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. Lyon, France: IARC Press; 2004. |
| 21. | Rabbani F, Cordon-Cardo C. Mutation of cell cycle regulators and their impact on superficial bladder cancer. Urol Clin North Am. 2000;27:83-102, ix. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Fitzgerald JM, Ramchurren N, Rieger K, Levesque P, Silverman M, Libertino JA, Summerhayes IC. Identification of H-Ras mutations in urine sediments complements cytology in the detection of bladder tumors. J Natl Cancer Inst. 1995;87:129-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 64] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 23. | Cattan N, Saison-Behmoaras T, Mari B, Mazeau C, Amiel JL, Rossi B, Gioanni J. Screening of human bladder carcinomas for the presence of Ha-ras codon 12 mutation. Oncol Rep. 2000;7:497-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 24. | Przybojewska B, Jagiello A, Jalmuzna P. H-RAS, K-RAS, and N-RAS gene activation in human bladder cancers. Cancer Genet Cytogenet. 2000;121:73-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 25. | Bartram CR, Ludwig WD, Hiddemann W, Lyons J, Buschle M, Ritter J, Harbott J, Fröhlich A, Janssen JW. Acute myeloid leukemia: analysis of ras gene mutations and clonality defined by polymorphic X-linked loci. Leukemia. 1989;3:247-256. [PubMed] |
| 26. | Ball NJ, Yohn JJ, Morelli JG, Norris DA, Golitz LE, Hoeffler JP. Ras mutations in human melanoma: a marker of malignant progression. J Invest Dermatol. 1994;102:285-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 140] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 27. | Pandith AA, Shah ZA, Khan NP, Bhat AY, Wani SM, Siddiqi MA. Screening of N-Ras gene mutations in urothelial cell carcinomas of the urinary bladder in the Kashmiri population. Asian Pac J Cancer Prev. 2009;10:1063-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 28. | Miller MS, Miller LD. RAS mutations and oncogenesis: Not all RAS mutations are created equally. Front Genet. 2011;2:100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 29. | Uchida T, Wada C, Ishida H, Egawa S, Ao T, Yokoyama E, Koshiba K. Infrequent involvement of mutations on neurofibromatosis type 1, H-Ras, K-Ras and N-Ras in urothelial tumors. Urol Int. 1995;55:63-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 30. | Olderøy G, Daehlin L, Ogreid D. Low-frequency mutation of Ha-Ras and Ki-Ras oncogenes in transitional cell carcinoma of the bladder. Anticancer Res. 1998;18:2675-2678. [PubMed] |
| 31. | Kawahara T, Kojima T, Kandori S, Kurobe M, Yoshino T, Kimura T, Nagumo Y, Ishituka R, Mitsuzuka K, Narita S, Kobayashi T, Matsui Y, Ogawa O, Sugimoto M, Miyazaki J, Nishiyama H. TP53 codon 72 polymorphism is associated with FGFR3 and RAS mutation in non-muscle-invasive bladder cancer. PLoS One. 2019;14:e0220173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |