Published online Jun 1, 2023. doi: 10.4291/wjgp.v14.i3.46
Peer-review started: March 27, 2023
First decision: April 8, 2023
Revised: May 19, 2023
Accepted: May 31, 2023
Article in press: May 31, 2023
Published online: June 1, 2023
Processing time: 65 Days and 23.9 Hours
In the world, hepatocellular carcinoma (HCC) is among the top 10 most prevalent malignancies. HCC formation has indeed been linked to numerous etiological factors, including alcohol usage, hepatitis viruses and liver cirrhosis. Among the most prevalent defects in a wide range of tumours, notably HCC, is the silencing of the p53 tumour suppressor gene. The control of the cell cycle and the preservation of gene function are both critically important functions of p53. In order to pinpoint the core mechanisms of HCC and find more efficient treatments, molecular research employing HCC tissues has been the main focus. Stimulated p53 triggers necessary reactions that achieve cell cycle arrest, genetic stability, DNA repair and the elimination of DNA-damaged cells’ responses to biological stressors (like oncogenes or DNA damage). To the contrary hand, the oncogene protein of the murine double minute 2 (MDM2) is a significant biological inhibitor of p53. MDM2 causes p53 protein degradation, which in turn adversely controls p53 function. Despite carrying wt-p53, the majority of HCCs show abnormalities in the p53-expressed apoptotic pathway. High p53 in-vivo expression might have two clinical impacts on HCC: (1) Increased levels of exogenous p53 protein cause tumour cells to undergo apoptosis by preventing cell growth through a number of biological pathways; and (2) Exogenous p53 makes HCC susceptible to various anticancer drugs. This review describes the functions and primary mechanisms of p53 in pathological mechanism, chemoresistance and therapeutic mechanisms of HCC.
Core Tip: This review articles covers all the points related to p53 gene suppression in the pathogenesis of hepatocellular carcinoma. The review manuscript also includes information about tumour suppressor P53, pathogenesis of P53-associated hepatocellular carcinoma (HCC), role of P53 in mechanisms of hepatocarcinogenesis, roles of P53 in extrinsic factor-induced liver carcinogenesis, relationship between hepatitis C virus core protein and P53 in HCC, other mechanisms of progression to cirrhosis and HCC.
