Published online Aug 22, 2024. doi: 10.4291/wjgp.v15.i4.93606
Revised: May 14, 2024
Accepted: July 23, 2024
Published online: August 22, 2024
Processing time: 173 Days and 11.4 Hours
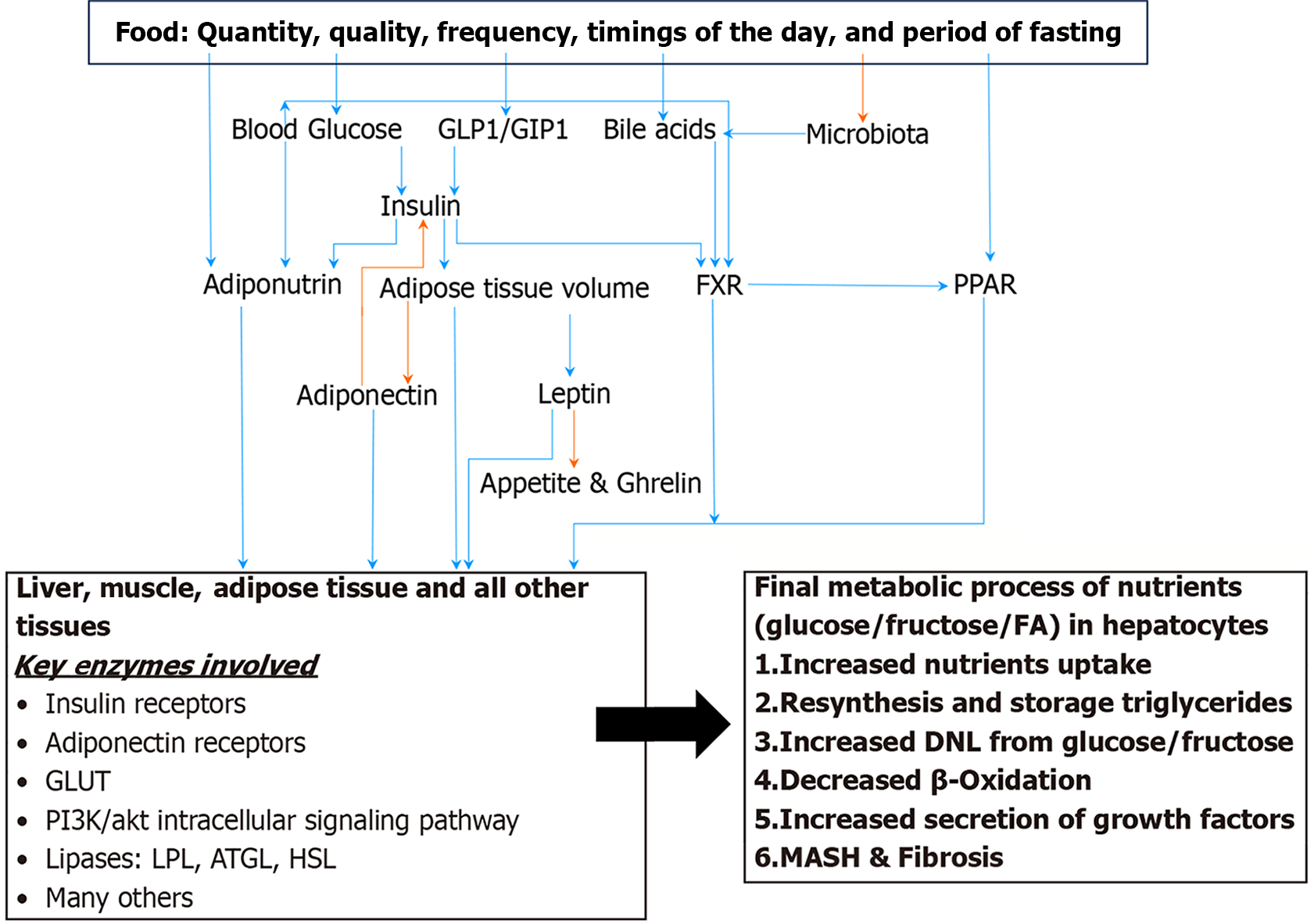
Nutrient metabolism is regulated by several factors. Social determinants of health with or without genetics are the primary regulator of metabolism, and an unhealthy lifestyle affects all modulators and mediators, leading to the adaptation and finally to the exhaustion of cellular functions. Hepatic steatosis is defined by presence of fat in more than 5% of hepatocytes. In hepatocytes, fat is stored as triglycerides in lipid droplet. Hepatic steatosis results from a combination of multiple intracellular processes. In a healthy individual nutrient metabolism is regulated at several steps. It ranges from the selection of nutrients in a grocery store to the last step of consumption of ATP as an energy or as a building block of a cell as structural component. Several hormones, peptides, and genes have been described that participate in nutrient metabolism. Several enzymes participate in each nutrient metabolism as described above from ingestion to generation of ATP. As of now several publications have revealed very intricate regulation of nutrient metabolism, where most of the regulatory factors are tied to each other bidirectionally, making it difficult to comprehend chronological sequence of events. Insulin hormone is the primary regulator of all nutrients’ metabolism both in prandial and fasting states. Insulin exerts its effects directly and indirectly on enzymes involved in the three main cellular function processes; metabolic, inflammation and repair, and cell growth and regeneration. Final regulators that control the enzymatic functions through stimulation or suppression of a cell are nuclear receptors in especially farnesoid X receptor and peroxisome proliferator-activated receptor/RXR ligands, adiponectin, leptin, and adiponutrin. Insulin hormone has direct effect on these final modulators. Whereas blood glucose level, serum lipids, incretin hormones, bile acids in conjunction with microbiota are intermediary modulators which are controlled by lifestyle. The purpose of this review is to overview the key players in the pathogenesis of metabolic dysfunction-associated steatotic liver disease (MASLD) that help us understand the disease natural course, risk stratification, role of lifestyle and pharmacotherapy in each individual patient with MASLD to achieve personalized care and target the practice of precision medicine. PubMed and Google Scholar databases were used to identify publication related to metabolism of carbohydrate and fat in states of health and disease states; MASLD, cardiovascular disease and cancer. More than 1000 publications including original research and review papers were reviewed.
Core Tip: The pathogenesis of metabolic dysfunctions associated steatotic liver disease (MASLD) is complex and a thorough analysis of the contributing factors to disease progression is pivotal to individualized patient care. Understanding and identifying the key pathogenic processes responsible for MASLD in each patient allows for the utilization of appropriate and effective pharmacotherapy. This review details the various key players in MASLD pathogenesis, through the lens of carbohydrate and fat metabolism, to divulge new areas of focus that could strengthen the capability of precision medicine for this population of patients.
- Citation: Habib S. Team players in the pathogenesis of metabolic dysfunctions-associated steatotic liver disease: The basis of development of pharmacotherapy. World J Gastrointest Pathophysiol 2024; 15(4): 93606
- URL: https://www.wjgnet.com/2150-5330/full/v15/i4/93606.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v15.i4.93606
Hepatic steatosis is defined by presence of fat in more than 5% of hepatocytes. Within hepatocytes, fat is stored in lipid droplets in the form of triglycerides. Hepatic steatosis results from a combination of multiple intracellular processes. These include increased fatty acid uptake, de novo lipogenesis (DNL), decreased excretion of very low-density lipoprotein (VLDL) and decreased beta oxidation of fatty acids. The source composition of fat in lipid droplet is albumin bound free fatty acid (FFA) derived from adipocyte lipolysis (59%), DNL (26%) and diet (15%). This composition may vary based upon pathogenesis of underlying key metabolic dysfunction. The triglycerides in lipid droplet are converted into VLDL by combining with Apo B100 and other lipoproteins. Subsequently, VLDL is secreted into systemic circulation. The type of fat depends upon eating habits and other factors[1]. The fatty acid composition of triglycerides in subjects with and without metabolic dysfunctions associated steatotic liver disease (MASLD), showing increased levels of saturated fatty acids in subjects with MASLD pointing toward DNL as the source, as saturated fatty acids are the major product of DNL[2,3]. This is consistent with the increased inclusion of DNL-derived triglycerides in VLDLs produced in MASLD, with roughly 15% of produced triglycerides derived from DNL compared to 2%–5% in normal subjects consuming a typical Western diet[4,5]. This is also true in carbohydrate overfeeding in healthy subjects, where DNL-derived triglycerides made up approximately 20% of secreted VLDL triglyceride content[6]. Thus, in both diseased and healthy states, the contribution of DNL-derived triglyceride is still the minority of total triglyceride content but correlates with the overall secretion of VLDL and may be a marker or regulator of relative fatty acid esterification vs oxidation, an important consideration in MASLD which is based on an imbalance of fatty acid oxidation and accumulation[7-9].
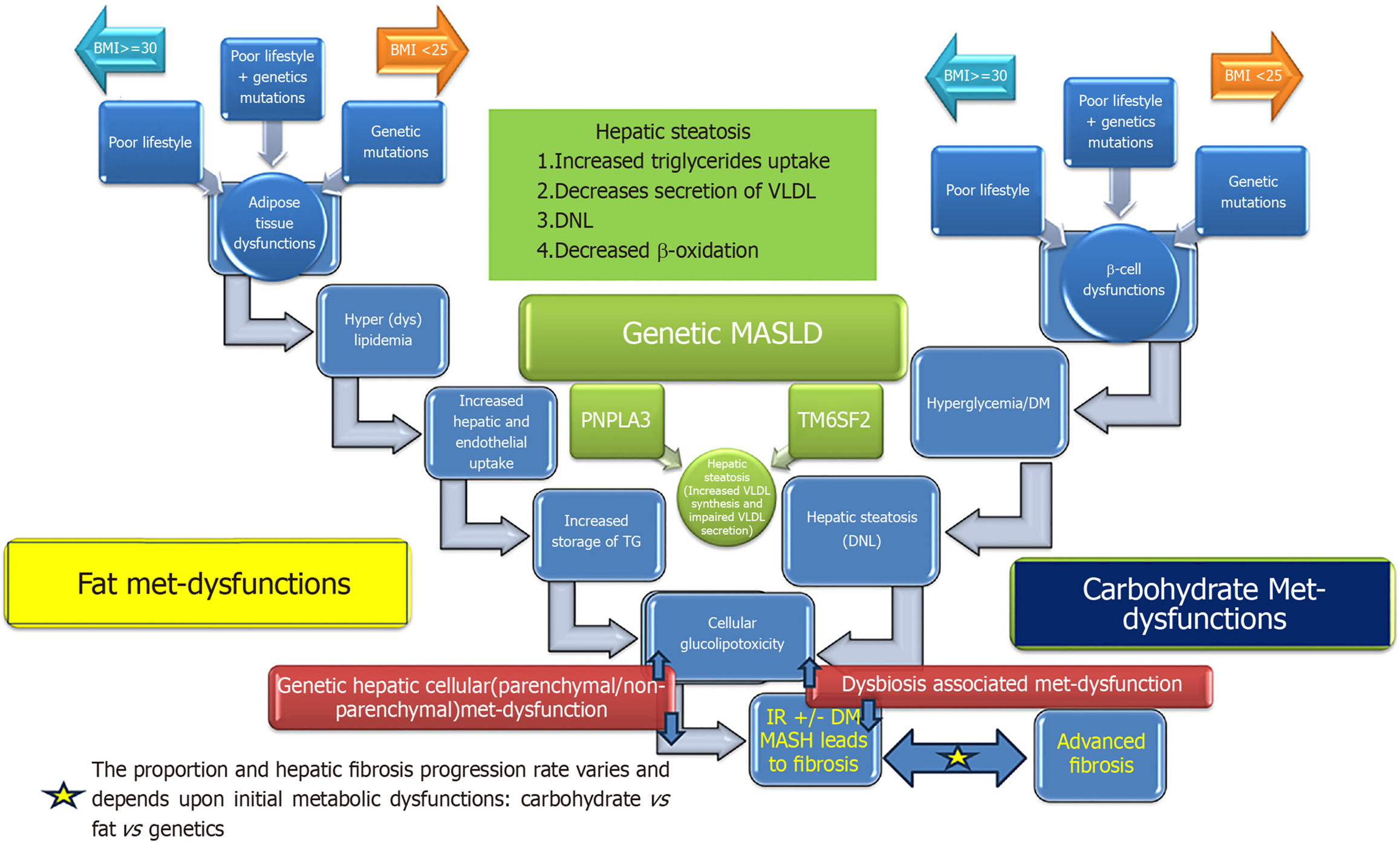
It is important to note that hepatic cellular metabolic dysfunction may arise as a consequence of one of the three or any combination of the three different metabolic derangements; carbohydrate metabolic dysfunctions resulting into hyperglycemia [diabetes mellitus (DM)], fat metabolic (adipose tissue) dysfunction resulting into excessive turnover of fatty acids between hepatocytes and adipose tissue or hepatocytes metabolic dysfunction of lipoprotein (VLDL) synthesis and secretion (Figure 1). Even though the end of the metabolic derangements is the same as hepatic steatosis but the natural course and consequences such as risk of steatohepatitis, hepatic fibrosis, cirrhosis, decompensation, carcinogenesis, and cardiovascular disease (CVD) is probably different in each pathogenic process. As of now we have very limited data on natural course of hepatic steatosis arising from each pathogenic process. Prospective longitudinal studies are needed to explore the impact of differential contribution of pathogenic processes in the natural history of hepatic steatosis.
Steatohepatitis and hepatic fibrosis are two independent processes that result from failure of hepatic adaptation to store excess fat. Hepatic steatosis is a benign process. Cardiometabolic risk is associated with onset of steatohepatitis defined by presence of any degree of inflammation or hepatic fibrosis. Severity and extent of inflammation does affect the subsequent course of progression. Hepatic fibrosis may start as a sequalae of steatohepatitis or independent of steatohe
The aim of this review is to comprehensively review the literature to understand the pathophysiology of MASLD and the factors involved in the pathogenesis of MASLD in order to help create a strategy for evaluation of a patient presenting with fatty liver. Moreover, it would help identify the therapeutic potentials.
Although this is not a formal systematic review with a prespecified question, however, a thorough literature review related to pathogenesis of MASLD was performed. PubMed and Google scholar are the main data bases that were used to identify all relevant English abstracts. Key search terms, used individually and in different combinations, included but not limited to “nonalcoholic fatty liver disease”, “non-alcoholic fatty liver disease”, “MASLD”, “pathogenesis”, “insulin resistance”, “diabetes mellitus” “obesity”, “visceral adipose tissue”, “glucagon”, “peroxisome proliferator-activated receptor”, “farnesoid X receptor”, “nuclear receptors”, “glucagon-like peptide-1”, “Glucose-dependent insulinotropic polypeptide (gastric inhibitory polypeptide-1) ”, “lipoproteins”, “fatty acid receptors”, “glucose and fructose receptors, ”, “signaling pathways”, β-oxidation”, “de novo lipogenesis”, “VLDL synthesis and secretion”, “adipokines”, “adult growth hormone deficiency”, “hypothyroidism” “insulin-like growth factor-1”, “management”, “replacement therapy”, “PNPLA3”, “genes”, and “medications”. Wherever possible question-based searches in Google were also performed. Moreover, databases were also searched by the name of key researchers in the field. A total of more than 1500 abstracts were identified, and key articles deemed to be relevant were reviewed in full. This review focuses on all original articles published until Jan 2024.
Nutrient metabolism processes that are regulated by the liver include: The production of lipoproteins, the secretion of triglycerides as VLDL, the conversion of excess glucose and other monosaccharides into triglycerides, glycogenosis, glycogenolysis, gluconeogenesis, cholesterol synthesis and the excretion of cholesterol through bile acids. 564 hepatocyte-secreted proteins, in humans, have been identified using mass spectroscopy. Fat accumulation and liver damage have been attributed to affecting the function of such proteins. Moreover, nutrient metabolism in the liver and other peripheral tissues is negatively impacted by the dysregulation of hepatokines[10-14].
Nutrient metabolism is regulated by several factors. Social determinants of health are the primary regulator of metabolism, and an unhealthy lifestyle affects all modulators, leading to the adaptation and finally to the exhaustion of cellular functions. The primary dysfunctions of individual modulators such as insulin, adipokines, incretins, nuclear receptors and other hormones involved are less common. The dysfunctions of enzymes and proteins involved in the final cellular processes of hepatic nutrient’s metabolic functions as described above are secondary to cellular stress. Though primary malfunctions of these proteins and enzymes have been described and linked to MASLD.
The clinical spectrum of MASLD ranges from mild simple hepatic steatosis to advanced fibrosis and hepatic malignancy. The presence of simple hepatic steatosis is a risk factor for progression to steatohepatitis in 20% of MASLD patients. Transitioning of hepatic steatosis into steatohepatitis sets the stage for progressive liver disease and creates a higher risk of CVD. Simple intracellular hepatic fat deposition is an adaptation phenomenon to maintain blood glucose and triglycerides homeostasis. Hepatic nuclear receptors play a fundamental role in maintaining cellular homeostasis while maintaining crosstalk with adipose tissue and pancreatic islet β cells. The transition of hepatic steatosis into steatohepatitis can be induced by several factors that affect these modulators. Insulin resistance acts as a cause and effect of cellular glucolipotoxicity. Here we discuss the role of nutrients, visceral adiposity, insulin resistance, DM, peroxisome proliferator-activated receptor (PPAR), farnesoid X receptor (FXR), bile acids, adiponectin, dysbiosis and genetics in the pathogenesis of MASLD and metabolic dysfunction-associated steatohepatitis (MASH) (Figure 2).
Nutrient’s quantity, quality, number of meals, the timing of food intake and duration of fasting in 24-hours period play significant role in nutrients’ metabolism and health of an individual. Fructose and saturated fat consumption have been associated with MASLD. The type of ingested and stored fat (saturated as opposed to polyunsaturated) influences cellular regulatory mechanisms. Dietary nutrients directly modulate the secretion of Insulin, glucagonlike peptide-1 glucagon-like peptide-1 (GLP1), gastric inhibitory peptide-1 gastric inhibitory polypeptide-1 (GIP1), PPAR, FXR and bile acids. Moreover, composition of diet and eating habits has an impact on the composition of microbiota in any given individual (discussed below). The harmful effects of a high saturated fatty acid diet on the liver, as opposed to a high unsaturated fatty acid diet, is due to the enhanced activity of SIRT3 in hepatocytes. SIRT3 is a manganese superoxide dismutase mainly localized in the mitochondria. The activation of deacetylation-induced manganese superoxide dismutase, inhibition of adenosine monophosphate-activated protein kinase (AMPK), and mammalian target of rapamycin C1–related autophagy inhibition are all processes that are induced by the enhanced activity of SIRT3[15,16].
The rate of carbohydrate absorption after a meal, as quantified by glycemic index, has significant effects on postprandial hormonal and metabolic responses. Though its contribution in the pathogenesis of glucotoxicity and cardiometabolic risk remains controversial[17-19]. High fructose consumption is source for DNL as it escapes rate limiting step. Moreover, it affect other mediators such as PPAR.
Depending upon location of fat in the body, white adipose tissue (WAT), can be further subcategorized into subcutaneous, visceral, and ectopic adipose tissue. Ectopic adipose tissue is located within the internal organs and found in lower amounts than the other two types of adipose tissue. Different types of ectopic fat include intrahepatocellular fat, intrapancreatic fat, intramyocellular fat, and intra cardio-myocellular fat. Visceral adipose tissue (VAT) is fat that surrounds the internal organs. Two types of VAT are epicardial fat and the abdominal visceral fat which surround the myocardia and gastrointestinal organs, respectively[20-22]. There are four types of intra-abdominal adipose depot: Subcutaneous, pre-peritoneal, peritoneal/mesenteric, and retroperitoneal. As the total body fat stores increase, regional fat depot increases accordingly. It is the mesenteric adipose tissue (MAT) only that significantly correlates with cardiometabolic health risk[20].
The key functions of all adipose depot include fat storage and secretion of adipokines. However, significant variability in these functions have been reported among fat depot based upon their regional location. MAT also plays a role in the intestinal physiological functions. In addition to holding intestine in its place, mesenteric fat acts as a physical barrier to inflammation and engages in controlling host immune response to translocation of gut bacteria. There are significant differences in cell morphology, gene expression profiles, cell components, biological characteristics, and immune and microbiota regulation roles between regional MAT, small bowel, and large bowel MAT. Several factors have been implicated that affect the fat deposition in MAT. Among them are eating habits, inactivity, male gender, and aging. In men, it has been estimated that about 21% of the ingested fat is stored in the intraperitoneal adipose depot and about 6% of it is stored in the retroperitoneal adipose depot[23]. In contrast, women only have about 5% of the ingested fat stored in the intraperitoneal adipose depot[24]. These gender differences in visceral adiposity may explain the gender differences in MASLD. These studies further support the notion that regional body fat distribution is predominantly determined by fat uptake and not lipolysis when considering gender. Leaky lymphatics are responsible for the VAT accumulation. Unhealthy lifestyle is the principal factor among all the factors that influence the accumulation of abdominal VAT.