- Citation: Choudhary HB, Mandlik SK, Mandlik DS. Role of p53 suppression in the pathogenesis of hepatocellular carcinoma. World J Gastrointest Pathophysiol 2023; 14(3): 46-70
- URL: https://www.wjgnet.com/2150-5330/full/v14/i3/46.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v14.i3.46
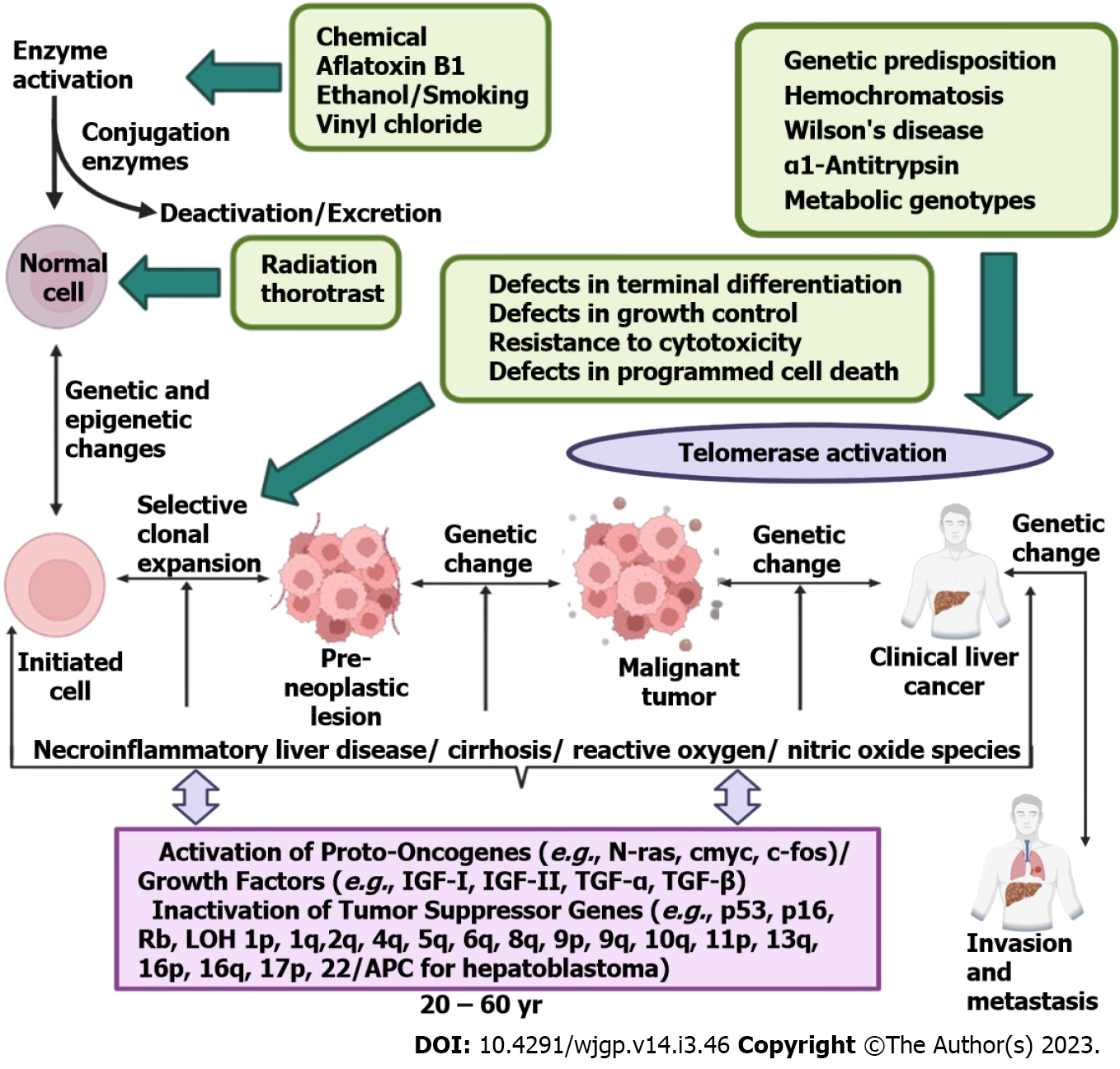
Hepatocellular carcinoma (HCC) is the most prevalent form of primary liver cancer and typically brought on by chronic liver diseases. HCC every year has 841000 new cases and 781000 deaths which makes HCC the fourth leading cause of cancer-related death globally[1]. By 2025 the annual diagnosed cases of HCC are expected to outstretch up to 1 million. HCC can appear on its own or more usually results from chronic liver illnesses of various etiological factors, like cirrhosis, alcoholic steatohepatitis (ASH) and non-ASH (NASH), viral infections [hepatitis C virus (HCV) and hepatitis B virus (HBV)] and hepatitis B and C[2] (Figure 1). As transplantation, ablation and resection can only be used for the treatment of early-stage HCC patients and since the majority of patients are diagnosed with severe stages or intrinsic chronic hepatic disease, discovering novel therapies to increase survival time and lower the likelihood of tumour recurrence is an important objective in the domain of HCC. According to the Barcelona clinic liver cancer staging system, Sorafenib has been utilized for several years as a treatment for HCC that has progressed to an advanced stage[2]. Nevertheless, concerns regarding its effectiveness and harmful effects have been raised[3], including the activation of metabolizing enzymes[4]. While the use of immunotherapy has altered the approach to treating advanced HCC, not all patients with HCC respond positively to these recent therapies. Consequently, there are ongoing investigations into various strategies to enhance the effectiveness of HCC medication by incorporating chemosensitizers[5] and/or personalized combination treatments[6]. Additionally, patients who receive immunotherapy may experience an upsurge in immune-related adverse events[7]. In this situation, tailoring HCC therapy to each patient is becoming an essential priority in order to achieve the greatest possible clinical advantage. Along with this, there is a need to find new biomarkers that can identify potential responders to pharmacological treatment by accurately predicting the clinical response[2].
The wild-type protein of the tumour suppressor gene p53 is responsible for regulating the cell cycle and inducing apoptosis after DNA damage[8]. However, if p53 undergoes a mutation, cells with DNA damage can avoid apoptosis and develop into cancerous cells[9]. Additionally, the mutant p53 protein, which loses the function of the wild-type protein, can amass in the nuclei of cells and is considered to be a highly specific marker for malignancy[10]. Studies have indicated that alterations in p53 are linked to tumour differentiation, vascular invasion, tumour stage, Child-Pugh class and serum alpha fetoprotein in HCC[11]. Nevertheless, the prognostic significance of p53 alterations in HCC is not yet fully understood and conflicting results have been reported in various studies[8]. While some studies have suggested that tumour p53 upregulation and serum anti-p53 antibody elevation are linked to recurrence-free survival and overall survival in HCC patients, other studies have not confirmed these findings[12]. Furthermore, most studies have indicated that HCC patients with a mutant p53 phenotype experience poor survival, but the prognostic effect has varied widely with a broad range of hazard ratios due to small and diverse studies[13].
Despite the progress made in treating certain other types of cancer, there isn’t any effective treatment available for HCC and presently, chemotherapy and surgical resection are the two most common treatment options[14]. As a result, learning more about the molecular basis for the etiology of HCC may help researchers to find novel therapeutic approaches[14]. This review seeks to examine the biological mechanisms implicated in development of HCC, with a specific focus upon the murine double minute 2 (MDM2)-p53 networks and the hypothesised function of MDM2 in suppressing p53 expression. By knowing how MDM2 contributes in the development of HCC, we may potentially find methods to inhibit HCC as well as discover novel therapeutic approaches[14]. We also explore the function of cellular biology in identifying novel HCC biomarkers, utilizing MDM2-p53 linkages as prognostic indicators and considering future treatment strategies for HCC[14].
Nuclear protein that is produced by the human p53 gene is essential for cell division. It takes up roughly 20 kb of DNA. It was located on the short arm of chromosome 17 (17 p. 13.1) via southern filter hybridization of DNA from a hybrid of human-rodent[15]. There are 11 exons in the gene. The human p53 protein has 4 primary functional domains and 393 amino acids, according to earlier studies. There are more discussions of the p53 gene's and protein's molecular structure in the literature[16]. The discovery that Simian virus 40 (SV40)-transformed cells produce novel protein type which might be extracted by serum of anti-SV40 tumour led to the detection of the p53 protein, a protein having a noticeable molecular mass of 53 kDa[17]. Additional research showed that fluorophenylalanine therapy of SV40-infected monkey cells resulted in an increment in cellular thymidine absorption and a progressive suppression of viral and host DNA synthesis[14]. Transformed mouse cells, such as leukemias, murine sarcoma virus and spontaneously transformed fibroblasts, have been shown to express p53 in various cellular studies[18,19]. In-vitro virus sequence-specific DNA linkage is blocked by the wild-type p53 protein's complex formation with the hepatitis B virus X protein (HBx). The broad range of p53 function changes caused by HBx contributes to the emergence of human HCC[20]. The discovery of p53 in human leukaemia cells and transformed cell lines raises the possibility that p53 might influence the gene makeup of some types of leukaemia[21]. The production of p53 protein seems towards being essential for the mechanism of physiological cycling cells, in addition to its control over how dormant normal cells move from the G0-phase to the S-phase of the cell cycle. While p53 in experimentally altered (Meth A) mice cells appeared to have entered the stage of translation of RNA, transcription of p53 in growth inhibited healthy cells is regulated at the transcriptional or post-transcriptional stage[22]. Additionally, p53 takes involvement in the conversion of healthy embryonic cells and can work alongside Ha-ras oncogene to accomplish this[23]. In untransformed cells, it displayed a short half-life of 5 to 10 min.