Hyperplasia and inflammatory infiltrate of adipose tissue correlate with MASLD. It is the MAT that is most strongly associated with metabolic disease and adverse outcomes[25,26]. Higher levels of FFA are noted in portal blood in patients with MASLD associated with MAT deposition. Indirect evidence of the inflammatory output of VAT has been highlighted by the demonstration of increased levels of circulating cytokines and acute phase reactants in patients with visceral adiposity[27,28]. Thus far, published data indicate that B lymphocytes infiltrate early in the MAT during the development of MASLD, which may not only promote MAT inflammation by regulating macrophages but also migrate to the liver and induce hepatocytes inflammation. MAT thickness is a risk factor of MASLD, regardless of body mass index (BMI), age, gender, insulin resistance, fasting blood glucose, lipid, and blood pressure. The risk of MASLD increases with every 1mm increase in the MAT thickness. MAT directly affects hepatic inflammation and fibrosis with or without the presence of insulin resistance and hepatic steatosis. The severity of hepatic inflammation and fibrosis increases incrementally with increases in VAT. For each 1% increase in VAT, the risk of hepatic inflammation and fibrosis rises by 2.4-fold. and 3.5-fold, respectively. VAT is an independent predictor of advanced steatohepatitis even when adjusted for insulin resistance and hepatic steatosis. Enhanced secretion of interleukin-6 (IL-6), by VAT, is most likely a mechanism that leads to an increase in hepatic inflammation and fibrosis. VAT has been associated with all components of metabolic syndrome and higher serum IL-6 levels[29].
Gut microbiota consists of bacteria, viruses, fungi, and archaea. In humans, four main phyla of bacteria (Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria) represent more than 95% of the total microbiota. The gut microbiota is enriched with several genes important for biosynthesis of vitamins, xenobiotic metabolism, glycan and amino acid metabolism, and methanogenesis. The metabolic role of gut microbiota explains it's contribution to host nutrition, bone health, xenobiotic metabolism, integrity of the immune system, intestinal epithelial health, and protection against pathogens. Several factors play a role in the causation of dysbiosis. Among them, dietary factors are key players. These include excessive consumption of sugar, consumption of saturated fat, calorie-dense foods, and alcohol consumption. Gastric acid suppression by proton pump inhibitors, or due to atrophic gastritis, has been implicated in the pathogenesis of dysbiosis. Environmental factors contribute to the healthiness of microbiota, and these factors include geography, climate, the pattern of antibiotic usage in the region, sanitary conditions, and personal hygiene. A decrease of total bile acids inflow from the liver to the gut in the setting of advanced liver disease and cirrhosis may cause outgrowth of Firmicutes, particularly Clostridium, with concurrent decline in beneficial Firmicutes taxa (i.e., Lachnospiraceae, Roseburia, Rumminococcaceae and Blautia)[30-32].
The effects of dysbiosis are impaired production of the short-chain fatty acids butyrate, propionate, and acetate, which normally serve as sources of energy for colon mucosa. Butyrate, mainly produced by Firmicutes, is protective and enhances the secretion of leptin. On the other hand, acetate is produced by Bacteroides and is obesogenic. Other effects of dysbiosis include modulation of bile acids. Dysbiosis, in particular outgrowth of clostridium increases the ratio of conjugated to unconjugated bile acids. Dysbiosis affects bile acid detergent and signaling properties through the enzyme bile salt hydrolase of Lactobacillus and Bifidobacteria species, and through bile acid inducible operon. As a result of dysbiosis, microbiota produces ethanol from undigested sugars, and this ethanol production has been associated with damage to the intestinal epithelium. The bacteria/endotoxins termed as pathogen-associated molecular patterns (PAMPs) include microbial components, lipopolysaccharide (an endotoxin), flagellin, Lipoteichoic acid and peptidoglycan. These are translocated into portal circulation and bring about liver damage via toll-like receptor signaling. PAMPs induce damage-associated molecular patterns (DAMPs), also known as alarmins, are released by necrotic cells. DAMPs function as endogenous danger signals to promote and exacerbate the inflammatory response (Figure 3). Consequentially decline of VLDL liver export will result in hepatic steatosis as a result of reduction of choline metabolism. Finally, dysbiosis interferes with the intestinal adaptive immune system, enhancing the growth of abnormal and pathogenic bacteria and viruses[30].
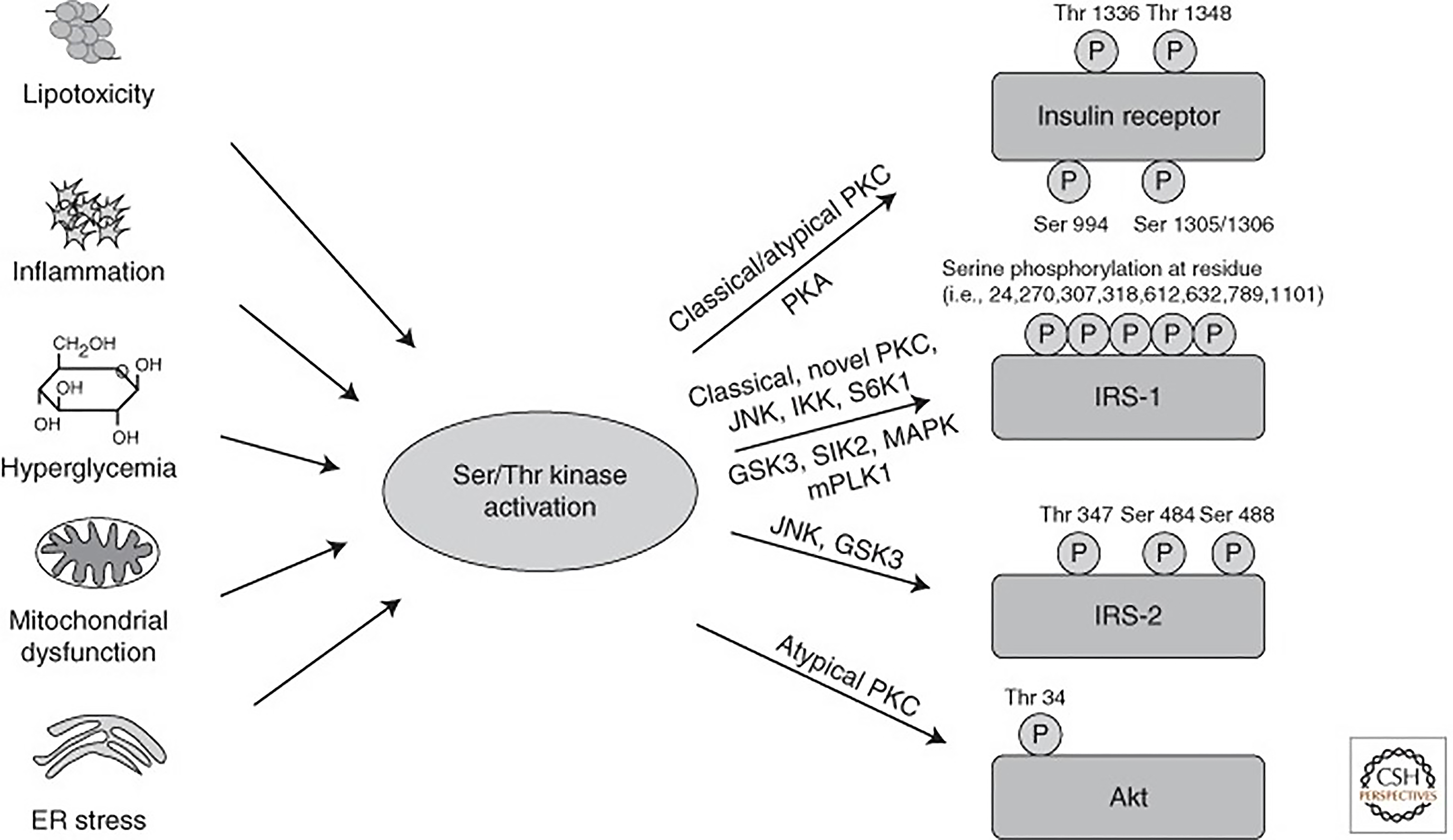
The hormone insulin plays a leading role in glucose and lipid metabolism. In the liver, it mediates its effect through liver specific insulin receptor (tyrosine kinase superfamily) and activation of downstream signal transduction. The receptor activates a complex intracellular signaling network through insulin receptor substrate (IRS) proteins that regulate two key functions: Metabolism and cell growth. The magnitude of intracellular signaling network induction required to perform metabolic function is different compared to the cell growth and depends upon insulin concentrations. While induction of the metabolic response requires lower insulin amounts, the mitogenic response requires higher concentrations[33]. The regulation of glucose homeostasis involves hepatic IRS1 and IRS2. The phosphoinositide 3-kinases (PI3K) activity associated with IRS2 increases during fasting, reaches its peak immediately after refeeding, and decreases rapidly thereafter. By contrast, the PI3K activity associated with IRS1 increases a few hour safter refeeding and reaches its peak thereafter. The key function of insulin is to transport glucose into the cell through glucose transporters (GLUT) and mediate intracellular metabolic and mitogenic functions. Among the 14 different types of glucose transporters, GLUT-4 is insulin sensitive. Insulin mediates glucose transport into adipocytes and myocytes via GLUT-4. Whereas glucose is also transported through GLUT-2 in hepatocytes, pancreatic β-cell, and other tissues independent of insulin. The critical pathway linking insulin receptors proteins to the metabolic actions of insulin is the PI3-kinase (PI3K) and protein-kinase B (Akt) pathway. Insulin also mediates its effect via other intracellular signaling pathways to control cellular homeostasis and cell cycle (Supplementary Figure 1). Insulin signaling is tightly controlled as uncontrolled activity of the downstream pathways could lead to severe perturbations in metabolism and tumorigenesis. Both intensity and duration of the signal play a significant role. Therefore, the ability to turn off the insulin signal in a rapid manner at various levels is critical. Nevertheless, impairment of either liver-specific insulin receptors or impairment of the PI3K/Akt or other intracellular signaling pathway leads to severe glucose intolerance and impaired lipid metabolism[34,35]. Also, some of the inhibitory mechanisms can be altered in pathophysiological conditions and participate in the development of insulin resistance[34].
Insulin sensitivity is defined as the rate at which the insulin hormone disposes blood glucose into muscle and fat tissue. Insulin resistance is a condition in which cells cannot respond adequately and efficiently to insulin. This occurs primarily at the level of so-called insulin-sensitive tissues, such as liver, muscle, and fat tissue, and can be caused by multiple mechanisms (Figure 4). Insulin resistance is the major pathogenic mechanism leading to DM type II. In addition to metabolic stress caused by hepatic steatosis and other conditions such as hepatitis C infection, genetic modifiers also contribute to the pathogenesis of hepatic insulin resistance. Several genetic mutations have been described affecting insulin receptors and insulin signaling molecules. Thus, the onset of insulin resistance is a multifactorial process, and this explains its’ heterogeneity among patients with MASLD[34-37]. Under the schema of selective insulin resistance, insulin receptor signaling via insulin receptor substrate (IRS)/PI3K/PKB/Foxo to suppress gluconeogenesis is dysfunctional but signaling via sterol regulatory element binding proteins (SREBP1c) is maintained, which is the key mechanism DNL[37-39]. In summary, MASLD-related insulin resistance is a steppingstone in the path toward DM type II in the subset of patients with diabetes (hepatogenous diabetes), which perpetuates with advancing fibrosis. Improvement in MASLD reduces the risk of DM type II[40-44].
Unger et al[43] defines DM as a bi-hormonal disease where insulin to glucagon ratio is critical in onset of diabetes[42-44]. Inadequate compensatory secretion of insulin in the setting of insulin resistance, pancreatic β-cell dysfunction or β-cell failure is the fundamental abnormality in the pathogenesis of diabetes[45-47]. However, in DM type II, β-cell loss is accompanied by increase in α-cell activity thereby increase in glucagon level. Low insulin to glucagon ratio has also been reported in DM type I, as glucagon secretion in response to meal remains unaffected[45]. Insulin to glucagon ratio plays significant role in DM related complications. In DM related MASLD, both increased uptake of fatty acids derived from adipose tissue lipolysis and DNL play role in causation of hepatic steatosis. Decreased insulin to glucagon ratio enhances lipolysis of adipose triglycerides by increased hormone sensitive lipase (HSL) activity. Whereas hepatic DNL is the product of increased gluconeogenesis and glycogenolysis along with increased lipogenesis from pyruvic acid, the product of fructolysis/glycolysis (Supplementary Figure 2). Hormone signaling and transcription factors, such as SREBP1c and carbohydrate response element binding protein (ChREBP) tightly regulate DNL. They regulate the expression of the key lipogenic genes acetyl CoA carboxylase; fatty acid synthase and ATP-citrate lyase. Moreover, insulin contributes to lipogenesis and adiposity. Both gluconeogenesis and glycogenolysis are affected by low insulin to glucagon ratio, whereas insulin signaling (PI3K/AKt/mTOR/SREBP) mediated lipogenesis from pyruvate remains unaffected by glucagon, thereby creating a state of selective intra-hepatic insulin resistance[37]. Treatment of DM type II with GLP1 agonist improves insulin to glucagon ratio by enhancing insulin and decreasing glucagon secretion. The rapid increase in trioses in the setting of high fructose consumption is among other mechanisms that contribute to DNL. Bypass of the rate-limiting step of glycolysis catalyzed by phosphofructokinase 1 allows for unrestricted hepatic fructolysis. Hence, fructose metabolism occurs at a significantly faster rate than glucose metabolism[48]. Pyruvic acid is the product of fructolysis/glycolysis, which enters into Kreb citric acid cycle and generates acetyl-CoA, which is a substrate for lipogenesis. Furthermore, fructose has been suggested to activate PPARγ coactivator-1β which acts as a co-activator of SREBP1c. This leads to increased expression of enzymes crucial to DNL[49,50]. Additionally, fructose has been suggested to inhibit the transcriptional activities of PPARα, thus reducing the levels of mitochondrial fatty acid oxidative enzymes regulated by PPARα[51,52]. This shift toward lipogenesis over fatty acid oxidation may contribute to hepatic steatosis and hence insulin resistance[53]. Fructose is also considered an activator of mitogen-activated protein kinase kinase-7 in rat primary hepatocytes, an upstream activator of c-Jun N-terminal kinase, which is considered also to inhibit insulin signaling[54,55].
Nevertheless, the pathogenesis of MASLD in the setting of obesity, DM and insulin resistance is not straightforward. Significant degree of heterogeneity exists among each of above-mentioned risk factors[56]. Despite presence of one or more of the above-mentioned risks factors, a proportion of patients do not develop MASLD. Genetic dysfunctions at multiple levels in metabolism of lipids and carbohydrates probably play a role[56,57]. Moreover, an intricate relationship has been reported among above-mentioned risk factors and hepatic steatosis (Supplementary Figure 3). There is a bidirectional relationship between DM type II and hepatic steatosis. Also, there is a bidirectional relationship between beta cell failure and IR. Moreover, a bidirectional relationship has been reported between IR and hepatic steatosis. In nutshell all of them are intertwined very closely and set into a vicious cycle.
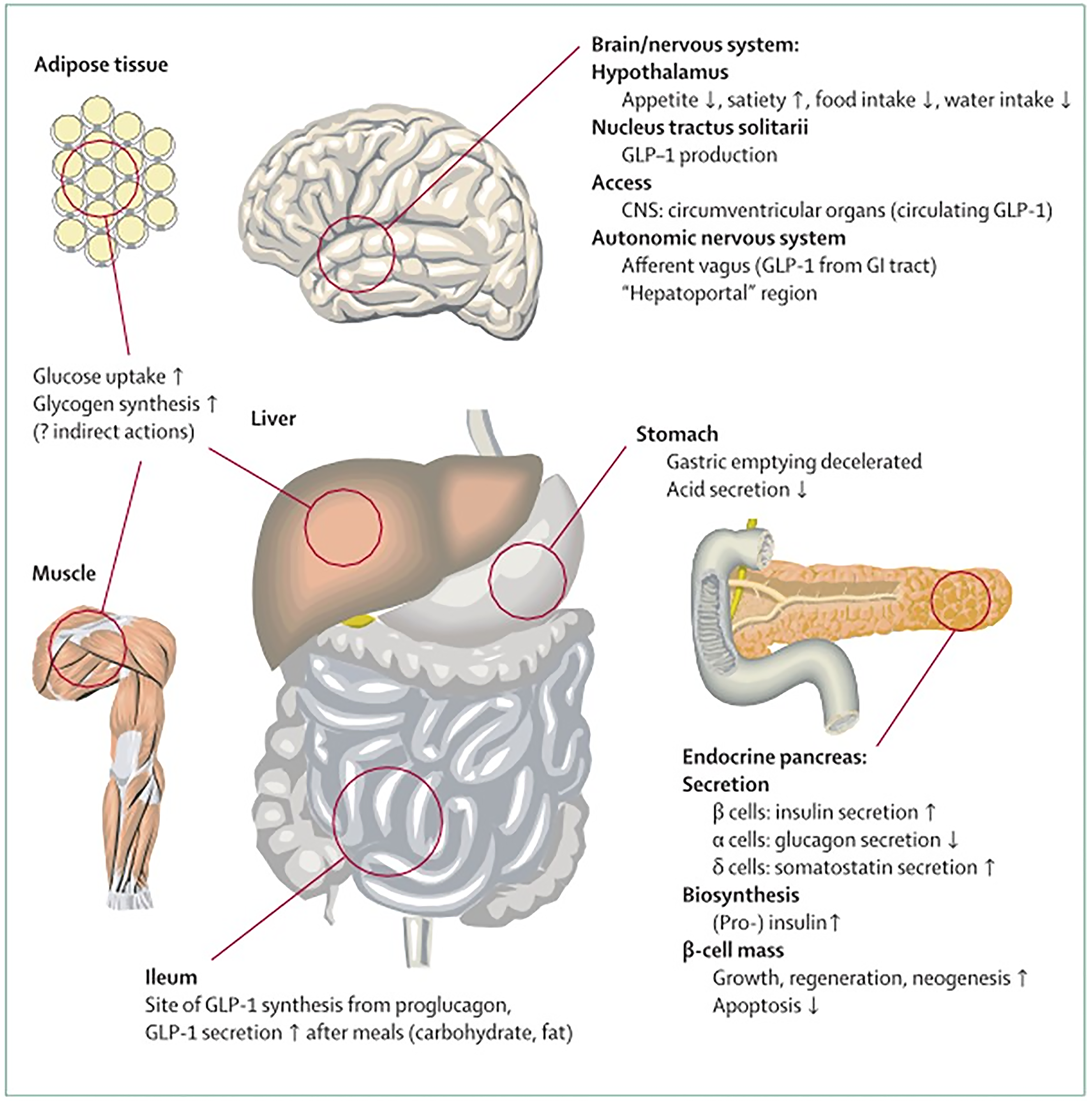
The incretin hormones GLP-1 and glucose-dependent insulinotropic polypeptide 1 play a role in both glucose and fat metabolism. Their secretion from intestinal cells is directly mediated by food intake.
The primary stimuli for the secretion of GLP-1 are glucose, other sugars, sweeteners, fatty acids, amino acids, and dietary fiber[58]. GLP 1 is produced in intestinal L cells to the most part in response to stimuli described as above. After synthesis, it is stored in granules within L cells and subsequently released after ingestion of food. Although small fraction of functional GLP-1has been found in tastebuds and the brain. Intestinal GLP 1 account for the most peripherally measured GLP 1 in the blood.
Additionally, non-nutrient factors, such as insulin and leptin, have also been linked as stimulators of GLP-1 secretion. On the contrary, intestinal and pancreatic somatostatin, whose secretion is increased by GLP-1, is the inhibitor of GLP-1 secretion, which serves as a negative local feedback loop in the gut[59-64].
Besides their role in glucose homoeostasis, the incretin hormones GIP and GLP-1 have additional biological functions: GLP-1 at pharmacological concentrations reduces appetite, food intake, and eventually, body weight, and a similar role is evolving for GIP.GIP stimulates insulin secretion and interacts with GIP receptors and regional blood vessels. Both events are responsible for increasing triglyceride storage in WAT and GLP-1 is not implicated in this mechanism. GIP and GLP-1, to a lesser extent, are involved in the bone remodeling process. Post-meal glycemic incretin secretion is reduced by GLP-1, but not GIP, due to its ability to slow gastric emptying. It has been shown that both GIP and GLP-1 have beneficial effects on cardiovascular and neurodegenerative central nervous system disorders (Figure 5). Moreover, it was found that co-agonists of GIP/GLP-1receptors have better efficacy compared to selective GLP-1 receptor agonists alone regarding glycemic control as well as body weight. Previously it was thought that GIP was without any therapeutic potential[65].