Previous research has indicated that p53 can alter normal fibroblasts so that they become tumour-bearing cells[24]. However, later research revealed that dominant-negative mutants were present in a conserved area of p53 in the initially cloned p53 cDNAs utilised in the earlier experiments. Its biological processes and conformation were improved by these changes. This idea was supported by research done in the late 1980s, which also demonstrated that the wild-type p53 functioned as a tumour-suppressing gene and prevented transformation of oncogene[14]. Also, it was demonstrated that the p53 gene commonly had mutations in human large intestine cancer. These results and subsequent research demonstrated that p53 functioned as a tumour-suppressing gene. In reality, numerous investigations have supported the widely accepted theory that authentic wild-type p53 functions as a tumour-suppressing gene and exhibits certain wild-type p53 features. The key points from p53 study in this field could be described in the following way:
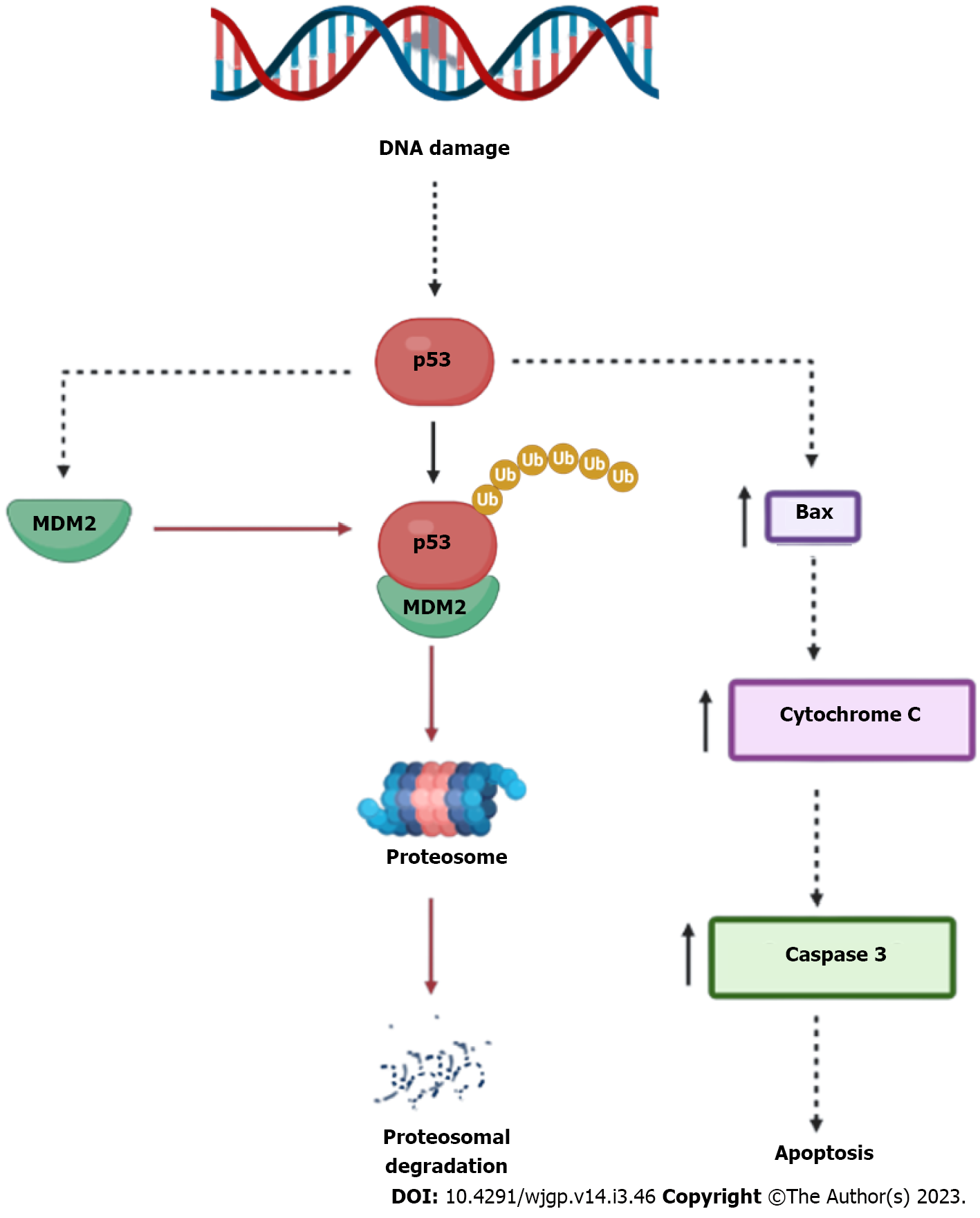
First, a mutant p53 gene combined with RAS activation can prevent transformation. Second, the p53 gene mutation is the most frequent gene change found in human cancers. There are regional differences in the p53 mutational profile[25] and disruption of normal p53 activity frequently promotes cancer inhibition and advancement[26]. Finally, mutant p53 proteins might function as suppressive proteins that decrease the action of wild-type p53 proteins by competing with them[27]. Additionally, MDM2-p53 binding can be blocked by modified p53, which kills tumour cells that overexpress MDM2. The combined approach for the destruction of MDM2-p53 connection with radiation revealed improved anticancer effects in-vitro and in-vivo, as proven by Koom et al[28]. As a transcription factor, p53 exhibits tumour-suppressing properties via controlling the transcription of genes[14]. p53 has a shallow level of activation in healthy cells. A variety of stress signals, such as DNA damage, oncogene activation and hypoxia, are detected by p53[14]. As a result, in response to stress signals, p53 is overexpressed and accumulates in high quantities. The expression of the relevant genes is then transcriptionally regulated by p53 after it attaches to a particular DNA sequence known as the p53 responsive element (RE) in the DNA. The cellular roles of p53 gene have been identified through studies, including regulation of cell cycle, apoptosis, senescence, DNA repair, alteration of metabolism and metastasis[29,30]. The interplay of certain p53 functions with particular oncoproteins, like MDM2. The following sections of this review go into further detail about these interactions. Posttranscriptional changes primarily regulate the levels of the p53 protein. Ubiquitination, phosphorylation, acetylation, methylation and SUMOylation are a few of these alterations. Ubiquitination is the primary modification that controls p53 in comparison to other changes[31] and a variety of deubiquitinating enzymes control p53 stability[32].
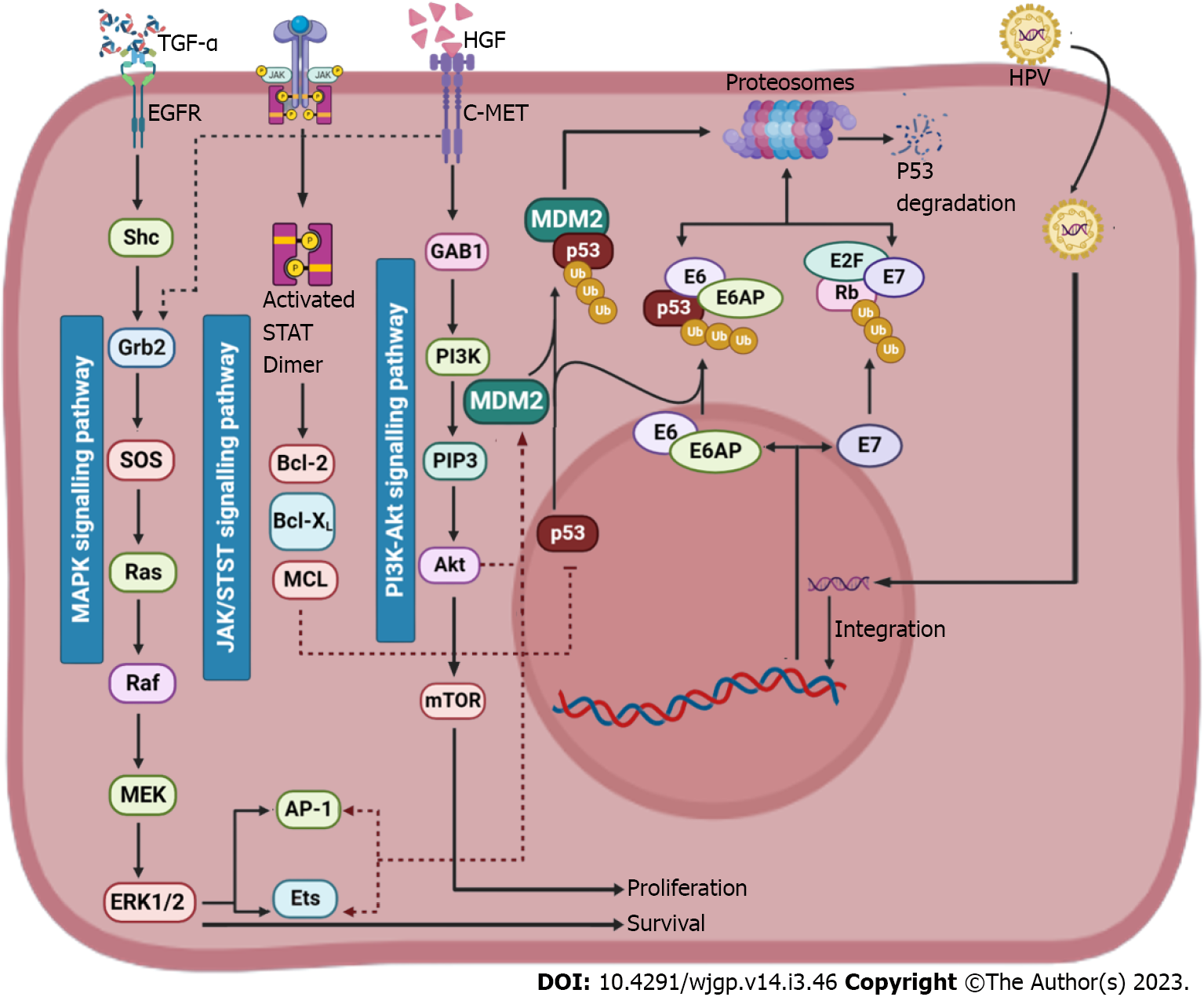
MDM2, a negative regulator of p53, is the primary controller of p53[14]. MDM2 participates in this mechanism by directly interacting with p53 and mediating its ubiquitination-dependent destruction (Figure 2). Fourth, a molecular similarity of MDM2, that attaches to the p53 transcriptional activation region and blocks it from activating transcription, has been discovered in the MDM4 (MDMX) protein[14]. MDM4 thereby aids in the development and operation of tumours. It's essential to note that MDM4 can prevent the breakdown of the MDM2 protein by interacting with the MDM2 protein through the RING finger domain, controlling the function of MDM2 in suppressing p53 activity. As a result, both MDM4 and MDM2 are regularly upregulated in several types of human malignancies. Schlaeger et al[33] have demonstrated that MDM4 and EFF1A2 could function as etiology-independent oncogenes in a sizable portion of human HCC.