It has been long known that besides insulin and glucagon, other hormones also modulate macronutrient and energy metabolism. In fact, based upon their role they have been classified as anabolic and catabolic. Anabolic hormones include insulin, growth hormone, insulin-like growth factor and androgen and catabolic hormones are glucagon, cortisol, and catecholamines. It is the balance between anabolic and catabolic hormones functions, which is required to maintain homoeostasis. The imbalance by either decrease in the anabolic functions or increase in catabolic functions will result in impaired homeostasis and hepatic steatosis. Thyroid hormone has a unique role in energy metabolism and modulates basal energy expenditure and metabolic rate. Itis beyond the scope of this review to discuss each of these hormones. However, few helpful review articles are cited here[66-69].
MASLD has been studied in different endocrinopathies. Both clinical and experimental studies have suggested a strong correlation that hypothyroidism, hypopituitarism, and growth hormone deficiency can cause MASLD by affecting hepatic fatty acids uptake, DNL, β-oxidation, VLDL synthesis and VLDL secretion. Whereas the cause-and-effect relationship remains debatable between MASLD and polycystic ovarian syndrome, and hypogonadism in particular testosterone deficiency. Such an involvement of other hormones has been evaluated as a therapeutic potential for treatment of MASLD. There is some data on utility of growth hormone and testosterone replacement therapy for the treatment of MASLD. Nonetheless, the benefit of such hormone replacement in the setting of hormone deficiency for the treatment of MASLD remains questionable and therefore it is not recommended[66-85].
The concept of enhancing basal energy metabolism by thyroid hormone for MASLD and weight loss in euthyroid patient is very interesting and has been explored extensively (Supplementary Figure 4). Both T3 and T4 affect hepatic lipases and decreased activity in hypothyroidism leads to decreased clearance of VLDL. Both T3 and T4 regulate the synthesis of Apo B to promote VLDL) secretion[86]. More importantly, T3 regulates hepatic mitochondrial β-oxidation, which is impaired in MASLD and DM independent of thyroid status. T3 regulates the expression of both nuclear and mitochondrial genes involved in β-oxidation[87]. T3 exerts it is effect through by activating PPARγ and liver X receptor (LXR), which involves PPARγ coactivator 1-alpha (PGC-1α), acyl-CoA oxidase and fibroblast growth factor 21 (FGF 21)[88-90]. Also, T3 regulates mitochondrial biogenesis by modulating the rate limiting enzyme of fatty acid oxidation, carnitine O-palmitoyl transferase 1, PGC-1α-nuclear respiratory factor 1-transcription factor A, and phosphatase and tens in-induced kinase 1[91-94]. Moreover, T3 is also involved in regulation of mitochondrial turnover by selective degradation of mitochondria (mitophagy) via estrogen-related receptor α[42] as well as via induction of DAPK2 (death-associated protein kinase 2; a serine/threonine protein kinase)-SQSTM1/p62 (sequestosome 1)[43]. T3 can also activate mediator complex subunit 1-mediated autophagy and lipophagy in hepatocytes[44.].In conclusion, such a selective role of T3 in mitochondrial functions makes it a potential therapeutic target for treatment of MASLD[95,96].
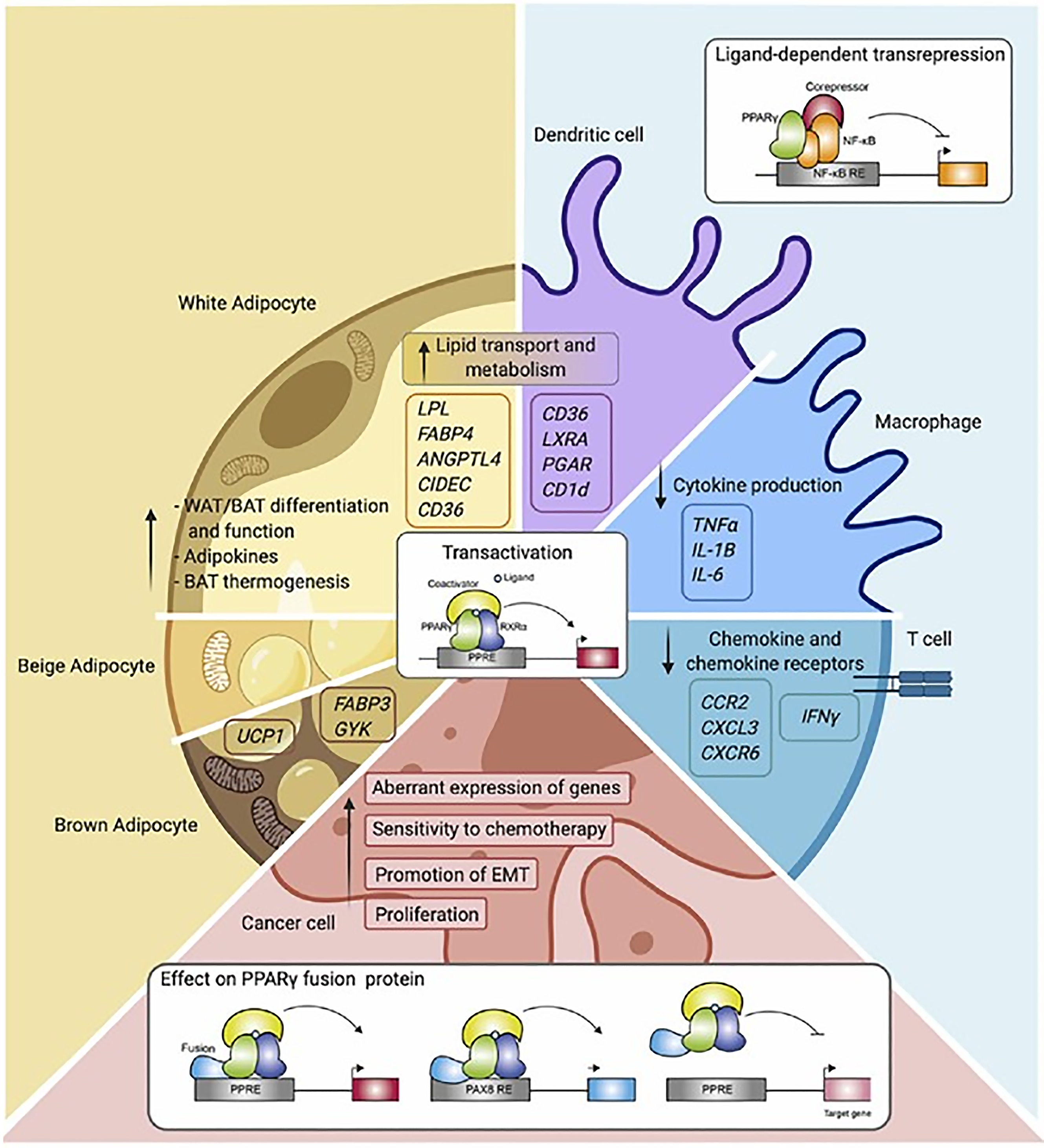
PPARs α, β/δ, and γ all modulate lipid homeostasis. PPAR-α regulates lipid metabolism in the liver. PPAR-β/δ promotes fatty acid β-oxidation largely in extrahepatic organs, and PPAR-γ stores triacylglycerol in adipocytes (Figure 6)[97].
PPAR-α functions as a lipid sensor in the liver[98]. It recognizes and responds to the influx of fatty acids by stimulating the transcription of specific genes. PPAR-α gene regulates transcription proteins, which perform triglycerides uptake, de novo lipogenesis and beta oxidation (Table 1)[99]. In addition, PPAR-α exerts anti-inflammatory activities (Table 1)[100]. Lipid metabolism is controlled by many steps including transport, binding, uptake, synthesis, mitochondrial and peroxisomal degradation, storage, lipoprotein metabolism and ketogenesis in fasting. PPAR-α activation induces the expression of a wide range of genes that are involved in the execution of the aforementioned processes[101]. PPAR-α expression is increased by polyunsaturated fat and decreased by saturated fat in hepatocytes. PPAR-α activation enhances hepatic fatty acid uptake and conversion into acyl-CoA derivatives. It enhances combustion of fatty acids via the β-oxidation pathways in mitochondria and peroxisomes and it also enhances microsomal omega-oxidation. PPAR-α increases hepatic fatty acid uptake by lipolysis via induction of the lipoprotein lipase, which hydrolyzes lipoprotein triglycerides into FFA and monoacylglycerol. PPAR-α controls lipoprotein lipase mRNA by binding to a domain PPAR response element. PPAR-α increases plasma high-density lipoproteins (HDL)-C by stimulating the synthesis of its major apolipoproteins: Apolipoprotein-AI and apolipoprotein-AII.
| Organ/tissue | Positive regulation | Negative regulation |
| Liver | ↑ ApoAI; ↑ ApoAII; ↑ ApoAV | ↓ Apoptosis |
| ↑ Cholesterol catabolism | ↓ HL | |
| ↑ FA oxidation and activation | ↓ Inflammation | |
| ↑ Fatty acid oxidation genes | ↓ Oxidative stress | |
| ↑ FATP; ↑ FAT/CD3 | ↓ Proteolysis of SREBF1 | |
| ↑ Gluconeogenesis and glycolysis | ↓ SREBF2 | |
| ↑ Insig1 | ||
| ↑ Ketogenesis | ||
| ↑ Lipogenesis | ||
| ↑ LPL and triglyserides clearance | ||
| Skeletal muscle | ↑ FA oxidation gene | ↓ Glucose intolerance |
| ↑ FA oxidation | ||
| ↑ Insulin sensitivity | ||
| Cardiac muscle | ↑ AT2 receptor | ↓ AT1 receptor |
| ↑ FA uptake | ↓ Cardiac hypertrophy and inflammation | |
| ↑ FA oxidation | ↓ Genes for glucose uptake and oxidation | |
| ↓ ED-1 (CD68) expression | ||
| ↓ NF-κB activity | ||
| ↓ VCAM-1, ICAM-1 | ||
| ↓ Platelet and endothelial cell adhesion | ||
| Plasma | ↑ HDL | ↓ ApoC3 |
| ↑ RCT | ↓ Dyslipidemia | |
| ↓ Inflammation | ||
| ↓ IFN-γ | ||
| ↓ TNFα | ||
| ↓ sdLDL | ||
| ↓ VLDL-triglyserides |
Ineffective PPAR-α sensing can lead to reduced energy burning, resulting in hepatic steatosis and steatohepatitis. Down regulation of PPAR-α is seen in the setting of saturated fat accumulation in hepatocytes and due to genetic abnormalities. In human liver, a, dysfunctional, and less-expressed isoform in small amount has also been identified, which is the result of introduction of a premature stop codon (exon 6 Lacking)[102]. There is no difference in the expression of PPAR-α in patients with suspected MASLD (free of MASH) compared to healthy subjects. On the other hand, lower levels of PPAR-α expression were found in the livers of MASH patients compared to those of MASLD patients. Liver PPAR-α expression correlated negatively with the presence of MASH and also with fibrosis. Further analysis revealed similar correlation with each histological component of MASH; steatosis, hepatocyte ballooning, and the MASH activity score. This correlation has been confirmed on histological examination of liver biopsies by immunohistochemical staining of PPAR-α[103,104]. It showed that, compared to normal liver tissues, PPAR-α expression was decreased in steatosis and reduced even more so in the inflammatory state of MASH[105]. In an animal model, PPAR-α expression is also decreased in MASH caused by a methionine- and choline-deficient diet in conjunction with a reduction in peroxisomal and mitochondrial fatty acid oxidation[102]. PPAR-α activation upregulates adiponectin receptors while reducing obesity-related inflammation adipose tissue. This ultimately contributes to the resolution of MASLD[106,107].
Food, fasting, activity and stress are the key regulators of glucose and fatty acid metabolism in adipose tissue to maintain nutrient homeostasis. Adipose tissue volume is controlled by adipokines. Chronic exposure to high-calorie food results in the expansion of adipose tissue by means of hypertrophy of adipocytes and the hyperplasia of adipose tissue to adapt to the physiological need. As adipose tissue grows, it initiates the process of satiety to control food consumption through the leptin–hypothalamus–ghrelin axis. The nuclear receptor PPAR-γ is a key transcriptional factor implicated in adipokine gene expression. PPAR-γ is highly expressed in adipose tissue and macrophages, playing a pivotal role in adipogenesis, lipid metabolism, insulin sensitivity, and immune regulation. PPAR-γ expression has been observed to be relatively high in the placenta while being much lower in the liver and muscle. PPAR-γ is regulated by CCAAT/enhancer-binding proteins (C/EBP-α). However, the function of PPAR-γ is dependent upon other transcription factors to transform preadipocytes into mature adipocytes with metabolic and secretory functions[97].
Adipose tissue failure is characterized by a failure to expand adequately to meet the demand to store excessive calories and a failure to maintain adipokine secretory functions. Adipocyte tissue fails to grow due to inadequate activation or loss of function of C/EBP-α and/or PPAR-γ. Factors affecting C/EBP-α include hormones (insulin, glucocorticoid, growth hormone, thyroid hormone, and noradrenaline) and several inflammatory cytokines including tumor necrosis factor-alpha (TNF-α), IL-6, IL-1, epidermal growth factor, and protein-kinase C (PKC) activators. Above all, amino acid deprivation and endoplasmic reticulum stress due to the enhanced need to grow also regulate the C/EBP-α. Several genetic mutations have also been described as causes of a loss of transcription factor function. Restoring PPAR-γ functions helps adipose tissue grow and achieve homeostasis of metabolism[108,109].
In addition to inadequate function of C/EBP-α, inadequate function of HSL results in a loss of lipolytic function of adipose tissue, which in turn leads to the decreased expression of lipogenic genes in adipocytes and reduced expression of PPAR-α target genes. S-nitrosylation of insulin-signaling proteins increases the hypertrophy of adipocytes, leading to an inhibition of the effects of insulin and causing insulin resistance. Additionally, lipolysis can be induced by endoplasmic reticulum stress stimuli via the activation of cyclic adenosine monophosphate/Protein kinase A and extracellular signal-regulated kinase ½ signaling in adipocytes. In accordance with the inhibition of insulin-mediated anti-lipolytic action, plasma FFA levels rise.
Taken together, these data demonstrate the requirement of HSL-mediated adipocyte lipolysis for the maintenance of proper adipose tissue function and activation of PPAR-α in the liver, thereby inhibiting the accumulation of hepatic TAGs and the development of steatotic liver disease[110]. Furthermore, there is ample evidence to support the fact that decreased PPAR-γ activity leads to decreased levels of adiponectin, which in turn causes insulin resistance, atherosclerosis, fatty liver disease and impaired fatty acid oxidation and energy expenditure in muscle[111,112].
Adiponectin secretion is decreased once adipose tissue fails to meet the ongoing need to expand, and reduced adiponectin leads to insulin resistance, atherosclerosis, and steatohepatitis. Adiponectin secretion is also modulated by other factors independent of PPAR-γ. BMI correlates inversely with adiponectin secretion. Adiponectin levels are higher in non-obese than obese people. Adiponectin has protective effects against metabolic syndromes and DM type II. After bariatric surgery, improvement in adiponectin levels suggest that its expression is subject to feedback inhibition in obesity, which is a proinflammatory state with a resulting increased secretion of inflammatory cytokines TNF-α, IL-6, IL-10, and nitric oxide adipose tissue macrophages. Proinflammatory markers lead to a reduction in the expression of adiponectin mRNA and the release of adiponectin from adipocytes. Adiponectin expression is suppressed by IL-6 and enhanced by the expression of resistin[111,113].
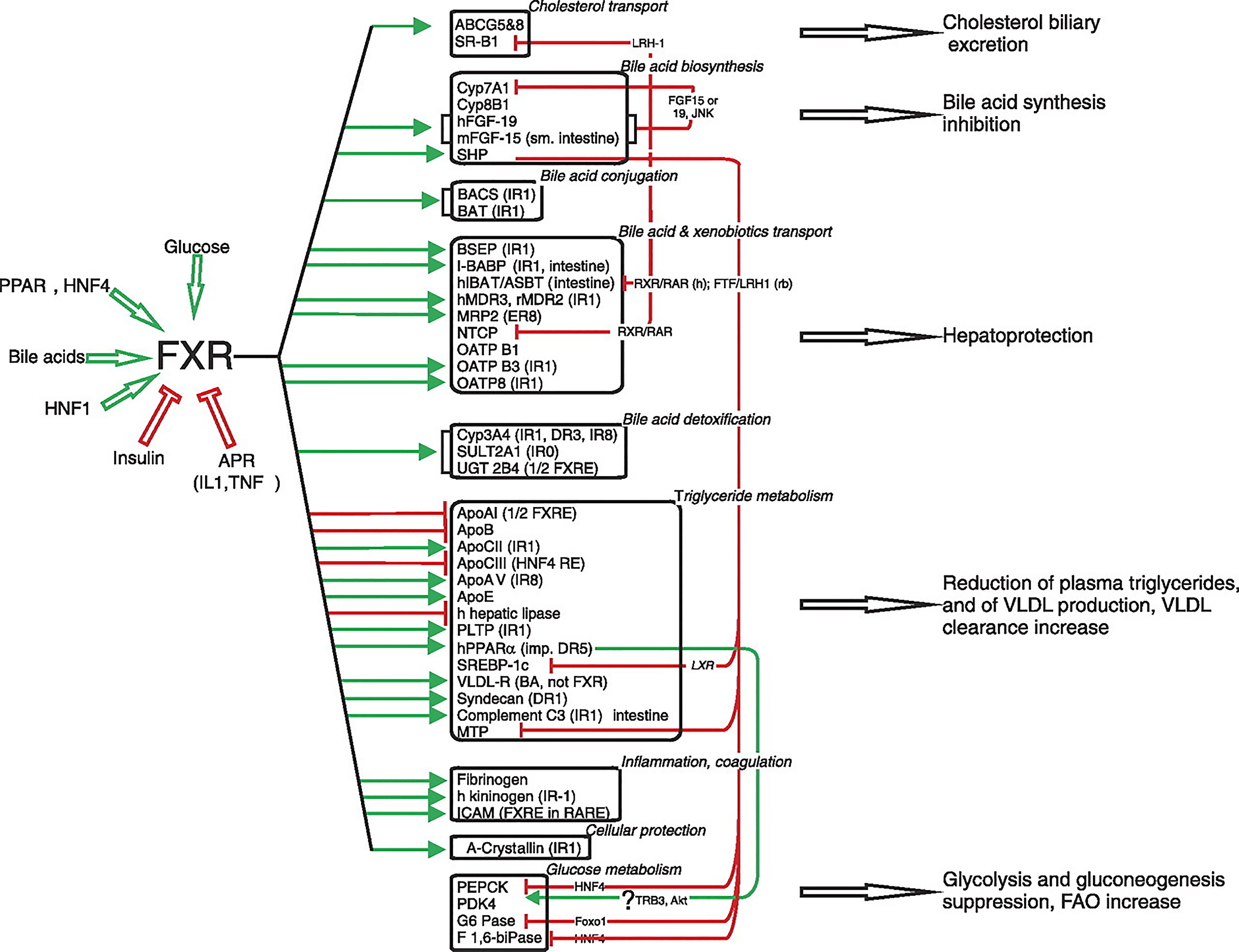
Bile acids are synthesized from cholesterol in the pericentral hepatocytes of the hepatic acini. Bile acids, in addition to their role in the digestion and absorption of triglycerides, also promote the hepatic secretion of cholesterol into bile by inducing bile flow and solubilizing biliary cholesterol, thereby promoting cholesterol to move from the hepatocyte into the intestinal lumen for excretion. Moreover, bile acids function as hormones regulating enterohepatic circulation in addition to fat, glucose, and energy homeostasis by signaling through nuclear and G protein–coupled receptors[114]. In 1999, it was discovered that bile acids are directly involved in regulating gene expression in the liver and intestine via interaction with FXR, also known as bile acid receptor[115]. FXR is expressed in many tissues in addition to liver tissue, especially intestinal endothelial cells. FXR is not expressed in the brain, heart, or lungs, or in skeletal muscle. FXR functions as a sensor, thus maintaining cellular functional homeostasis. Bile acids [primary bile acids: Cheno-deoxy cholic acid, cholic acid, deoxy cholic acid (DCA), and lithocholic acid], glucose/carbohydrates, PPAR, HNF1 and HNF4 all stimulate FXR activity, whereas insulin and APR (IL1 and TNF) inhibits FXR (Figure 7). Urso DCA does not influence FXR. In a physiological setting, fasting specifically increases hepatic FXR α3/4 expression, and a high-carbohydrate refeeding subsequently reduces FXR mRNA levels. White adipose tissue FXR-α and FXR-β expression also varies with circadian variation[116]. FXR modulates a considerable number of gene-encoding proteins involved in BA synthesis, metabolism, and transport in enterohepatic circulation[116,117]. FXR prevents the development of high-fat-induced obesity and insulin resistance. FXR inhibits hepatic lipogenesis through the interference of glucose-regulated gene promoters and by the downregulation of SREBP-1c in a SHP-dependent manner. Conversely, FXR may amplify FFA catabolism by increasing PPAR-α activity[118]. FXR’s control of VLDL assembly is due to its repression of MTP. The stimulation of LPL activity and hepatic uptake of remnants by FXR and its stimulation of LDL by its induction of syndecan-1 and VLDL-R increases the clearance of triglycerides. Finally, FXR may promote adipose triglyceride storage by stimulating adipocyte differentiation.