The mechanisms described in this section, which may also apply to other cancer types, have been shown to promote the progression of HCC by changing the MDM2-p53 pathway[34]. As numerous ribosomal proteins (RP) that link to MDM2 are secreted from the nucleolus when ribosome biogenesis is hindered, it has already been demonstrated that p53 stabilized by ribosome biogenesis disruption[35]. There is further proof that a decrease in rRNA synthesis can stabilize p53 by blocking MDM2-mediated p53 damage by binding to RPs released from the nucleolus in the regenerating rat liver following partial hepatectomy[34,36]. It has been established that the p53-binding HBx, which localizes p53 to the cytoplasm, is crucial for the growth of HCC[34]. In cells that possess the HBx protein, doxorubicin therapy has been found to raise p53 levels; in addition, doxorubicin therapy recovers p53-mediated transcriptional function by lowering MDM2 amounts and raising p53 nuclear accumulation[34,37]. Similar to this, the tumour suppressor KLF6 has been found to be inactivated in response to HCV infection, and KLF6 expression has been linked to a poor prognosis for HCC[38]. The Kruppel-like €C2H2 zinc finger family, which has been linked to cell differentiation, signal transduction, and cell cycle control, includes KLF6[34]. According to Tarocchi and colleagues, reduced KLF6 expression results in increased MDM2 and decreased p53, and these disparities in the MDM2-p53 pathway is additionally linked to reduced survival in patients with surgically removed HCC[34]. In contrast, increased KLF6 expression decreases MDM2 expression and increases p53 expression in HCC cell lines[39]. In order to boost MDM2 ubiquitin ligase activity and p53 degradation, Jung et al[40] discovered the Enigma LIM domain protein, which plays a role in signal transduction via protein kinases[34,40].