Adipocytes express and release numerous protein, lipid, and nucleic acid factors that can act in a paracrine or endocrine manner (Figure 8). Leptin and adiponectin are discussed here as their role in the pathogenesis of MASLD is well established[119-121]. However, there is emerging data revealing the role of resistin in the pathogenesis of MASLD.
Adiponectin is highly expressed in hepatocytes than other tissues such as endothelial cells. The key function of adiponectin is improving insulin sensitivity and decreasing blood glucose levels. Adiponectin acts via two adiponectin receptors: AdipoR1 (low affinity, ubiquitous but abundant in skeletal muscles) and AdipoR2 (high affinity, expressed in the liver). Adiponectin also acts via T-cadherin receptors (Figure 8). AdipoR1 and AdipoR2 serve as receptors for adiponectin and mediate increased PPAR-α, AMPK, glucose uptake, and fatty-acid oxidation by adiponectin. Adiponectin hormone exhibits anti-inflammatory properties and suppresses the development of atherosclerosis and liver fibrosis. It also plays a significant role in preserving hepatic metabolic functions to continue to adapt chronic metabolic stress, thereby preventing the development of steatohepatitis from simple hepatic steatosis[122].
Loss of adiponectin function is the fundamental pathogenic mechanism that turns simple hepatic steatosis, which is an adaptive phenomenon, to steatohepatitis, which marks the failure of hepatic metabolic system and may be termed a state of metabolic decompensation. It may reflect a stage of significant health risk for hepatic and extrahepatic diseases. It is supported by evidence from healthy, obese, and fatty liver disease studies.
In healthy people, adiponectin levels correlate negatively with alanine aminotransferase (ALT) and gamma glutamyl transferase levels before and after adjusting for gender, age, BMI, and insulin resistance[121,123]. This draws attention to the role of adiponectin in the maintenance of hepatocyte metabolic homeostasis through the regulation of both insulin sensitivity and/or inflammatory responses. Serum leptin and TNF levels are significantly higher and the adiponectin and ghrelin levels lower in patients with MASH compared with controls[121,123]. A meta-analysis of 27 studies evaluating a total of 2,243 subjects revealed that serum adiponectin was higher in controls than in MASLD or MASH patients, whereas adiponectin levels were significantly lower in MASH patients than in MASLD patients[124]. In fact, adiponectin levels were similar between the control and MASLD groups when analyzed based on histopathology. These findings suggest an important pathophysiological role of hypoadiponectinemia in the progression from MASLD to MASH. These findings support the role of low circulating adiponectin in the pathogenesis of MASLD. Additionally, they confirm the strong association among reduced adiponectin production by adipose tissue, MASLD, and IR. Interestingly, the findings reinforce the hypothesis that an imbalance between proinflammatory and anti-inflammatory cytokines may have a pathogenic role in the development of liver damage in MASLD[125]. Other studies also confirmed that low adiponectin was associated with MASH independent of BMI, IR and other adipokines such as TNF. Adiponectin levels also correlate with the severity of MASH[126-129]. Nonetheless, the relationship between low levels of adiponectin and fibrosis in patients with MASH remains controversial[122].
Leptin is another adipokine secreted by adipocytes that plays a role in diet-related metabolic and hormone regulation and that is affected by energy status and adipocyte fat mass. Leptin’s role in metabolic regulation includes hunger and satiety, body composition, energy expenditure, and body weight. It has been suggested that leptin plays a role in the pathogenesis of nonalcoholic steatohepatitis (NASH); however, the precise mechanism remains unclear and debated[130,131].
Leptin and other inflammatory adipokines such as IL-6 or TNF-α promote insulin resistance. Specifically, leptin antagonizes insulin functions by modifying the sensitivity of adipocytes, thereby reducing lipogenesis in adipose tissue, decreasing the binding capacity of insulin receptors in the liver, and inhibiting insulin secretion in pancreatic islets[132-134]. Hyperleptinemia damages pancreatic β-cells and inhibits JAK2/PI3K signaling in obese patients with T2DM and MASLD. This signaling pathway is known as the “leptin–insulin pathway” and under normal conditions itis activated to regulate glucose metabolism. Initial reports suggested that high leptin levels were associated with DM type II, but subsequent studies clarified that elevated levels of leptin were instead attributable to obesity[135]. The current data suggests that leptin is elevated in cholestatic mice, and excess leptin is responsible for increased ductular reaction, hepatic fibrosis, and inflammation via leptin receptor–mediated phosphorylation of Akt in cholangiocytes and hepatic stellate cells (HSC)[136,137].
Adipokines not only help the liver to adapt by storing excess nutrients in the form of lipid droplets, but they also mediate fibrogenesis[138-140]. Leptin in particular promotes stellate cell fibrogenesis and enhances the expression of tissue inhibitor of metalloproteinases, which promotes fibrosis in the injured liver through the inhibition of matrix metalloproteases and degradation of the extracellular matrix[138,141-143]. Concurrently, adiponectin, a counter-regulatory hormone that antagonizes the fibrogenic activity of leptin, is reduced in hepatic fibrosis[144]. Both of these adipokines are associated with insulin resistance and thus equally contribute to fibrogenesis[145,146].
In the liver, fatty acid uptake occurs mainly through the family of slc27a fatty acid transport proteins and the scavenger receptor CD36 (fatty acid translocase). CD36 is a scavenger receptor cell membrane surface glycoprotein, also known as fatty acid translocase. Its expression has been documented in a plethora of tissues including but not limited to adipose tissue, macrophages, platelets, endothelial cells, heart muscles, skeletal muscles, and liver cells. CD36 is a member of the SR-B1 subtype, which binds HDL particles and plays a role in the reverse cholesterol pathway. CD36 performs several functions. Related to nutrient metabolism, it facilitates lipid transport and binds various lipids, e.g., long chain fatty acids (LCFA) and oxidized low-density lipoprotein. Palmitoylation of CD36 molecule helps transport LCFA from cell membrane to intracellular organelles such as Golgi apparatus, and mitochondria[147].
Several factors affect the expression of CD36. In a normal healthy state, expression level is low, however it is expression increases with aging, high fat diet consumption, and in the setting of elevated FFA in circulation[147-149]. CD36 is regulated by nuclear receptors PPARγ, LXR and pregnane X receptor[150,151]. Upregulation leads to hepatic steatosis and fibrosis whereas down regulation improves steatoses and insulin resistance. After transport into the hepatocytes, fatty acids are esterified in triglycerides and subsequently packaged into VLDL for storage and secretion, respectively. If rate of fatty acid uptake is higher than the rate of packaging, it initiates inflammatory process as FFA are very toxic. CD36 plays a key role in the development of MASLD under conditions of elevated free FAs by modulating the rate of FA uptake by hepatocytes[150,151]
Several reports have shown its role in pathogenesis of insulin resistance, MASLD and atherosclerosis and also plays an important role in the pathogenesis of hepatic fibrogenesis. Increased hepatic uptake of LCFA through CD 36 upregulates hepatocytes stress, thereby promoting hepatic fibrosis[152,153]. Maréchal et al[152] in 2022 reported CD36 as a potential therapeutic target. Their work revealed that the inhibition of FAT/CD36 palmitoylation results in the localization of FAT/CD36 to the mitochondria of hepatocytes[152]. Mitochondrial FAT/CD36 functions as a molecular bridge between LCFAs and ACSL1 to augment the production of long-chain acyl-CoA, subsequently promoting fatty acid oxidation (FAO), and thereby avoiding lipid accumulation and overproduction of reactive oxygen species (ROS) in hepatocytes. De-palmitoylation of CD36 helps reverse the pathogenic mechanisms; reduced FAO in mitochondria of hepatocytes and excessive production of ROS and oxidative damage[152].
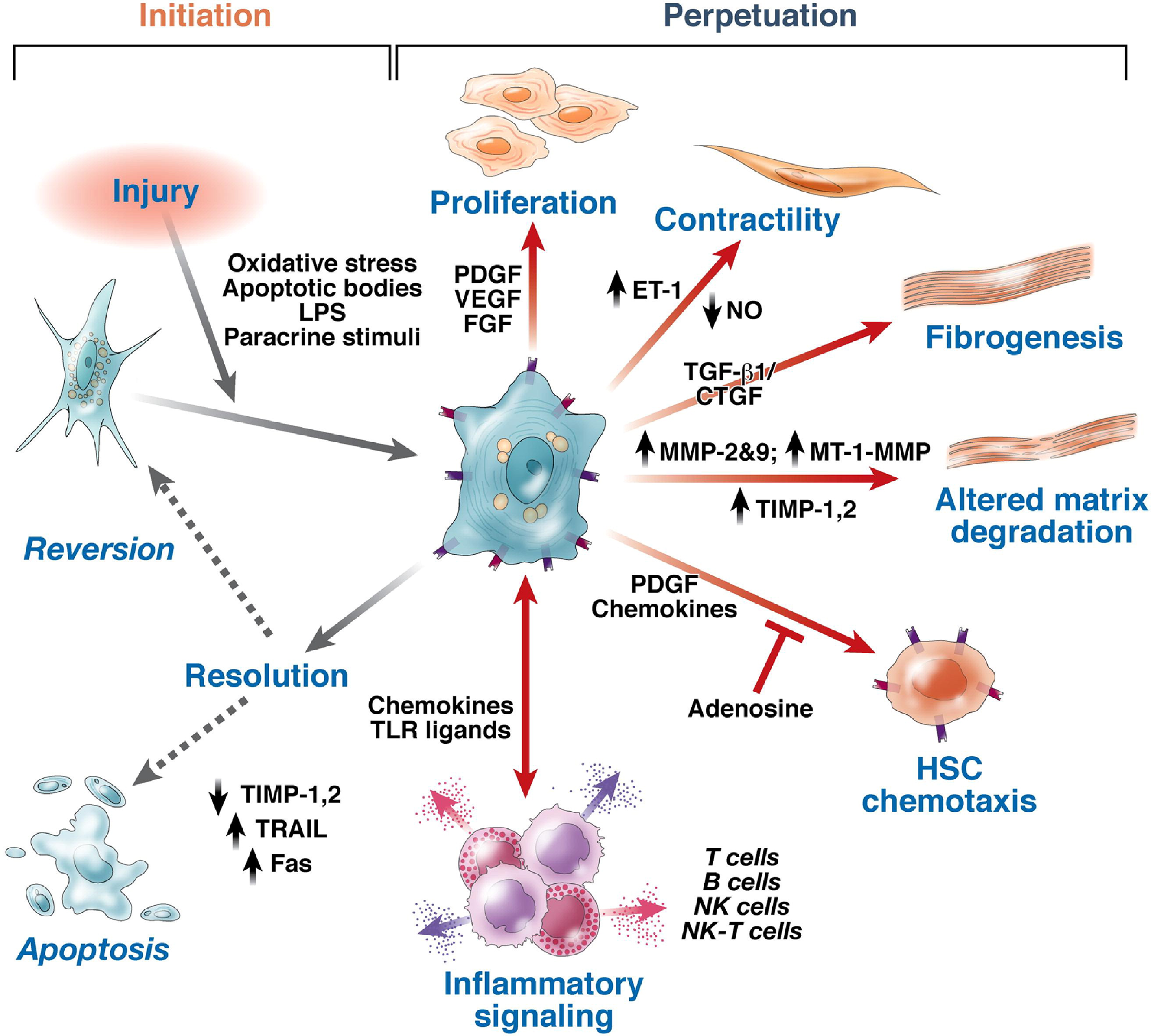
Hepatic steatosis and hepatocellular metabolic stress (with or without dysbiosis) initiate the processes of chronic inflammation and hepatic fibrosis. Subsequently, hepatic fibrotic tissue may be resorbed (fibro lysed) and replaced with regenerated hepatic tissue to restore normal hepatic tissue structure and functions. The balance between the progression of fibrosis and fibrotic resorption with hepatic regeneration depends on several factors. Elimination of the etiopathogenic process is the most principal factor in achieving reversal of fibrosis. Both environmental and genetic factors determine fibrosis progression or regression rate. It is imperative with ongoing efforts to define specific polymorphisms or relating with fibrosis progression rates. Studies have shown that hepatic fibrosis is a dynamic process and depends on many factors. Over the course of time it can progress, regress, or remain unchanged[154,155].
Several different cells, cellular receptors, signaling pathways, and secreted proteins participate in the pathogenesis of hepatic fibrosis. The major cell types involved appear to be HSC, liver sinusoidal endothelial cells, and macrophages (Kupffer cells). Transformation of hepatocytes into fibroblast (activated HSC) and loss of hepatocytes functions has also been reported[138]. The proteins secreted from these cells include connective tissue growth factor, transforming growth factor-β (TGF-β), platelet-derived growth factor, vascular endothelial growth factor, intercellular adhesion molecule-1, and adhesion molecules[138,156-159]. TGF-β is designated as “fibrogenic master cytokine” with multiple effects on extracellular matrix turnover[160,161].
Activation of HSC is the first step towards fibrogenesis. Factors activating HSC include hepatocellular necrosis due to oxidant stress, apoptosis, and soluble growth factors. Hepatic necrosis is considered a classical inflammatory and fibrogenic stimulus. Inflammatory cells such as neutrophils, Kupffer cells (the resident macrophages of the liver), and bone-marrow-derived monocytes and lymphocytes (Th17 cells) can promote HSC activation by secreting cytokines and growth factors[162-173]. Moreover, increased free cholesterol accumulation in HSCs plays a key role in the HSC activation, which results in further accumulation of free cholesterol and exaggerates liver fibrosis in a vicious cycle[163,174]. A study also demonstrated that LCFA facilitates hepatocyte activation by up-regulating oxidative stress through CD36[153]. Bechmann et al[165] revealed that LCFA (oleate: Palmitic acid 2:1) had profibrogenic effects on HSC via upregulation of a-SMA and TGF-β mRNA expression[164,165,175].
Activated HSCs are transformed into myofibroblasts; a key step in fibrogenesis and these myofibroblasts proliferate and migrate to the liver injury site, where they secrete extracellular matrix (Figure 9)[165]. Transformation of HSC into myofibroblast is controlled by a balanced activity of transforming growth factor and the bone-morphogenetic protein-7 (BMP-7). BMP-7 is a member of the TGF-β superfamily and is a natural antagonist of many TGF-βaction[166]. It counteracts TGF-β-induced epithelial mesenchymal transformation, extracellular matrix synthesis and inhibition of parenchymal cell proliferation[167-170]. Thus, the balance of both growth factors; TGF-β and BMP-7, is crucial.
HSCs express several receptors, including C-X-C Motif Chemokine Receptor 3, and C-C Motif Chemokine Receptors (CCR) 5 and 7, and they secrete numerous chemokines, including C-C motif ligands (CCL) 2, CCL3, CCL5 and C-X-C motif ligands (CXCL) 1, CXCL8, CXCL9, CXCL10[171,172]. These chemokines promote the migration of myofibroblasts to the site of injury, thereby enhancing fibrogenesis through proliferation and amplified inflammation. Notably, CCR5 is induced by nuclear factor kappa-light-chain-enhancer of activated B cells signaling and stimulates HSC migration and proliferation[172].
The chemokines secreted by inflammatory cells work in endocrine, paracrine, and autocrine fashion to stimulate activated HSC to produce, secrete and deposit collagen type I and III as an extracellular matrix. Mechanisms involved are the reduced expression of genes such as GFAP and PPARγ, and enhanced expression of fibrogenic genes such as alpha-smooth muscle actin (α-SMA) and collagen type I and III[162,173]. α-SMA immunohistochemical staining of hepatic tissue could be a valuable marker in the evaluation of stellate cell activation and fibrosis progression and a nearly indicator of the development of fibrosis[163,164,174,175].
In the past decades, significant developments have been made in the areas of tissue healing, repair, and regeneration. Growth factors provide chemical signals to stem cells, and in turn regulate their biological responses and tissue differentiation. The basic biological functions of growth factors and their endogenic roles in the tissue development and repair process have been investigated extensively. The regulatory roles of growth factors in cellular functions, including adhesion, proliferation, migration, and differentiation have made them potential agents for specific tissue reaction targeting. The FGF family encompasses a considerable number of factors that are involved in diverse actions such as cell growth, cell differentiation, and embryonic development[176-178]. FGF21 is a peptide hormone that plays a role in the regulation of energy homeostasis and is produced by several organs. The biology of FGF21 is exceedingly complicated due to its diverse metabolic functions in multiple target organs and its ability to function as an autocrine, paracrine, and endocrine factor. In the liver, FGF21 plays a key role in the regulation of fatty acid oxidation. FGF analogs have surfaced as a promising pharmco-therapy due to their capability to shift whole-body metabolism to a healthier state by acting directly in the liver.
Ligand binding induces several known signaling cascades, including Ras/MAPK, PI3K/Akt, and PKC. Hepatic FGF21 was identified as a downstream target of PPAR-α. Fatty liver resulting from either genetic obesity or diet induced obesity is associated with increased hepatic expression and serum levels of FGF21, presumably as a result of PPAR-α induction by fatty acids. In the liver, FGF21expression is also induced by protein insufficiency, secondary to amino acid deprivation, an effect that is downstream of activating transcription factor 4. The mechanisms of downstream acute signaling that might mediate the FGF21 response are largely unknown[179,180].
FGF19 plays an essential role in carbohydrate and lipid metabolism and has a favorable effect on MASLD[181]. FGF19 is a postprandial hormone, which is regulated by nutrients. It inhibits gluconeogenesis, while stimulating glycogenesis and protein synthesis without stimulating lipogenesis[182,183]. Abnormal signaling of the FGF19 is reported in MASLD patients, with resultant low levels in both fasting and postprandial states[184,185]. Fascinatingly, other reports have revealed low FGF19 Levels only in patients with MASH and fibrosis whereas there was no significant difference in basal FGF19 Levels between MASLD patients and healthy subjects[186]. Moreover, in longitudinal studies, a decline in FGF19 was detected in MASLD patients as the condition progressed to MASH. This FGF19 decline overtime was in parallel with increases in ALT, triglyceride, and other indicators[187]. The upregulation of FGF19 expression is associated with hepatocyte dysplasia, tumor formation, and a poor prognosis in individuals with hepatocellular carcinoma (HCC), which limits it’s utility as a therapeutic agent[188-191].
FGF21 generally acts directly or indirectly on multiple major organs, most notably on adipose tissue, liver and brain. It’s key function is to protect from obesity, insulin resistance, aberrant metabolisms, and irregular vascular homeostasis to a certain extent[192]. FGF 21 is synthesized in a variety of tissues, but liver and adipose tissue are the primary sources. Other sources are pancreatic islets, skeletal muscle, heart, kidneys, and certain other metabolic organs[193]. FGF21 usually recruits β-Klotho and FGFRs (FGFR1c, FGFR2c, or FGFR3c) as coreceptors for activation[194]. FGFRs are widely expressed in humans, whereas β-klotho is only expressed in specific tissues[195]. Both preclinical and clinical studies have demonstrated that FGF21 played a significant role in regulating energy homeostasis[196]. Physiological doses of FGF21 can decrease body weight and fat content, and mitigate insulin resistance, hyperglycemia, and dyslipidemia. FGF21 is needed for fasting liver metabolism that is responsible for blood glucose elevation through various pathways such as fatty acid oxidation[197]. Unlike FGF19, FGF21 does not induce the adverse effects of mitogenesis in vivo[198].
MASLD is a very heterogenous group of pathogenic processes. Inter-individual and inter-ethnic differences as well as differences in the severity and progression of liver disease are quite significant among patients with MASLD. Undoubtedly, cultural and socioeconomic factors play a role in MASLD acquisition and progression. Nevertheless, the observed variability of MASLD phenotypes and the variability of severity among individuals is best linked to the interaction between genetic risk factors and the environment[199]. Genetic and epigenetic factors play a significant role in the pathogenic process. Genetic involvement also explains the clustering of cases of MASLD among families[200-203]. The heritability of the disease has also been confirmed in twin studies. Evidence to support the considerable heritability of MASLD comes from familial aggregation and twin studies[204-207]. There is greater concordance of advanced disease between identical monozygotic twins than dizygotic twins, with an estimated 50% heritable contribution to variability[204,208,209]. Furthermore, the level of susceptibility differs in various ethnic groups: Hispanic people are more prone to advanced disease than White or Black people[204,210-214].