Serum response factor (SRF), which is likewise elevated in HCC and promotes additional MDM2 stabilization and p53 degradation, can also trigger Enigma[40,41]. By causing MDM2 phosphorylation via Akt signalling, Minand and colleagues demonstrated that one p53 target gene, phosphatase of regenerating liver 1 (PRL-1), which is overexpressed in a number of cancers via an unknown mec
In addition, Sirt3 overexpression reduced HCC cell growth and promoted apoptosis in HepG2 and HuH-7 cell lines by upregulating p53 protein activity, which is found to be downregulated in human HCC tissue[46]. The balance between MDM2 and p53 is thrown off in HCC[34]. Each of the elements demonstrated has a distinct part in promoting the growth of cancer, although defective p53 is invariably identified as the primary cause of HCC[34]. To provide a better understanding of these pathways and other as-yet-unidentified mechanisms that contribute to HCC formation through the modification of p53, further systematic and thorough research is required[34].
Hepatocarcinogenesis is a multi-step process that includes stages of initiation, promotion and adva
The p53 protein is known to act as a transcriptional factor, however, it may still be discovered to have additional roles[55-57]. It induces the transcription of several genes implicated in the management of the cell cycle, such as the growth arrest and DNA damage-inducible gene GADD45[58], the gene encoding p21WAF1/CIP1 (also known as p21), the gene encoding MDM2 (a negative regulator of p53) and the gene encoding 14-3-3 (a regulator of G2-M progression)[59]. Additionally, it stimulates a number of genes that are likely involved in apoptosis, such as the gene for Bax, a pro-apoptotic protein linked to Bcl-2 and a number of genes that encode proteins included in the production of reactive oxygen species (ROS)[55,57]. The regulation of the cell cycle, DNA synthesis and repair, cell differentiation, transcriptional repression, genomic plasticity and apoptosis have all been linked to p53[53,60]. This includes stopping the cell cycle at G1 and G2 to evaluate genomic integrity. It is crucial for the detection of DNA damage and the subsequent reaction[61]. Cells express more p53 when their DNA is compromised. The cell can then activate the DNA repair systems because this protein prevents the G1 phase. p53 overexpression triggers apoptosis when the damage is too severe to be repaired, which results in cell death[62,63].
Numerous levels of strict regulation are in place for the p53 protein. Protein activity, protein stability and subcellular distribution are recognized as the three main levels of control[61]. Recently, thorough reviews on additional p53 regulation topics, like p53 post-translational changes and how they affect p53 activities, have been reported[64]. The cell cycle regulates how the p53 protein moves among the cytoplasm and nuclear compartments[65]. For p53 to function as a tumour suppressor, it must build up in the nucleus. An effective way for tumour cells to remain to multiply in the existence of wild-type p53 is to avoid nuclear aggregation[61]. In fact, cytoplasmic sequestration of p53 has frequently been seen in a variety of tumours, including neuroblastomas, breast cancer and colon cancer[66,67]. MDM2 is the factor that causes p53 to accumulate in the cytoplasm of at least some of these tumours and other proteins have also been linked to cytoplasmic retention[68-70]. Many viral proteins, including the human papillomavirus E6 protein[71], the adenoviral E1B 55 kDa protein and the HBV HBx protein[72,73] affect the location of p53. Additionally, several tumour types have indeed been linked to p53 import/export abnormalities[74]. p53 is a protein that shuttles back and forth through the nuclear pore complex. A bipartite nuclear localization signal (NLS) found at the C-terminal domain of p53 and two nuclear export signals found in the N- and C-terminal areas are responsible for nucleocytoplasmic transport. NLS mutations impede MDM2-mediated cytoplasmic breakdown and inhibit p53 export[75].
A damaged p53 gene, which is typically rendered inactive via a single mutation or a gene deletion, is implicated in about 50% of all malignancies. These modifications are thought to hinder p53 from oligomerizing and forming the tetrameric complexes which link to particular DNA sequences, changing the way the wild-type protein behaves physiologically[53,63,76]. Human malignancies with a p53 mutation are more deadly, aggressive and likely to metastasize. Therefore, p53 abnormality detection may reveal details about the molecular pathogenesis and -etiology of human cancer[77,78].