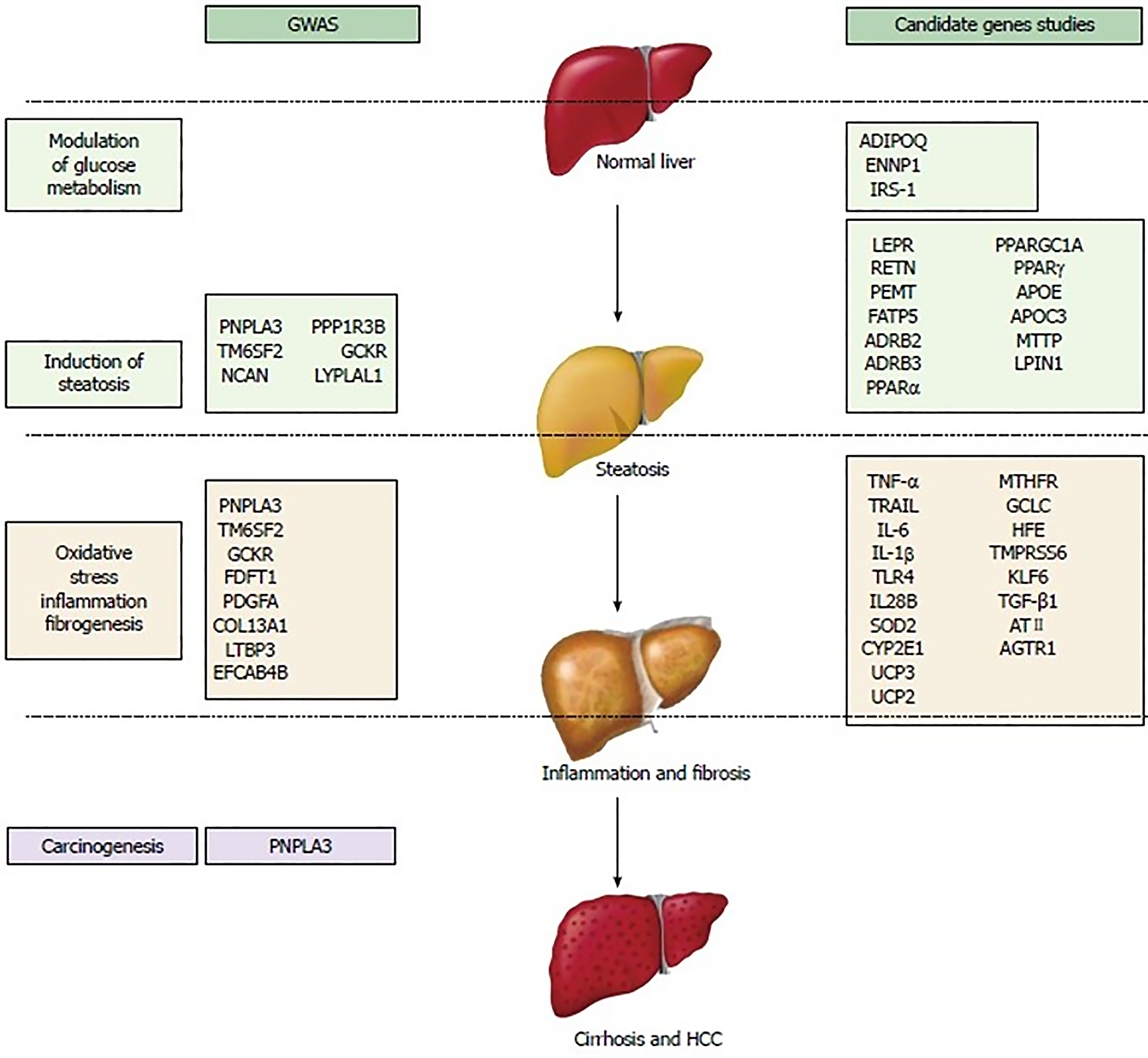
Thus far, numerous gene mutations have been implicated in the pathogenesis of MASLD. These genetic variants have been categorized based on their pathogenic contribution[207,215-217]. These SNP sand variants may affect glucose metabolism, insulin resistance, lipid metabolism, oxidative stress, inflammatory pathways, and fibrosis (Figure 9). Most of these genetic mutations were identified in genome-wide association study (GWAS) studies, which do not necessarily ensure the clinical relevance of the results, even though they show modest to large odds ratios and extreme statistical significance. Nonetheless, candidate gene studies have identified and validated a minority of loci associated with the risk of MASLD prevalence or progression. Several epigenetics factors such as microRNAs and DNA methylation could also modify the disease course in MASLD[200]. Apart from PNPLA3, none of the other genetic mutations have been shown to have a significant effect (independent of environmental factors) on large scale. Such genetic mutations increase the susceptibility to developing significant disease in the setting of other metabolic factors. A single genetic variant is unlikely to play a strong role in risk prediction. Nevertheless, a combination of several genetic variants may provide useful insight into disease progression. It is also important to recognize that these mutations are different from the already known mutations involved in the pathogenesis of monogenic obesity, DM, and dyslipidemia which can cause secondary MASLD in combination with acquired or other genetic/epigenetic factors. Genetic factors are estimated to contribute to 30%–50% of the risk for high-prevalence diseases such as obesity, DM type II, CVD, and cirrhosis[204,218].
The mere presence of a genetic mutation in either a homozygous or heterozygous allele, without confounding factors, may not increase the risk of disease. Clinical penetrance, defined as the proportion of people with the genotype that develops into the phenotype (clinical disease), varies among different genetic mutations. For example, it is estimated that only less than 5% of people with the C282Y homozygous hereditary haemochromatosis develop clinical disease, which is far less than its prevalence in the population. However, the precise estimate remains unknown[219]. Similarly, among patients with ZZ allele homozygotes, the lifetime risk of cirrhosis may be as high as 40%[220,221]. Nonetheless, the clinical penetrance of the MASLD-associated SNPs and variants remains to be determined. It is an undeniable fact that there are pronounced variations in clinical penetrance of each genetic heterozygous or homozygous mutation(s) involved in MASLD. Such an uncertainty of linking genetic mutations with clinical disease leads to the expansion of search for other genes, modifiers of genes, and environmental factors that might explain the observed variation in clinical penetrance. In conclusion, genetic susceptibility, which may make someone vulnerable to MASLD, is determined by the combined effects of many relatively common polymorphisms (minor allele frequency of 1%–5%), which each make a small contribution to the overall disease risk[204]. A polygenic risk score model was developed to estimate the risk of onset, severity, and progression of diseases such as MASLD, although the most appropriate method to evaluate the utility of genetic variant risk estimates is still under debate[217,222].
Description of each of these SNPs and variant is beyond the scope of this review. However, some important variants are summarized in Table 2[223,224].
| Gene | Variant | Impact on protein | Effect of the variant | Allelic frequency Europeans | Hispanics | Asians | Africans | Effect size | Direction of association (Ancestral allele) | Fat | NASH | Fibrosis | HCC | Mortality | Response to the therpaies |
| PNPLA3 | rs738409 C>G | I148M | Complex: Loss- plus gain-of-function | 0.23 | 0.57 | 0.38 | 0.14 | +++ | Up | + | + | + | + | + | + |
| TM6SF2 | rs58542926 C>T | E167K | Loss-of-function | 0.08 | 0.03 | 0.07 | 0.04 | +++ | Up | + | + | + | + | + | |
| GCKR | rs1260326 T>C | P446L | Loss-of-function | 0.6 | 0.67 | 0.5 | 0.86 | + | Up | + | + | + | + | ||
| MBOAT7 | rs641738 C>T | Linked to 3'-UTR | Reduced expression | 0.42 | 0.33 | 0.24 | 0.34 | + | Up | + | + | + | + | ||
| HSD17B13 | rs72613567 T>TA rs62305723 G>A | Alternate splicing P260S | Loss-of-function | 0.27 0.07 | 0.09 0.02 | 0.34 | 0.06 0.01 | ++ | Down | + | + | ||||
| IL28B (IFNL3/4) | rs368234815 TT>dG | Alternate protein translation site | Alternative protein | 0.27 | 0.09 | 0.34 | 0.06 | + | Down | + | + | ||||
| MERTK | rs4374383 G>A | Noncoding variant | Reduced expression | 0.37 | 0.37 | 0.73 | 0.47 | + | Down | + |
PNPLA3 is also known as adiponutrin (ADPN) or calcium-independent phospholipase A2 epsilon. In 2001, PNPLA3 was first described as a nutritionally regulated lipid-metabolizing enzyme. In 2008, the Dallas Heart Study, a genome-wide association study, was performed on a population that included a wide range of ethnicities. Dallas Heart Study reported a strong correlation between a genetic variant of PNPLA3, rs738409 C>G and the risk for developing hepatic steatosis and inflammation. Several independent reports subsequently confirmed the finding. PNPLA3, rs738409 C>G allele was most common in Hispanic people, the group most susceptible to MASLD; hepatic fat content was more than two times higher in PNPLA3-148M homozygotes than in noncarriers. Resequencing discovered another allele associated with lower hepatic fat content in African Americans, the group at lowest risk of MASLD. Such differences in the prevalence of PNPLA3 explain ethnic and inter-individual differences in hepatic fat content and susceptibility to MASLD. The PNPLA3 variant is a single-base polymorphism, which leads to a switch from a cytosine (C) to a guanine (G), resulting in an amino acid substitution of an isoleucine (I) for a methionine (M) at position148 (I148M) of the coding sequence. This results in loss of function of the native protein and eventually fat accumulation. The heritability of MASLD and hepatic fat accumulation is generally estimated to be between 20% and 70%, depending upon the ethnicity, the study design and methodology used to investigate it. The gene expression analysis showed that human PNPLA3 is expressed in a multitude of tissues, but the highest level was found in the liver, followed by the retinas, skin, adipose tissue, kidneys, brain, and spleen. The precise physiological role is unknown. PNPLA3 expression is impaired in adipose tissue and liver tissue as a result of acute and chronic nutritional and hormonal challenges, thereby supporting a potential role of PNPLA3 in the pathogenesis and/or physiological adaptation in two common disorders: Metabolic syndrome and MASLD[224-226].
Within the liver, PNPLA3 is expressed more than twice as much in human HSCs as in hepatocytes. It has also been shown that PNPLA3 protein mainly localizes to cellular membranes and lipid droplets. PNPLA3 linearly correlates with profibrogenic markers (Collagen1α1 and α-SMA) in HSCs. Additionally, PNPLA3-transient downregulation is accompanied by decreased expression of the two main profibrogenic markers, Collagen1α1 and α-SMA. Fascinatingly, it has been determined that the human PNPLA3 expression colocalizes with the profibrogenic marker α-SMA in pathological biopsies of MASH patients compared to patients with healthy livers. This finding further supports the theory that in humans, PNPLA3 probably has a more pivotal function in HSCs than in hepatocytes.
Nonetheless, experimental models suggest that the accumulation of I148M PNPLA3 on LDs is an initial sign of lipid metabolism impairment. Thereafter, the clearance of the protein is altered due to decreased ubiquitination, subsequently resulting in retarded proteasome degradation and diminished triacylglycerol mobilization from lipid droplets[227,228].
I148M PNPLA3 affects several molecular mechanisms regulating the phenotype of activated cells, such as decreased PPAR-γ signaling, and an enhanced c-Jun activated kinase/activator protein 1 pathway. The influence of variant carriage is clinically significant. A United States population study demonstrated by a high prevalence of dysmetabolism showed that it increased not only liver-related mortality, but also translated into increased overall mortality. The PNPLA3 genetic variant has been associated with a higher risk for simple steatosis, MASH, fibrosis and cirrhosis development in different populations and ethnicities. I148M allele acts as an independent risk factor in MASLD pathogenesis. Interestingly, there was no association between the PNPLA3 genotype and other risk factors for MASLD, such as obesity, DM type II and IR. Intriguingly, PNPLA3-related MASLD and MASH are different from “metabolic” MASLD and MASH. In metabolic MASLD and MASH, lipotoxic FFA (mainly saturated fatty acids and harmful ceramides) are the determinants of IR and DM type II-driven MASLD. On the other hand, the genetic “PNPLA3 MASLD” is characterized by polyunsaturated triglycerides, confirming a profound and marked discrepancy in origin of fatty liver diseases[229,230]. Moreover, PNPLA3 SNP has also been significantly associated with HCC in non-fibrotic liver[223,231].
TM6SF2 is the transmembrane 6 superfamily 2 human gene that codes for a protein by the same name. This gene is otherwise called KIAA1926[232]. Its exact function is currently unknown. A point mutation (rs58542926, c.499 C>T, P. Glu167 Lys, E167K) in the TM6SF2 gene is independently associated with elevated hepatic triglycerides, higher circulating levels of ALT, and lower levels of LDL-cholesterol[233-235]. TM6SF2 SNP has been shown to be significantly associated with a higher risk of hepatic steatosis, MASH, cirrhosis, HCC, as well with a lower risk of cardiovascular disease independent of other comorbidities[231,233-236].
TM6SF2 was identified in 2000[237]. The frequency of the Glu167 LysTM6SF2 variant was higher in individuals of European ancestry (7.2%), than in African-Americans (3.4%) or Hispanic-Americans (4.7%). Elevated mean and median hepatic triglyceride concentration, after adjusting for ethanol intake and HOMA-IR, were found among the carriers of the Glu167 LysTM6SF2 variant in all three ethnic groups. Moreover, the effect of the Glu167 LysTM6SF2 variant on hepatic triglyceride concentration was independent of the PNPLA3 rs738409 polymorphism. Homozygote TM6SF2 rs5854292 minor (T) allele carriage was shown to be associated with a significant increase hepatic triglyceride concentration (5.86% ± 0.25% in CC homozygotes vs 15.04% ± 2.23% in TT homozygotes)[233-235].
The TM6SF2 gene is located on chromosome 19p12, which functions as a lipid transporter. TM6SF2 is expressed predominantly in the liver and intestine. The TM6SF2 encodes a protein with transmembrane domains. Within human liver cells, TM6SF2 is localized in the endoplasmic reticulum and ER-Golgi intermediate compartment. In in-vitro studies, TM6SF2 inhibition was associated with reduced secretion of triglyceride-rich lipoproteins and increased cellular-triglyceride concentration and lipid droplet content, while TM6SF2 overexpression reduced steatosis[236,238,239]. TM6SF2 activity is required for normal VLDL secretion, and impaired TM6SF2 function causally contributes to MASLD[236,240]. TM6SF2 rs58542926 T-allele–mediated hepatic retention of triglycerides and cholesterol predispose patients to MASLD-related fibrosis, whereas C-allele carriage promotes VLDL excretion, thus increasing the risk of CVD or atherosclerosis while protecting the liver[236,241]. Looking at a large cohort (n = 1074) of MASLD patients, Liu et al[237] reported a significant association between TM6SF2 and advanced fibrosis/cirrhosis. This association was independent of age, BMI, T2DM and PNPLA3 rs738409 genotype[236].
The membrane-bound O-acyltransferase domain-containing 7 (MBOAT7) genetic variant is also associated with MASLD. Based on hepatic expression quantitative trait loci analysis, it has been suggested that MBOAT7 loss of function promotes liver disease progression, but this has never been formally evaluated. However, MBOAT7 loss in mice is sufficient to promote the progression of MASLD in the setting of a high-fat diet. MBOAT7 loss of function is associated with the accumulation of its substrate lysophosphatidylinositol (LPI) lipids, and direct administration of LPI promotes hepatic inflammatory and fibrotic transcriptional changes in an MBOAT7-dependent manner[242-244].
Although decreased MBOAT7 activity plays a role in NASH, MBOAT7 overexpression has not been shown to result in NASH pathology improvement, possibly due to the low abundance of its arachidonoyl-CoA substrate[242,245].
However, the different sample sizes, clinical features, and severity of the disease (i.e., obesity and T2D presence), and ethnicity of the cohorts enrolled in the studies are reasons the association between the rs641738 variant and liver injuries continues to be controversial and, ultimately, not fully replicated. The fact that the association has not been fully replicated could also be due to the diverse assessment of hepatic steatosis[246-251].
In a meta-analysis, data from 1066175 participants (9688 with liver biopsies) across 42 studies, the genetic variant rs641738 C>T was associated with higher liver fat on computed tomography/magnetic resonance imaging and diagnosis of MASLD in Caucasian adults. The variant was also positively associated with the presence of advanced fibrosis in Caucasian adults using a recessive model of inheritance (CC + CT vs TT). This analysis revealed rs641738 C>T near MBOAT7 as a risk factor for the presence and severity of MASLD in individuals of European descent[252,253].
Understanding the entire spectrum of physiological processes in carbohydrate and fat metabolism is pivotal to comprehend the pathogenesis of MASLD, which is quite complex. Owing to the participation of multiple factors in the pathogenesis, natural history is not quite predictable. Despite significant advancement in knowledge, we still lack adequate tools to stratify MASLD patients. Our review elaborates that several factors have to be assessed to comprehend the nature of MASLD in every individual patient. Clinically, it is important to evaluate each patient with MASLD to uncover the pathogenic process (es) to tailor the treatment to individual level. To characterize MASLD and predict the risk of MASH, cirrhosis, HCC, and CVD, we need to quantify hepatic fat, intra-abdominal VAT, baseline hepatic fibrosis, severity of insulin resistance, characterize dyslipidemia and DM, assess glycemic control of diabetes, conduct genetic testing for PNPLA3 and TM6SF2 gene mutations and obtain polygenic risk scores. Moreover, staining of liver tissue for CD36 and α-SMA may help assess the rate of hepatic uptake of adipose tissue derived fatty acid and the risk of fibrosis. Immunohistochemical staining of PPAR-α in liver biopsies might be a way to better assess PPARα activity in the liver to identify MASLD patients. Having detailed assessment of MASLD would help identify patients for individualized pharmacotherapy. At present we lack sophisticated validated tools to assess comprehensively for all the above features in clinical practice. Routine testing of adiponectin and leptin is debatable at present, nonetheless low adiponectin level favors aggressive disease behavior. We are not yet in a position to routinely assess genetic mutation in clinical practice. Lastly, addressing these metabolic dysfunctions by means of augmenting the functions of GLP1, PPARs, FXR, growth factors, or hormones is the key theory behind the development of novel therapeutic agent to treat MASH and reverse hepatic fibrosis. Selection of appropriate pharmaceutical agent (precision medicine) depends upon the key pathogenic process involved in a MASLD patient.
| 1. | Geidl-Flueck B, Gerber PA. Fructose drives de novo lipogenesis affecting metabolic health. J Endocrinol. 2023;257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (1)] |
| 2. | Kotronen A, Seppänen-Laakso T, Westerbacka J, Kiviluoto T, Arola J, Ruskeepää AL, Oresic M, Yki-Järvinen H. Hepatic stearoyl-CoA desaturase (SCD)-1 activity and diacylglycerol but not ceramide concentrations are increased in the nonalcoholic human fatty liver. Diabetes. 2009;58:203-208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 200] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 3. | Kotronen A, Velagapudi VR, Yetukuri L, Westerbacka J, Bergholm R, Ekroos K, Makkonen J, Taskinen MR, Oresic M, Yki-Järvinen H. Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia. 2009;52:684-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 158] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 4. | Diraison F, Moulin P, Beylot M. Contribution of hepatic de novo lipogenesis and reesterification of plasma non esterified fatty acids to plasma triglyceride synthesis during non-alcoholic fatty liver disease. Diabetes Metab. 2003;29:478-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 306] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 5. | Diraison F, Pachiaudi C, Beylot M. Measuring lipogenesis and cholesterol synthesis in humans with deuterated water: use of simple gas chromatographic/mass spectrometric techniques. J Mass Spectrom. 1997;32:81-86. [PubMed] [DOI] [Full Text] |
| 6. | Aarsland A, Chinkes D, Wolfe RR. Contributions of de novo synthesis of fatty acids to total VLDL-triglyceride secretion during prolonged hyperglycemia/hyperinsulinemia in normal man. J Clin Invest. 1996;98:2008-2017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 157] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 7. | Lewis GF, Carpentier A, Adeli K, Giacca A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr Rev. 2002;23:201-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 687] [Cited by in RCA: 771] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 8. | Kamagate A, Dong HH. FoxO1 integrates insulin signaling to VLDL production. Cell Cycle. 2008;7:3162-3170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 9. | Schwarz JM, Neese RA, Turner S, Dare D, Hellerstein MK. Short-term alterations in carbohydrate energy intake in humans. Striking effects on hepatic glucose production, de novo lipogenesis, lipolysis, and whole-body fuel selection. J Clin Invest. 1995;96:2735-2743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 183] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Baratta F, D'Erasmo L, Bini S, Pastori D, Angelico F, Del Ben M, Arca M, Di Costanzo A. Heterogeneity of non-alcoholic fatty liver disease (NAFLD): Implication for cardiovascular risk stratification. Atherosclerosis. 2022;357:51-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Gabriel-Medina P, Ferrer-Costa R, Rodriguez-Frias F, Ciudin A, Augustin S, Rivera-Esteban J, Pericàs JM, Selva DM. Influence of Type 2 Diabetes in the Association of PNPLA3 rs738409 and TM6SF2 rs58542926 Polymorphisms in NASH Advanced Liver Fibrosis. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Stender S, Loomba R. PNPLA3 Genotype and Risk of Liver and All-Cause Mortality. Hepatology. 2020;71:777-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 13. | Kanwar P, Nelson JE, Yates K. NASH Clinical Research Network. Available from: https://jhuccs1.us/nash/open/publicpublish.htm. |
| 14. | Kanwar P, Nelson JE, Yates K, Kleiner DE, Unalp-Arida A, Kowdley KV. Association between metabolic syndrome and liver histology among NAFLD patients without diabetes. BMJ Open Gastroenterol. 2016;3:e000114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Nascimbeni F, Bedossa P, Fedchuk L, Pais R, Charlotte F, Lebray P, Poynard T, Ratziu V; LIDO (Liver Injury in Diabetes and Obesity) Study Group. Clinical validation of the FLIP algorithm and the SAF score in patients with non-alcoholic fatty liver disease. J Hepatol. 2020;72:828-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (1)] |
| 16. | Mao T, Sun Y, Xu X, He K. Overview and prospect of NAFLD: Significant roles of nutrients and dietary patterns in its progression or prevention. Hepatol Commun. 2023;7. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Li S, Dou X, Ning H, Song Q, Wei W, Zhang X, Shen C, Li J, Sun C, Song Z. Sirtuin 3 acts as a negative regulator of autophagy dictating hepatocyte susceptibility to lipotoxicity. Hepatology. 2017;66:936-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 18. | Ludwig DS. The glycemic index: physiological mechanisms relating to obesity, diabetes, and cardiovascular disease. JAMA. 2002;287:2414-2423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1234] [Cited by in RCA: 1159] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 19. | Kalaitzopoulou I, Theodoridis X, Kotzakioulafi E, Evripidou K, Chourdakis M. The Effectiveness of a Low Glycemic Index/Load Diet on Cardiometabolic, Glucometabolic, and Anthropometric Indices in Children with Overweight or Obesity: A Systematic Review and Meta-Analysis. Children (Basel). 2023;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 20. | Chiavaroli L, Lee D, Ahmed A, Cheung A, Khan TA, Blanco S, Mejia, Mirrahimi A, Jenkins DJA, Livesey G, Wolever TMS, Rahelić D, Kahleová H, Salas-Salvadó J, Kendall CWC, Sievenpiper JL. Effect of low glycaemic index or load dietary patterns on glycaemic control and cardiometabolic risk factors in diabetes: systematic review and meta-analysis of randomised controlled trials. BMJ. 2021;374:n1651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 106] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 21. | Nauli AM, Matin S. Why Do Men Accumulate Abdominal Visceral Fat? Front Physiol. 2019;10:1486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 22. | Bertaso AG, Bertol D, Duncan BB, Foppa M. Epicardial fat: definition, measurements and systematic review of main outcomes. Arq Bras Cardiol. 2013;101:e18-e28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 23. | Frank AP, de Souza Santos R, Palmer BF, Clegg DJ. Determinants of body fat distribution in humans may provide insight about obesity-related health risks. J Lipid Res. 2019;60:1710-1719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 158] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 24. | Mårin P, Lönn L, Andersson B, Odén B, Olbe L, Bengtsson BA, Björntorp P. Assimilation of triglycerides in subcutaneous and intraabdominal adipose tissues in vivo in men: effects of testosterone. J Clin Endocrinol Metab. 1996;81:1018-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 45] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 25. | Votruba SB, Mattison RS, Dumesic DA, Koutsari C, Jensen MD. Meal fatty acid uptake in visceral fat in women. Diabetes. 2007;56:2589-2597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 54] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Kuk JL, Katzmarzyk PT, Nichaman MZ, Church TS, Blair SN, Ross R. Visceral fat is an independent predictor of all-cause mortality in men. Obesity (Silver Spring). 2006;14:336-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 439] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 27. | Després JP. Inflammation and cardiovascular disease: is abdominal obesity the missing link? Int J Obes Relat Metab Disord. 2003;27 Suppl 3:S22-S24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Lemieux I, Pascot A, Prud'homme D, Alméras N, Bogaty P, Nadeau A, Bergeron J, Després JP. Elevated C-reactive protein: another component of the atherothrombotic profile of abdominal obesity. Arterioscler Thromb Vasc Biol. 2001;21:961-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 369] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 29. | Rexrode KM, Carey VJ, Hennekens CH, Walters EE, Colditz GA, Stampfer MJ, Willett WC, Manson JE. Abdominal adiposity and coronary heart disease in women. JAMA. 1998;280:1843-1848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 785] [Cited by in RCA: 745] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 30. | van der Poorten D, Milner KL, Hui J, Hodge A, Trenell MI, Kench JG, London R, Peduto T, Chisholm DJ, George J. Visceral fat: a key mediator of steatohepatitis in metabolic liver disease. Hepatology. 2008;48:449-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 457] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 31. | Wiest R, Albillos A, Trauner M, Bajaj JS, Jalan R. Targeting the gut-liver axis in liver disease. J Hepatol. 2017;67:1084-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 307] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 32. | Chen J, Vitetta L. Gut Microbiota Metabolites in NAFLD Pathogenesis and Therapeutic Implications. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 206] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 33. | De Angelis M, Garruti G, Minervini F, Bonfrate L, Portincasa P, Gobbetti M. The Food-gut Human Axis: The Effects of Diet on Gut Microbiota and Metabolome. Curr Med Chem. 2019;26:3567-3583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 34. | Bedinger DH, Adams SH. Metabolic, anabolic, and mitogenic insulin responses: A tissue-specific perspective for insulin receptor activators. Mol Cell Endocrinol. 2015;415:143-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 35. | Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014;6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 994] [Article Influence: 90.4] [Reference Citation Analysis (0)] |
| 36. | Banday MZ, Sameer AS, Nissar S. Pathophysiology of diabetes: An overview. Avicenna J Med. 2020;10:174-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 266] [Article Influence: 53.2] [Reference Citation Analysis (1)] |
| 37. | Mattis KK, Gloyn AL. From Genetic Association to Molecular Mechanisms for Islet-cell Dysfunction in Type 2 Diabetes. J Mol Biol. 2020;432:1551-1578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 38. | Sanders FW, Griffin JL. De novo lipogenesis in the liver in health and disease: more than just a shunting yard for glucose. Biol Rev Camb Philos Soc. 2016;91:452-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 367] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 39. | Stefan N, Cusi K. A global view of the interplay between non-alcoholic fatty liver disease and diabetes. Lancet Diabetes Endocrinol. 2022;10:284-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 349] [Article Influence: 116.3] [Reference Citation Analysis (0)] |
| 40. | Tian J, Goldstein JL, Brown MS. Insulin induction of SREBP-1c in rodent liver requires LXRα-C/EBPβ complex. Proc Natl Acad Sci U S A. 2016;113:8182-8187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 41. | Heo NY. Nonalcoholic Fatty Liver Disease Is a Stepping Stone in the Path toward Diabetes Mellitus. Gut Liver. 2019;13:383-384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Cho HJ, Hwang S, Park JI, Yang MJ, Hwang JC, Yoo BM, Lee KM, Shin SJ, Lee KJ, Kim JH, Cheong JY, Cho SW, Kim SS. Improvement of Nonalcoholic Fatty Liver Disease Reduces the Risk of Type 2 Diabetes Mellitus. Gut Liver. 2019;13:440-449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 43. | Unger RH, Aguilar-Parada E, Müller WA, Eisentraut AM. Studies of pancreatic alpha cell function in normal and diabetic subjects. J Clin Invest. 1970;49:837-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 553] [Cited by in RCA: 574] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 44. | Müller WA, Faloona GR, Aguilar-Parada E, Unger RH. Abnormal alpha-cell function in diabetes. Response to carbohydrate and protein ingestion. N Engl J Med. 1970;283:109-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 422] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 45. | Jiang S, Young JL, Wang K, Qian Y, Cai L. Diabeticinduced alterations in hepatic glucose and lipid metabolism: The role of type 1 and type 2 diabetes mellitus (Review). Mol Med Rep. 2020;22:603-611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 46. | Unger RH. Glucagon and the insulin: glucagon ratio in diabetes and other catabolic illnesses. Diabetes. 1971;20:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 208] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 47. | Halban PA, Polonsky KS, Bowden DW, Hawkins MA, Ling C, Mather KJ, Powers AC, Rhodes CJ, Sussel L, Weir GC. β-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care. 2014;37:1751-1758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 353] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 48. | Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150:1223-1234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 939] [Cited by in RCA: 1116] [Article Influence: 85.8] [Reference Citation Analysis (0)] |
| 49. | Dholariya SJ, Orrick JA. Biochemistry, Fructose Metabolism. 2022 Oct 17. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024. [PubMed] |
| 50. | Nagai Y, Yonemitsu S, Erion DM, Iwasaki T, Stark R, Weismann D, Dong J, Zhang D, Jurczak MJ, Löffler MG, Cresswell J, Yu XX, Murray SF, Bhanot S, Monia BP, Bogan JS, Samuel V, Shulman GI. The role of peroxisome proliferator-activated receptor gamma coactivator-1 beta in the pathogenesis of fructose-induced insulin resistance. Cell Metab. 2009;9:252-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 163] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 51. | Linden AG, Li S, Choi HY, Fang F, Fukasawa M, Uyeda K, Hammer RE, Horton JD, Engelking LJ, Liang G. Interplay between ChREBP and SREBP-1c coordinates postprandial glycolysis and lipogenesis in livers of mice. J Lipid Res. 2018;59:475-487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 170] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 52. | Dekker MJ, Su Q, Baker C, Rutledge AC, Adeli K. Fructose: a highly lipogenic nutrient implicated in insulin resistance, hepatic steatosis, and the metabolic syndrome. Am J Physiol Endocrinol Metab. 2010;299:E685-E694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 293] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 53. | Ament Z, Masoodi M, Griffin JL. Applications of metabolomics for understanding the action of peroxisome proliferator-activated receptors (PPARs) in diabetes, obesity and cancer. Genome Med. 2012;4:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 54. | Lim JS, Mietus-Snyder M, Valente A, Schwarz JM, Lustig RH. The role of fructose in the pathogenesis of NAFLD and the metabolic syndrome. Nat Rev Gastroenterol Hepatol. 2010;7:251-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 555] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 55. | Wei Y, Wang D, Pagliassotti MJ. Fructose selectively modulates c-jun N-terminal kinase activity and insulin signaling in rat primary hepatocytes. J Nutr. 2005;135:1642-1646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 56. | Hirosumi J, Tuncman G, Chang L, Görgün CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2445] [Cited by in RCA: 2457] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 57. | Redondo MJ, Hagopian WA, Oram R, Steck AK, Vehik K, Weedon M, Balasubramanyam A, Dabelea D. The clinical consequences of heterogeneity within and between different diabetes types. Diabetologia. 2020;63:2040-2048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 58. | Unger RH. Lipotoxicity in the pathogenesis of obesity-dependent NIDDM. Genetic and clinical implications. Diabetes. 1995;44:863-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 563] [Cited by in RCA: 612] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 59. | Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology. 2007;132:2131-2157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2461] [Cited by in RCA: 2660] [Article Influence: 147.8] [Reference Citation Analysis (0)] |
| 60. | Anini Y, Brubaker PL. Muscarinic receptors control glucagon-like peptide 1 secretion by human endocrine L cells. Endocrinology. 2003;144:3244-3250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 93] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 61. | Anini Y, Brubaker PL. Role of leptin in the regulation of glucagon-like peptide-1 secretion. Diabetes. 2003;52:252-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 200] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 62. | Lim GE, Brubaker PL. Glucagon-like peptide 1 secretion by the L-cell. Diabetes. 2006;55:S70-S77. [DOI] [Full Text] |
| 63. | Brubaker PL. Regulation of intestinal proglucagon-derived peptide secretion by intestinal regulatory peptides. Endocrinology. 1991;128:3175-3182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 101] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 64. | Chisholm C, Greenberg GR. Somatostatin-28 regulates GLP-1 secretion via somatostatin receptor subtype 5 in rat intestinal cultures. Am J Physiol Endocrinol Metab. 2002;283:E311-E317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 65. | Hansen L, Hartmann B, Bisgaard T, Mineo H, Jørgensen PN, Holst JJ. Somatostatin restrains the secretion of glucagon-like peptide-1 and -2 from isolated perfused porcine ileum. Am J Physiol Endocrinol Metab. 2000;278:E1010-E1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 93] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 66. | Nauck MA, Quast DR, Wefers J, Pfeiffer AFH. The evolving story of incretins (GIP and GLP-1) in metabolic and cardiovascular disease: A pathophysiological update. Diabetes Obes Metab. 2021;23 Suppl 3:5-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 203] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 67. | Clemmons DR. Metabolic actions of insulin-like growth factor-I in normal physiology and diabetes. Endocrinol Metab Clin North Am. 2012;41:425-443, vii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 208] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 68. | Doycheva I, Erickson D, Watt KD. Growth hormone deficiency and NAFLD: An overlooked and underrecognized link. Hepatol Commun. 2022;6:2227-2237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |
| 69. | Zitzmann M. Testosterone deficiency, insulin resistance and the metabolic syndrome. Nat Rev Endocrinol. 2009;5:673-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 246] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 70. | Vidal-Cevallos P, Murúa-Beltrán Gall S, Uribe M, Chávez-Tapia NC. Understanding the Relationship between Nonalcoholic Fatty Liver Disease and Thyroid Disease. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 71. | Zeng X, Li B, Zou Y. The relationship between non-alcoholic fatty liver disease and hypothyroidism: A systematic review and meta-analysis. Medicine (Baltimore). 2021;100:e25738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 72. | Mauras N, O'Brien KO, Welch S, Rini A, Helgeson K, Vieira NE, Yergey AL. Insulin-like growth factor I and growth hormone (GH) treatment in GH-deficient humans: differential effects on protein, glucose, lipid, and calcium metabolism. J Clin Endocrinol Metab. 2000;85:1686-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 73. | He W, An X, Li L, Shao X, Li Q, Yao Q, Zhang JA. Relationship between Hypothyroidism and Non-Alcoholic Fatty Liver Disease: A Systematic Review and Meta-analysis. Front Endocrinol (Lausanne). 2017;8:335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 74. | Bikeyeva V, Abdullah A, Radivojevic A, Abu Jad AA, Ravanavena A, Ravindra C, Igweonu-Nwakile EO, Ali S, Paul S, Yakkali S, Teresa Selvin S, Thomas S, Hamid P. Nonalcoholic Fatty Liver Disease and Hypothyroidism: What You Need to Know. Cureus. 2022;14:e28052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (1)] |
| 75. | Isley WL, Underwood LE, Clemmons DR. Dietary components that regulate serum somatomedin-C concentrations in humans. J Clin Invest. 1983;71:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 260] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 76. | Lowe WL Jr, Adamo M, Werner H, Roberts CT Jr, LeRoith D. Regulation by fasting of rat insulin-like growth factor I and its receptor. Effects on gene expression and binding. J Clin Invest. 1989;84:619-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 135] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Boisclair YR, Rhoads RP, Ueki I, Wang J, Ooi GT. The acid-labile subunit (ALS) of the 150 kDa IGF-binding protein complex: an important but forgotten component of the circulating IGF system. J Endocrinol. 2001;170:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 182] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 78. | Mauras N, Haymond MW. Metabolic effects of recombinant human insulin-like growth factor-I in humans: comparison with recombinant human growth hormone. Pediatr Nephrol. 1996;10:318-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 79. | Møller N, Jørgensen JO. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr Rev. 2009;30:152-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 677] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 80. | Vijayakumar A, Yakar S, Leroith D. The intricate role of growth hormone in metabolism. Front Endocrinol (Lausanne). 2011;2:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 81. | Yakar S, Liu JL, Fernandez AM, Wu Y, Schally AV, Frystyk J, Chernausek SD, Mejia W, Le Roith D. Liver-specific igf-1 gene deletion leads to muscle insulin insensitivity. Diabetes. 2001;50:1110-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 259] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 82. | Yakar S, Setser J, Zhao H, Stannard B, Haluzik M, Glatt V, Bouxsein ML, Kopchick JJ, LeRoith D. Inhibition of growth hormone action improves insulin sensitivity in liver IGF-1-deficient mice. J Clin Invest. 2004;113:96-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 93] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 83. | Shen M, Shi H. Sex Hormones and Their Receptors Regulate Liver Energy Homeostasis. Int J Endocrinol. 2015;2015:294278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 159] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 84. | Corona G, Monami M, Rastrelli G, Aversa A, Sforza A, Lenzi A, Forti G, Mannucci E, Maggi M. Type 2 diabetes mellitus and testosterone: a meta-analysis study. Int J Androl. 2011;34:528-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 247] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 85. | Völzke H, Aumann N, Krebs A, Nauck M, Steveling A, Lerch MM, Rosskopf D, Wallaschofski H. Hepatic steatosis is associated with low serum testosterone and high serum DHEAS levels in men. Int J Androl. 2010;33:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 86. | Hermoso DAM, Bizerra PFV, Constantin RP, Ishii-Iwamoto EL, Gilglioni EH. Association between metabolic syndrome, hepatic steatosis, and testosterone deficiency: evidences from studies with men and rodents. Aging Male. 2020;23:1296-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 87. | Mukhopadhyay D, Plateroti M, Anant S, Nassir F, Samarut J, Davidson NO. Thyroid hormone regulates hepatic triglyceride mobilization and apolipoprotein B messenger ribonucleic Acid editing in a murine model of congenital hypothyroidism. Endocrinology. 2003;144:711-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 88. | Hatziagelaki E, Paschou SA, Schön M, Psaltopoulou T, Roden M. NAFLD and thyroid function: pathophysiological and therapeutic considerations. Trends Endocrinol Metab. 2022;33:755-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 48] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 89. | Araki O, Ying H, Furuya F, Zhu X, Cheng SY. Thyroid hormone receptor beta mutants: Dominant negative regulators of peroxisome proliferator-activated receptor gamma action. Proc Natl Acad Sci U S A. 2005;102:16251-16256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 90. | Erion MD, Cable EE, Ito BR, Jiang H, Fujitaki JM, Finn PD, Zhang BH, Hou J, Boyer SH, van Poelje PD, Linemeyer DL. Targeting thyroid hormone receptor-beta agonists to the liver reduces cholesterol and triglycerides and improves the therapeutic index. Proc Natl Acad Sci U S A. 2007;104:15490-15495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 179] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 91. | Mullur R, Liu YY, Brent GA. Thyroid hormone regulation of metabolism. Physiol Rev. 2014;94:355-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 965] [Cited by in RCA: 1395] [Article Influence: 126.8] [Reference Citation Analysis (0)] |
| 92. | Chi HC, Chen SL, Lin SL, Tsai CY, Chuang WY, Lin YH, Huang YH, Tsai MM, Yeh CT, Lin KH. Thyroid hormone protects hepatocytes from HBx-induced carcinogenesis by enhancing mitochondrial turnover. Oncogene. 2017;36:5274-5284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 93. | Singh BK, Sinha RA, Tripathi M, Mendoza A, Ohba K, Sy JAC, Xie SY, Zhou J, Ho JP, Chang CY, Wu Y, Giguère V, Bay BH, Vanacker JM, Ghosh S, Gauthier K, Hollenberg AN, McDonnell DP, Yen PM. Thyroid hormone receptor and ERRα coordinately regulate mitochondrial fission, mitophagy, biogenesis, and function. Sci Signal. 2018;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 94. | Chi HC, Chen SL, Tsai CY, Chuang WY, Huang YH, Tsai MM, Wu SM, Sun CP, Yeh CT, Lin KH. Thyroid hormone suppresses hepatocarcinogenesis via DAPK2 and SQSTM1-dependent selective autophagy. Autophagy. 2016;12:2271-2285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 95. | Zhou J, Singh BK, Ho JP, Lim A, Bruinstroop E, Ohba K, Sinha RA, Yen PM. MED1 mediator subunit is a key regulator of hepatic autophagy and lipid metabolism. Autophagy. 2021;17:4043-4061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 96. | Li L, Song Y, Shi Y, Sun L. Thyroid Hormone Receptor-β Agonists in NAFLD Therapy: Possibilities and Challenges. J Clin Endocrinol Metab. 2023;108:1602-1613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 21] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 97. | Harrison SA, Taub R, Neff GW, Lucas KJ, Labriola D, Moussa SE, Alkhouri N, Bashir MR. Resmetirom for nonalcoholic fatty liver disease: a randomized, double-blind, placebo-controlled phase 3 trial. Nat Med. 2023;29:2919-2928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 208] [Article Influence: 104.0] [Reference Citation Analysis (0)] |
| 98. | Wang Y, Nakajima T, Gonzalez FJ, Tanaka N. PPARs as Metabolic Regulators in the Liver: Lessons from Liver-Specific PPAR-Null Mice. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 335] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 99. | Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol. 2015;62:720-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 793] [Cited by in RCA: 1143] [Article Influence: 114.3] [Reference Citation Analysis (0)] |
| 100. | Contreras AV, Torres N, Tovar AR. PPAR-α as a key nutritional and environmental sensor for metabolic adaptation. Adv Nutr. 2013;4:439-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 179] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 101. | Han L, Shen WJ, Bittner S, Kraemer FB, Azhar S. PPARs: regulators of metabolism and as therapeutic targets in cardiovascular disease. Part I: PPAR-α. Future Cardiol. 2017;13:259-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 102. | Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, De Bosscher K. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocr Rev. 2018;39:760-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 556] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 103. | Todisco S, Santarsiero A, Convertini P, De Stefano G, Gilio M, Iacobazzi V, Infantino V. PPAR Alpha as a Metabolic Modulator of the Liver: Role in the Pathogenesis of Nonalcoholic Steatohepatitis (NASH). Biology (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 104. | Francque S, Verrijken A, Caron S, Prawitt J, Paumelle R, Derudas B, Lefebvre P, Taskinen MR, Van Hul W, Mertens I, Hubens G, Van Marck E, Michielsen P, Van Gaal L, Staels B. PPARα gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J Hepatol. 2015;63:164-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 105. | Kersten S, Stienstra R. The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie. 2017;136:75-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 288] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 106. | Park HS, Jang JE, Ko MS, Woo SH, Kim BJ, Kim HS, Park HS, Park IS, Koh EH, Lee KU. Statins Increase Mitochondrial and Peroxisomal Fatty Acid Oxidation in the Liver and Prevent Non-Alcoholic Steatohepatitis in Mice. Diabetes Metab J. 2016;40:376-385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 134] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 107. | Marinovic MP, Sousa-Filho CPB, Batista FAH, Avelino TM, Cogliati B, Figueira ACM, Otton R, Rodrigues AC. Green tea extract increases adiponectin and PPAR α levels to improve hepatic steatosis. J Nutr Biochem. 2022;103:108957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 108. | Tsuchida A, Yamauchi T, Takekawa S, Hada Y, Ito Y, Maki T, Kadowaki T. Peroxisome proliferator-activated receptor (PPAR)alpha activation increases adiponectin receptors and reduces obesity-related inflammation in adipose tissue: comparison of activation of PPARalpha, PPARgamma, and their combination. Diabetes. 2005;54:3358-3370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 324] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 109. | Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1019] [Cited by in RCA: 1093] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 110. | Moseti D, Regassa A, Kim WK. Molecular Regulation of Adipogenesis and Potential Anti-Adipogenic Bioactive Molecules. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 373] [Cited by in RCA: 532] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 111. | Yang A, Mottillo EP. Adipocyte lipolysis: from molecular mechanisms of regulation to disease and therapeutics. Biochem J. 2020;477:985-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 154] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 112. | Astapova O, Leff T. Adiponectin and PPARγ: cooperative and interdependent actions of two key regulators of metabolism. Vitam Horm. 2012;90:143-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 113. | Maeda N, Takahashi M, Funahashi T, Kihara S, Nishizawa H, Kishida K, Nagaretani H, Matsuda M, Komuro R, Ouchi N, Kuriyama H, Hotta K, Nakamura T, Shimomura I, Matsuzawa Y. PPARgamma ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes. 2001;50:2094-2099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1280] [Cited by in RCA: 1252] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 114. | Basantani MK, Sitnick MT, Cai L, Brenner DS, Gardner NP, Li JZ, Schoiswohl G, Yang K, Kumari M, Gross RW, Zechner R, Kershaw EE. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52:318-329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 196] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 115. | Li Z, Yuan H, Chu H, Yang L. The Crosstalk between Gut Microbiota and Bile Acids Promotes the Development of Non-Alcoholic Fatty Liver Disease. Microorganisms. 2023;11. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 116. | Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362-1365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2012] [Cited by in RCA: 2091] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 117. | Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1046] [Cited by in RCA: 1235] [Article Influence: 77.2] [Reference Citation Analysis (0)] |
| 118. | Hashimoto N, Han KH, Wakagi M, Ishikawa-Takano Y, Ippoushi K, Fukushima M. Bile acids induced hepatic lipid accumulation in mice by inhibiting mRNA expression of patatin-like phospholipase domain containing 3 and microsomal triglyceride transfer protein. Nutr Res. 2021;92:12-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 119. | Tschuck J, Theilacker L, Rothenaigner I, Weiß SAI, Akdogan B, Lam VT, Müller C, Graf R, Brandner S, Pütz C, Rieder T, Schmitt-Kopplin P, Vincendeau M, Zischka H, Schorpp K, Hadian K. Farnesoid X receptor activation by bile acids suppresses lipid peroxidation and ferroptosis. Nat Commun. 2023;14:6908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 42] [Reference Citation Analysis (0)] |
| 120. | Meier U, Gressner AM. Endocrine regulation of energy metabolism: review of pathobiochemical and clinical chemical aspects of leptin, ghrelin, adiponectin, and resistin. Clin Chem. 2004;50:1511-1525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 648] [Cited by in RCA: 623] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 121. | Richard AJ, White U, Elks CM, Stephens JM. Adipose Tissue: Physiology to Metabolic Dysfunction. Endotext. . |
| 122. | Pan J, Ding Y, Sun Y, Li Q, Wei T, Gu Y, Zhou Y, Pang N, Pei L, Ma S, Gao M, Xiao Y, Hu D, Wu F, Yang L. Associations between Adipokines and Metabolic Dysfunction-Associated Fatty Liver Disease Using Three Different Diagnostic Criteria. J Clin Med. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 123. | Silva TE, Colombo G, Schiavon LL. Adiponectin: A multitasking player in the field of liver diseases. Diabetes Metab. 2014;40:95-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 124. | Al-Jiffri O, Alsharif F. Levels of circulating adipokines and their relation with glycemic control and insulin resistance in Saudi patients with non-alcoholic fatty liver disease. Eur J Gen Med. 14:99. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 125. | Polyzos SA, Toulis KA, Goulis DG, Zavos C, Kountouras J. Serum total adiponectin in nonalcoholic fatty liver disease: a systematic review and meta-analysis. Metabolism. 2011;60:313-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 243] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 126. | Pagano C, Soardo G, Esposito W, Fallo F, Basan L, Donnini D, Federspil G, Sechi LA, Vettor R. Plasma adiponectin is decreased in nonalcoholic fatty liver disease. Eur J Endocrinol. 2005;152:113-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 127. | Targher G, Bertolini L, Rodella S, Zoppini G, Scala L, Zenari L, Falezza G. Associations between plasma adiponectin concentrations and liver histology in patients with nonalcoholic fatty liver disease. Clin Endocrinol (Oxf). 2006;64:679-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 147] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 128. | Hui JM, Hodge A, Farrell GC, Kench JG, Kriketos A, George J. Beyond insulin resistance in NASH: TNF-alpha or adiponectin? Hepatology. 2004;40:46-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 671] [Cited by in RCA: 692] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 129. | Musso G, Gambino R, Biroli G, Carello M, Fagà E, Pacini G, De Michieli F, Cassader M, Durazzo M, Rizzetto M, Pagano G. Hypoadiponectinemia predicts the severity of hepatic fibrosis and pancreatic Beta-cell dysfunction in nondiabetic nonobese patients with nonalcoholic steatohepatitis. Am J Gastroenterol. 2005;100:2438-2446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 148] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 130. | Savvidou S, Hytiroglou P, Orfanou-Koumerkeridou H, Panderis A, Frantzoulis P, Goulis J. Low serum adiponectin levels are predictive of advanced hepatic fibrosis in patients with NAFLD. J Clin Gastroenterol. 2009;43:765-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 131. | Jiménez-Cortegana C, García-Galey A, Tami M, Del Pino P, Carmona I, López S, Alba G, Sánchez-Margalet V. Role of Leptin in Non-Alcoholic Fatty Liver Disease. Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 132. | Martínez-Uña M, López-Mancheño Y, Diéguez C, Fernández-Rojo MA, Novelle MG. Unraveling the Role of Leptin in Liver Function and Its Relationship with Liver Diseases. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 133. | Hossain IA, Akter S, Rahman MK, Ali L. Gender Specific Association of Serum Leptin and Insulinemic Indices with Nonalcoholic Fatty Liver Disease in Prediabetic Subjects. PLoS One. 2015;10:e0142165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 134. | Cernea S, Roiban AL, Both E, Huţanu A. Serum leptin and leptin resistance correlations with NAFLD in patients with type 2 diabetes. Diabetes Metab Res Rev. 2018;34:e3050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 135. | Angulo P, Alba LM, Petrovic LM, Adams LA, Lindor KD, Jensen MD. Leptin, insulin resistance, and liver fibrosis in human nonalcoholic fatty liver disease. J Hepatol. 2004;41:943-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 141] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 136. | Bandaru P, Shankar A. Association between plasma leptin levels and diabetes mellitus. Metab Syndr Relat Disord. 2011;9:19-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 137. | Petrescu AD, Grant S, Williams E, An SY, Seth N, Shell M, Amundsen T, Tan C, Nadeem Y, Tjahja M, Weld L, Chu CS, Venter J, Frampton G, McMillin M, DeMorrow S. Leptin Enhances Hepatic Fibrosis and Inflammation in a Mouse Model of Cholestasis. Am J Pathol. 2022;192:484-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 138. | Saxena NK, Ikeda K, Rockey DC, Friedman SL, Anania FA. Leptin in hepatic fibrosis: evidence for increased collagen production in stellate cells and lean littermates of ob/ob mice. Hepatology. 2002;35:762-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 305] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 139. | Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655-1669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2139] [Cited by in RCA: 2163] [Article Influence: 127.2] [Reference Citation Analysis (0)] |
| 140. | London RM, George J. Pathogenesis of NASH: animal models. Clin Liver Dis. 2007;11:55-74, viii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 80] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 141. | Ikejima K, Okumura K, Kon K, Takei Y, Sato N. Role of adipocytokines in hepatic fibrogenesis. J Gastroenterol Hepatol. 2007;22 Suppl 1:S87-S92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 142. | Ikejima K, Takei Y, Honda H, Hirose M, Yoshikawa M, Zhang YJ, Lang T, Fukuda T, Yamashina S, Kitamura T, Sato N. Leptin receptor-mediated signaling regulates hepatic fibrogenesis and remodeling of extracellular matrix in the rat. Gastroenterology. 2002;122:1399-1410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 300] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 143. | Leclercq IA, Farrell GC, Schriemer R, Robertson GR. Leptin is essential for the hepatic fibrogenic response to chronic liver injury. J Hepatol. 2002;37:206-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 297] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 144. | Lin S, Saxena NK, Ding X, Stein LL, Anania FA. Leptin increases tissue inhibitor of metalloproteinase I (TIMP-1) gene expression by a specificity protein 1/signal transducer and activator of transcription 3 mechanism. Mol Endocrinol. 2006;20:3376-3388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 145. | Marra F, Aleffi S, Bertolani C, Petrai I, Vizzutti F. Adipokines and liver fibrosis. Eur Rev Med Pharmacol Sci. 2005;9:279-284. [PubMed] |
| 146. | Fartoux L, Poujol-Robert A, Guéchot J, Wendum D, Poupon R, Serfaty L. Insulin resistance is a cause of steatosis and fibrosis progression in chronic hepatitis C. Gut. 2005;54:1003-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 278] [Article Influence: 13.9] [Reference Citation Analysis (1)] |
| 147. | Moucari R, Asselah T, Cazals-Hatem D, Voitot H, Boyer N, Ripault MP, Sobesky R, Martinot-Peignoux M, Maylin S, Nicolas-Chanoine MH, Paradis V, Vidaud M, Valla D, Bedossa P, Marcellin P. Insulin resistance in chronic hepatitis C: association with genotypes 1 and 4, serum HCV RNA level, and liver fibrosis. Gastroenterology. 2008;134:416-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 377] [Article Influence: 22.2] [Reference Citation Analysis (1)] |
| 148. | Guan X, Fierke CA. Understanding Protein Palmitoylation: Biological Significance and Enzymology. Sci China Chem. 2011;54:1888-1897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 149. | Sheedfar F, Sung MM, Aparicio-Vergara M, Kloosterhuis NJ, Miquilena-Colina ME, Vargas-Castrillón J, Febbraio M, Jacobs RL, de Bruin A, Vinciguerra M, García-Monzón C, Hofker MH, Dyck JR, Koonen DP. Increased hepatic CD36 expression with age is associated with enhanced susceptibility to nonalcoholic fatty liver disease. Aging (Albany NY). 2014;6:281-295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 150. | Wilson CG, Tran JL, Erion DM, Vera NB, Febbraio M, Weiss EJ. Hepatocyte-Specific Disruption of CD36 Attenuates Fatty Liver and Improves Insulin Sensitivity in HFD-Fed Mice. Endocrinology. 2016;157:570-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 344] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 151. | Zhou J, Febbraio M, Wada T, Zhai Y, Kuruba R, He J, Lee JH, Khadem S, Ren S, Li S, Silverstein RL, Xie W. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology. 2008;134:556-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 478] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 152. | Maréchal L, Laviolette M, Rodrigue-Way A, Sow B, Brochu M, Caron V, Tremblay A. The CD36-PPARγ Pathway in Metabolic Disorders. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 153. | Zeng S, Wu F, Chen M, Li Y, You M, Zhang Y, Yang P, Wei L, Ruan XZ, Zhao L, Chen Y. Inhibition of Fatty Acid Translocase (FAT/CD36) Palmitoylation Enhances Hepatic Fatty Acid β-Oxidation by Increasing Its Localization to Mitochondria and Interaction with Long-Chain Acyl-CoA Synthetase 1. Antioxid Redox Signal. 2022;36:1081-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 70] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 154. | Liu J, Yang P, Zuo G, He S, Tan W, Zhang X, Su C, Zhao L, Wei L, Chen Y, Ruan X, Chen Y. Long-chain fatty acid activates hepatocytes through CD36 mediated oxidative stress. Lipids Health Dis. 2018;17:153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 155. | Iredale JP, Benyon RC, Pickering J, McCullen M, Northrop M, Pawley S, Hovell C, Arthur MJ. Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J Clin Invest. 1998;102:538-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 785] [Cited by in RCA: 828] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 156. | Issa R, Zhou X, Trim N, Millward-Sadler H, Krane S, Benyon C, Iredale J. Mutation in collagen-1 that confers resistance to the action of collagenase results in failure of recovery from CCl4-induced liver fibrosis, persistence of activated hepatic stellate cells, and diminished hepatocyte regeneration. FASEB J. 2003;17:47-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 151] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 157. | Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best Pract Res Clin Gastroenterol. 2011;25:195-206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 735] [Cited by in RCA: 737] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 158. | Kim H, Son S, Shin I. Role of the CCN protein family in cancer. BMB Rep. 2018;51:486-492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 159. | Jia Q, Xu B, Zhang Y, Ali A, Liao X. CCN Family Proteins in Cancer: Insight Into Their Structures and Coordination Role in Tumor Microenvironment. Front Genet. 2021;12:649387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 160. | Gressner OA, Gressner AM. Connective tissue growth factor: a fibrogenic master switch in fibrotic liver diseases. Liver Int. 2008;28:1065-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 213] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 161. | Bissell DM, Roulot D, George J. Transforming growth factor beta and the liver. Hepatology. 2001;34:859-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 284] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 162. | Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci. 2002;7:d793-d807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 483] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 163. | Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 741] [Article Influence: 74.1] [Reference Citation Analysis (0)] |
| 164. | Tomita K, Teratani T, Suzuki T, Shimizu M, Sato H, Narimatsu K, Okada Y, Kurihara C, Irie R, Yokoyama H, Shimamura K, Usui S, Ebinuma H, Saito H, Watanabe C, Komoto S, Kawaguchi A, Nagao S, Sugiyama K, Hokari R, Kanai T, Miura S, Hibi T. Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology. 2014;59:154-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 247] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 165. | Bechmann LP, Zahn D, Gieseler RK, Fingas CD, Marquitan G, Jochum C, Gerken G, Friedman SL, Canbay A. Resveratrol amplifies profibrogenic effects of free fatty acids on human hepatic stellate cells. Hepatol Res. 2009;39:601-608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 166. | Kisseleva T, Brenner DA. Mechanisms of fibrogenesis. Exp Biol Med (Maywood). 2008;233:109-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 393] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 167. | Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1556] [Cited by in RCA: 1569] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 168. | Zavadil J, Böttinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764-5774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1207] [Cited by in RCA: 1310] [Article Influence: 65.5] [Reference Citation Analysis (0)] |
| 169. | Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1051] [Cited by in RCA: 1072] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 170. | Kinoshita K, Iimuro Y, Otogawa K, Saika S, Inagaki Y, Nakajima Y, Kawada N, Fujimoto J, Friedman SL, Ikeda K. Adenovirus-mediated expression of BMP-7 suppresses the development of liver fibrosis in rats. Gut. 2007;56:706-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 126] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 171. | Sugimoto H, Yang C, LeBleu VS, Soubasakos MA, Giraldo M, Zeisberg M, Kalluri R. BMP-7 functions as a novel hormone to facilitate liver regeneration. FASEB J. 2007;21:256-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 91] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 172. | Sahin H, Trautwein C, Wasmuth HE. Functional role of chemokines in liver disease models. Nat Rev Gastroenterol Hepatol. 2010;7:682-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 173. | Schwabe RF, Bataller R, Brenner DA. Human hepatic stellate cells express CCR5 and RANTES to induce proliferation and migration. Am J Physiol Gastrointest Liver Physiol. 2003;285:G949-G958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 200] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 174. | Zisser A, Ipsen DH, Tveden-Nyborg P. Hepatic Stellate Cell Activation and Inactivation in NASH-Fibrosis-Roles as Putative Treatment Targets? Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 68] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 175. | Akpolat N, Yahsi S, Godekmerdan A, Yalniz M, Demirbag K. The value of alpha-SMA in the evaluation of hepatic fibrosis severity in hepatitis B infection and cirrhosis development: a histopathological and immunohistochemical study. Histopathology. 2005;47:276-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 176. | Yamaoka K, Nouchi T, Marumo F, Sato C. Alpha-smooth-muscle actin expression in normal and fibrotic human livers. Dig Dis Sci. 1993;38:1473-1479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 177. | Tian H, Zhang S, Liu Y, Wu Y, Zhang D. Fibroblast Growth Factors for Nonalcoholic Fatty Liver Disease: Opportunities and Challenges. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 178. | Yun YR, Won JE, Jeon E, Lee S, Kang W, Jo H, Jang JH, Shin US, Kim HW. Fibroblast growth factors: biology, function, and application for tissue regeneration. J Tissue Eng. 2010;2010:218142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 329] [Cited by in RCA: 433] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 179. | Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1242] [Cited by in RCA: 1465] [Article Influence: 91.6] [Reference Citation Analysis (0)] |
| 180. | Fisher FM, Maratos-Flier E. Understanding the Physiology of FGF21. Annu Rev Physiol. 2016;78:223-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 656] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 181. | Tillman EJ, Rolph T. FGF21: An Emerging Therapeutic Target for Non-Alcoholic Steatohepatitis and Related Metabolic Diseases. Front Endocrinol (Lausanne). 2020;11:601290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 147] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 182. | Sciarrillo CM, Keirns BH, Koemel NA, Anderson KL, Emerson SR. Fibroblast Growth Factor 19: Potential modulation of hepatic metabolism for the treatment of non-alcoholic fatty liver disease. Liver Int. 2021;41:894-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 183. | Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, Mastrandrea L, Buck MJ, Baker RD, Genco RJ, Zhu R, Zhu L. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018;67:1881-1891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 514] [Article Influence: 73.4] [Reference Citation Analysis (1)] |
| 184. | Fu L, John LM, Adams SH, Yu XX, Tomlinson E, Renz M, Williams PM, Soriano R, Corpuz R, Moffat B, Vandlen R, Simmons L, Foster J, Stephan JP, Tsai SP, Stewart TA. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology. 2004;145:2594-2603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 457] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 185. | Eren F, Kurt R, Ermis F, Atug O, Imeryuz N, Yilmaz Y. Preliminary evidence of a reduced serum level of fibroblast growth factor 19 in patients with biopsy-proven nonalcoholic fatty liver disease. Clin Biochem. 2012;45:655-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 186. | Stejskal D, Karpísek M, Hanulová Z, Stejskal P. Fibroblast growth factor-19: development, analytical characterization and clinical evaluation of a new ELISA test. Scand J Clin Lab Invest. 2008;68:501-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 187. | Schreuder TC, Marsman HA, Lenicek M, van Werven JR, Nederveen AJ, Jansen PL, Schaap FG. The hepatic response to FGF19 is impaired in patients with nonalcoholic fatty liver disease and insulin resistance. Am J Physiol Gastrointest Liver Physiol. 2010;298:G440-G445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 188. | Alisi A, Ceccarelli S, Panera N, Prono F, Petrini S, De Stefanis C, Pezzullo M, Tozzi A, Villani A, Bedogni G, Nobili V. Association between Serum Atypical Fibroblast Growth Factors 21 and 19 and Pediatric Nonalcoholic Fatty Liver Disease. PLoS One. 2013;8:e67160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 189. | Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, Zhao A, Busuttil RW, Yee H, Stein L, French DM, Finn RS, Lowe SW, Powers S. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19:347-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 357] [Cited by in RCA: 361] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 190. | Zhou M, Yang H, Learned RM, Tian H, Ling L. Non-cell-autonomous activation of IL-6/STAT3 signaling mediates FGF19-driven hepatocarcinogenesis. Nat Commun. 2017;8:15433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 191. | Zhou M, Luo J, Chen M, Yang H, Learned RM, DePaoli AM, Tian H, Ling L. Mouse species-specific control of hepatocarcinogenesis and metabolism by FGF19/FGF15. J Hepatol. 2017;66:1182-1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 192. | Wu X, Ge H, Lemon B, Vonderfecht S, Baribault H, Weiszmann J, Gupte J, Gardner J, Lindberg R, Wang Z, Li Y. Separating mitogenic and metabolic activities of fibroblast growth factor 19 (FGF19). Proc Natl Acad Sci U S A. 2010;107:14158-14163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 193. | Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA, Gromada J, Brozinick JT, Hawkins ED, Wroblewski VJ, Li DS, Mehrbod F, Jaskunas SR, Shanafelt AB. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115:1627-1635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1485] [Cited by in RCA: 1721] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 194. | Hondares E, Iglesias R, Giralt A, Gonzalez FJ, Giralt M, Mampel T, Villarroya F. Thermogenic activation induces FGF21 expression and release in brown adipose tissue. J Biol Chem. 2011;286:12983-12990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 499] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 195. | BonDurant LD, Ameka M, Naber MC, Markan KR, Idiga SO, Acevedo MR, Walsh SA, Ornitz DM, Potthoff MJ. FGF21 Regulates Metabolism Through Adipose-Dependent and -Independent Mechanisms. Cell Metab. 2017;25:935-944.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 252] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 196. | Ding X, Boney-Montoya J, Owen BM, Bookout AL, Coate KC, Mangelsdorf DJ, Kliewer SA. βKlotho is required for fibroblast growth factor 21 effects on growth and metabolism. Cell Metab. 2012;16:387-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 337] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 197. | Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, Goetz R, Mohammadi M, Finck BN, Mangelsdorf DJ, Kliewer SA, Burgess SC. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci U S A. 2009;106:10853-10858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 594] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 198. | Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1246] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 199. | Geng L, Lam KSL, Xu A. The therapeutic potential of FGF21 in metabolic diseases: from bench to clinic. Nat Rev Endocrinol. 2020;16:654-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 389] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 200. | Huang G, Wallace DF, Powell EE, Rahman T, Clark PJ, Subramaniam VN. Gene Variants Implicated in Steatotic Liver Disease: Opportunities for Diagnostics and Therapeutics. Biomedicines. 2023;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 201. | Choudhary NS, Duseja A. Genetic and epigenetic disease modifiers: non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD). Transl Gastroenterol Hepatol. 2021;6:2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 202. | Brouwers MC, Cantor RM, Kono N, Yoon JL, van der Kallen CJ, Bilderbeek-Beckers MA, van Greevenbroek MM, Lusis AJ, de Bruin TW. Heritability and genetic loci of fatty liver in familial combined hyperlipidemia. J Lipid Res. 2006;47:2799-2807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 203. | Struben VM, Hespenheide EE, Caldwell SH. Nonalcoholic steatohepatitis and cryptogenic cirrhosis within kindreds. Am J Med. 2000;108:9-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 195] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 204. | Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, Shiehmorteza M, Yokoo T, Chavez A, Middleton MS, Sirlin CB. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585-1592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 358] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 205. | Anstee QM, Seth D, Day CP. Genetic Factors That Affect Risk of Alcoholic and Nonalcoholic Fatty Liver Disease. Gastroenterology. 2016;150:1728-1744.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 194] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 206. | Willner IR, Waters B, Patil SR, Reuben A, Morelli J, Riely CA. Ninety patients with nonalcoholic steatohepatitis: insulin resistance, familial tendency, and severity of disease. Am J Gastroenterol. 2001;96:2957-2961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 281] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 207. | Niltwat S, Limwongse C, Charatcharoenwitthaya N, Bunditvorapoom D, Bandidniyamanon W, Charatcharoenwitthaya P. Familial clustering of nonalcoholic fatty liver disease in first-degree relatives of adults with lean nonalcoholic fatty liver disease. Liver Int. 2023;43:2713-2726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 208. | Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J Hepatol. 2018;68:268-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 732] [Cited by in RCA: 691] [Article Influence: 98.7] [Reference Citation Analysis (0)] |
| 209. | Makkonen J, Pietiläinen KH, Rissanen A, Kaprio J, Yki-Järvinen H. Genetic factors contribute to variation in serum alanine aminotransferase activity independent of obesity and alcohol: a study in monozygotic and dizygotic twins. J Hepatol. 2009;50:1035-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 210. | Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, Nguyen P, Hernandez C, Richards L, Salotti J, Lin S, Seki E, Nelson KE, Sirlin CB, Brenner D; Genetics of NAFLD in Twins Consortium. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology. 2015;149:1784-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 302] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 211. | Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2633] [Cited by in RCA: 2695] [Article Influence: 128.3] [Reference Citation Analysis (3)] |
| 212. | Browning JD, Kumar KS, Saboorian MH, Thiele DL. Ethnic differences in the prevalence of cryptogenic cirrhosis. Am J Gastroenterol. 2004;99:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 158] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 213. | Guerrero R, Vega GL, Grundy SM, Browning JD. Ethnic differences in hepatic steatosis: an insulin resistance paradox? Hepatology. 2009;49:791-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 283] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 214. | Bambha K, Belt P, Abraham M, Wilson LA, Pabst M, Ferrell L, Unalp-Arida A, Bass N; Nonalcoholic Steatohepatitis Clinical Research Network Research Group. Ethnicity and nonalcoholic fatty liver disease. Hepatology. 2012;55:769-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 171] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 215. | Szczepaniak LS, Nurenberg P, Leonard D, Browning JD, Reingold JS, Grundy S, Hobbs HH, Dobbins RL. Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab. 2005;288:E462-E468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1119] [Cited by in RCA: 1184] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 216. | Macaluso FS, Maida M, Petta S. Genetic background in nonalcoholic fatty liver disease: A comprehensive review. World J Gastroenterol. 2015;21:11088-11111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 68] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 217. | Wood KL, Miller MH, Dillon JF. Systematic review of genetic association studies involving histologically confirmed non-alcoholic fatty liver disease. BMJ Open Gastroenterol. 2015;2:e000019. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 218. | Trépo E, Valenti L. Update on NAFLD genetics: From new variants to the clinic. J Hepatol. 2020;72:1196-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 274] [Article Influence: 54.8] [Reference Citation Analysis (1)] |
| 219. | Hirschhorn JN, Gajdos ZK. Genome-wide association studies: results from the first few years and potential implications for clinical medicine. Annu Rev Med. 2011;62:11-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 220. | Rossi E, Jeffrey GP. Clinical penetrance of C282Y homozygous HFE haemochromatosis. Clin Biochem Rev. 2004;25:183-190. [PubMed] |
| 221. | Patel D, McAllister SL, Teckman JH. Alpha-1-antitrypsin deficiency: natural course and therapeutic strategies. Transl Gastroenterol Hepatol. 2021;6: 23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 222. | Wang J, Conti DV, Bogumil D, Sheng X, Noureddin M, Wilkens LR, Le Marchand L, Rosen HR, Haiman CA, Setiawan VW. Association of Genetic Risk Score With NAFLD in An Ethnically Diverse Cohort. Hepatol Commun. 2021;5:1689-1703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 223. | Arnold N, Koenig W. Polygenic Risk Score: Clinically Useful Tool for Prediction of Cardiovascular Disease and Benefit from Lipid-Lowering Therapy? Cardiovasc Drugs Ther. 2021;35:627-635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 224. | Kim DY, Park JY. Genetic risk factors associated with NAFLD. Hepatoma Res. 2020;6:85. |
| 225. | Carlsson B, Lindén D, Brolén G, Liljeblad M, Bjursell M, Romeo S, Loomba R. Review article: the emerging role of genetics in precision medicine for patients with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2020;51:1305-1320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 226. | Caimari A, Oliver P, Palou A. Regulation of adiponutrin expression by feeding conditions in rats is altered in the obese state. Obesity (Silver Spring). 2007;15:591-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 227. | Cherubini A, Casirati E, Tomasi M, Valenti L. PNPLA3 as a therapeutic target for fatty liver disease: the evidence to date. Expert Opin Ther Targets. 2021;25:1033-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 228. | Bruschi FV, Tardelli M, Claudel T, Trauner M. PNPLA3 expression and its impact on the liver: current perspectives. Hepat Med. 2017;9:55-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 229. | Winberg ME, Motlagh MK, Stenkula KG, Holm C, Jones HA. Adiponutrin: a multimeric plasma protein. Biochem Biophys Res Commun. 2014;446:1114-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 230. | Unalp-Arida A, Ruhl CE. Patatin-Like Phospholipase Domain-Containing Protein 3 I148M and Liver Fat and Fibrosis Scores Predict Liver Disease Mortality in the U.S. Population. Hepatology. 2020;71:820-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 231. | Kershaw EE, Hamm JK, Verhagen LA, Peroni O, Katic M, Flier JS. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55:148-157. [PubMed] |
| 232. | Yang J, Trépo E, Nahon P, Cao Q, Moreno C, Letouzé E, Imbeaud S, Gustot T, Deviere J, Debette S, Amouyel P, Bioulac-Sage P, Calderaro J, Ganne-Carrié N, Laurent A, Blanc JF, Guyot E, Sutton A, Ziol M, Zucman-Rossi J, Nault JC. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int J Cancer. 2019;144:533-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 233. | GeneCards®. GeneCards®: The Human Gene Database. Available from: https://www.genecards.org/. |
| 234. | Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 724] [Cited by in RCA: 930] [Article Influence: 84.5] [Reference Citation Analysis (0)] |
| 235. | Holmen OL, Zhang H, Fan Y, Hovelson DH, Schmidt EM, Zhou W, Guo Y, Zhang J, Langhammer A, Løchen ML, Ganesh SK, Vatten L, Skorpen F, Dalen H, Zhang J, Pennathur S, Chen J, Platou C, Mathiesen EB, Wilsgaard T, Njølstad I, Boehnke M, Chen YE, Abecasis GR, Hveem K, Willer CJ. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46:345-351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 243] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 236. | Mahdessian H, Taxiarchis A, Popov S, Silveira A, Franco-Cereceda A, Hamsten A, Eriksson P, van't Hooft F. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111:8913-8918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 286] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 237. | Liu YL, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JB, Allison ME, Alexander GJ, Piguet AC, Anty R, Donaldson P, Aithal GP, Francque S, Van Gaal L, Clement K, Ratziu V, Dufour JF, Day CP, Daly AK, Anstee QM. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 362] [Cited by in RCA: 460] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 238. | Carim-Todd L, Escarceller M, Estivill X, Sumoy L. Cloning of the novel gene TM6SF1 reveals conservation of clusters of paralogous genes between human chromosomes 15q24-->q26 and 19p13.3-->p12. Cytogenet Cell Genet. 2000;90:255-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 239. | Liu J, Ginsberg HN, Reyes-Soffer G. Basic and translational evidence supporting the role of TM6SF2 in VLDL metabolism. Curr Opin Lipidol. 2024;35:157-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 240. | Surakka I, Horikoshi M, Mägi R, Sarin AP, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, Kettunen J, Pirinen M, Karjalainen J, Thorleifsson G, Hägg S, Hottenga JJ, Isaacs A, Ladenvall C, Beekman M, Esko T, Ried JS, Nelson CP, Willenborg C, Gustafsson S, Westra HJ, Blades M, de Craen AJ, de Geus EJ, Deelen J, Grallert H, Hamsten A, Havulinna AS, Hengstenberg C, Houwing-Duistermaat JJ, Hyppönen E, Karssen LC, Lehtimäki T, Lyssenko V, Magnusson PK, Mihailov E, Müller-Nurasyid M, Mpindi JP, Pedersen NL, Penninx BW, Perola M, Pers TH, Peters A, Rung J, Smit JH, Steinthorsdottir V, Tobin MD, Tsernikova N, van Leeuwen EM, Viikari JS, Willems SM, Willemsen G, Schunkert H, Erdmann J, Samani NJ, Kaprio J, Lind L, Gieger C, Metspalu A, Slagboom PE, Groop L, van Duijn CM, Eriksson JG, Jula A, Salomaa V, Boomsma DI, Power C, Raitakari OT, Ingelsson E, Järvelin MR, Thorsteinsdottir U, Franke L, Ikonen E, Kallioniemi O, Pietiäinen V, Lindgren CM, Stefansson K, Palotie A, McCarthy MI, Morris AP, Prokopenko I, Ripatti S; ENGAGE Consortium. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 259] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 241. | Kawaguchi T, Sumida Y, Umemura A, Matsuo K, Takahashi M, Takamura T, Yasui K, Saibara T, Hashimoto E, Kawanaka M, Watanabe S, Kawata S, Imai Y, Kokubo M, Shima T, Park H, Tanaka H, Tajima K, Yamada R, Matsuda F, Okanoue T; Japan Study Group of Nonalcoholic Fatty Liver Disease. Genetic polymorphisms of the human PNPLA3 gene are strongly associated with severity of non-alcoholic fatty liver disease in Japanese. PLoS One. 2012;7:e38322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 212] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 242. | Kahali B, Liu YL, Daly AK, Day CP, Anstee QM, Speliotes EK. TM6SF2: catch-22 in the fight against nonalcoholic fatty liver disease and cardiovascular disease? Gastroenterology. 2015;148:679-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 243. | Helsley RN, Varadharajan V, Brown AL, Gromovsky AD, Schugar RC, Ramachandiran I, Fung K, Kabbany MN, Banerjee R, Neumann CK, Finney C, Pathak P, Orabi D, Osborn LJ, Massey W, Zhang R, Kadam A, Sansbury BE, Pan C, Sacks J, Lee RG, Crooke RM, Graham MJ, Lemieux ME, Gogonea V, Kirwan JP, Allende DS, Civelek M, Fox PL, Rudel LL, Lusis AJ, Spite M, Brown JM. Obesity-linked suppression of membrane-bound O-acyltransferase 7 (MBOAT7) drives non-alcoholic fatty liver disease. Elife. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 244. | Varadharajan V, Massey WJ, Brown JM. Membrane-bound O-acyltransferase 7 (MBOAT7)-driven phosphatidylinositol remodeling in advanced liver disease. J Lipid Res. 2022;63:100234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 245. | Massey WJ, Varadharajan V, Banerjee R, Brown AL, Horak AJ, Hohe RC, Jung BM, Qiu Y, Chan ER, Pan C, Zhang R, Allende DS, Willard B, Cheng F, Lusis AJ, Brown JM. MBOAT7-driven lysophosphatidylinositol acylation in adipocytes contributes to systemic glucose homeostasis. J Lipid Res. 2023;64:100349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 246. | Sharpe MC, Pyles KD, Hallcox T, Kamm DR, Piechowski M, Fisk B, Albert CJ, Carpenter DH, Ulmasov B, Ford DA, Neuschwander-Tetri BA, McCommis KS. Enhancing Hepatic MBOAT7 Expression in Mice With Nonalcoholic Steatohepatitis. Gastro Hep Adv. 2023;2:558-572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 247. | Meroni M, Longo M, Fracanzani AL, Dongiovanni P. MBOAT7 down-regulation by genetic and environmental factors predisposes to MAFLD. EBioMedicine. 2020;57:102866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 248. | Xia Y, Huang CX, Li GY, Chen KH, Han L, Tang L, Luo HQ, Bao MH. Meta-analysis of the association between MBOAT7 rs641738, TM6SF2 rs58542926 and nonalcoholic fatty liver disease susceptibility. Clin Res Hepatol Gastroenterol. 2019;43:533-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 249. | Sookoian S, Flichman D, Garaycoechea ME, Gazzi C, Martino JS, Castaño GO, Pirola CJ. Lack of evidence supporting a role of TMC4-rs641738 missense variant-MBOAT7- intergenic downstream variant-in the Susceptibility to Nonalcoholic Fatty Liver Disease. Sci Rep. 2018;8:5097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 250. | Basyte-Bacevice V, Skieceviciene J, Valantiene I, Sumskiene J, Petrenkiene V, Kondrackiene J, Petrauskas D, Lammert F, Kupcinskas J. TM6SF2 and MBOAT7 Gene Variants in Liver Fibrosis and Cirrhosis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 251. | Koo BK, Joo SK, Kim D, Bae JM, Park JH, Kim JH, Kim W. Additive effects of PNPLA3 and TM6SF2 on the histological severity of non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2018;33:1277-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 252. | Anstee QM, Darlay R, Cockell S, Meroni M, Govaere O, Tiniakos D, Burt AD, Bedossa P, Palmer J, Liu YL, Aithal GP, Allison M, Yki-Järvinen H, Vacca M, Dufour JF, Invernizzi P, Prati D, Ekstedt M, Kechagias S, Francque S, Petta S, Bugianesi E, Clement K, Ratziu V, Schattenberg JM, Valenti L, Day CP, Cordell HJ, Daly AK; EPoS Consortium Investigators. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort(☆). J Hepatol. 2020;73:505-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 341] [Article Influence: 68.2] [Reference Citation Analysis (0)] |
| 253. | Teo K, Abeysekera KWM, Adams L, Aigner E, Anstee QM, Banales JM, Banerjee R, Basu P, Berg T, Bhatnagar P, Buch S, Canbay A, Caprio S, Chatterjee A, Ida Chen YD, Chowdhury A, Daly AK, Datz C, de Gracia Hahn D, DiStefano JK, Dong J, Duret A; EU-PNAFLD Investigators, Emdin C, Fairey M, Gerhard GS; GOLD Consortium, Guo X, Hampe J, Hickman M, Heintz L, Hudert C, Hunter H, Kelly M, Kozlitina J, Krawczyk M, Lammert F, Langenberg C, Lavine J, Li L, Lim HK, Loomba R, Luukkonen PK, Melton PE, Mori TA, Palmer ND, Parisinos CA, Pillai SG, Qayyum F, Reichert MC, Romeo S, Rotter JI, Im YR, Santoro N, Schafmayer C, Speliotes EK, Stender S, Stickel F, Still CD, Strnad P, Taylor KD, Tybjærg-Hansen A, Umano GR, Utukuri M, Valenti L, Wagenknecht LE, Wareham NJ, Watanabe RM, Wattacheril J, Yaghootkar H, Yki-Järvinen H, Young KA, Mann JP. rs641738C>T near MBOAT7 is associated with liver fat, ALT and fibrosis in NAFLD: A meta-analysis. J Hepatol. 2021;74:20-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 95] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 254. | Habib S. Metabolic dysfunction-associated steatotic liver disease heterogeneity: Need of subtyping. World J Gastrointest Pathophysiol. 2024;15:92791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 255. | Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368:1696-1705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2787] [Cited by in RCA: 2842] [Article Influence: 149.6] [Reference Citation Analysis (1)] |
| 256. | Hernandez-Quiles M, Broekema MF, Kalkhoven E. PPARgamma in Metabolism, Immunity, and Cancer: Unified and Diverse Mechanisms of Action. Front Endocrinol (Lausanne). 2021;12:624112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 263] [Article Influence: 65.8] [Reference Citation Analysis (0)] |