The two most common genetic changes associated with human HCC are mutations of p53 gene and HBV DNA incorporation into the host gene. An abnormality in the p53 gene has been found in 30%-60% of HCC patients[79] and a hotspot of mutations in the gene has been identified in HCC patients from locations with significant aflatoxin exposure[80]. Aflatoxin B1, which contaminates foods in regions where it is endemic, clearly plays a part in the development of hepatocarcinogenesis because its metabolites encourage the formation of apurinic sites and G to T mutations in chromosomal DNA and the presence of a codon 249 G to T transversion in p53 in 50% of HCC patients from high aflatoxin exp
The transactivation domain of wild-type p53 is polymorphic at residue 72, where a single-base mutation results in the substitution of proline for arginine (CCCCGC)[81]. Numerous research has been conducted on the prevalence of these two alleles and their potential association with the chances of developing cancer, even though the clinical significance of the variants is unknown[61]. According to one study, hepatocarcinogenesis may be influenced by the frequent loss of the proline allele in HCV-associated liver cancer. To clarify this claim, however, further research on this topic has to be done[81]. Furthermore, Okada et al[82] discovered a strong association between HCV type 1b infection and male homozygotes for p53Pro. Therefore, there may be a connection between this p53 polymorphism and HCV infection.
Humans can consume aflatoxin B1 (AFB1), a very known mutagen of the liver made by the Aspergillus fungus, through contaminated food products[83,84]. According to one research, the population inhabitants’ risk of HCC caused by AFB1 is 17% in some regions of the world[85]. According to the mechanism, CYP40s convert AFB1 into AFB1-8,9-epoxide, which then interacts with DNA to generate 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 (AFB1-N7-guanine) adducts[86]. If these adducts are not repaired, they cause G>T transversions to occur while DNA replication[87,88]. The DNA binding domain of p53 is known to be altered by an arginine to serine missense mutation at codon 249 (R249S), that is brought on by a G>T transversion at codon 249's third base[89]. About 50% of HCC cases in places where AFB1 Levels are high, like Qidong City in China, contain the p53 R249S mutation[90], suggesting that p53 might have a role in AFB1-induced HCC[86].
In addition to the guanines at codon 248, AFB1-8,9-epoxide combine with the p53 gene, although those guanine adducts need not produce cancer-inducing alterations as often as p53 R249S[88,89,91]. While p53 is first activated by AFB1-mediated DNA damage to cause cell cycle arrest during the S to G2/M phases[92], liver cells which acquire p53 R249S will indeed bypass this cellular defense process with a preferential benefit for proliferation, that might further progress towards liver cancer[86]. In fact, p53R249S is demonstrated to boost insulin-like growth factor 2 (IGF-2) transcription in Hep3B (p53 null) cells, indicating a potential gain-of-function activity[86]. IGF-2 has been linked to accelerating the course of human HCC and is overexpressed in 16%-40% of cases[93]. Additionally, there is a strong association between the genotype IGF-2 +3580 AA and the threat of HCC. It's interesting to note that HepG2 cell viability and proliferation are reduced when IGF-2 is silenced[86]. As a result, p53 mutation caused by AFB1 is critical for the development of HCC, presumably through increased IGF-2 signalling[86].
It is debatable whether vinyl chloride (VC), a carcinogenic gas involved in the production of polyvinyl chloride, can cause human HCC, but it primarily causes angiosarcomas of the liver (ASL)[94-96]. The liver's CYP2E1 enzyme converts VC from the lungs into chloroethylene oxide, which causes liver cancer by forming large DNA adducts[97,98]. 7-(2-oxoethyl)-deoxyguanosine, 3,N4-etheno-deoxycytidine, 1,N6-ethenodeoxyadenosine and N2,3-etheno-deoxyguanosine are four VC-associated DNA adducts that have been identified in-vivo[97].
According to reports, the p53 gene's codons 179, 249 and 255 had more A>T transversions in VC-induced human ASLs[99,100]. According to a study employing Sprague Dawley rats, the most of p53 mutations in HCC and ASL after exposure of VC, are A>T transversions; the A>T transversions in ASLs are found at 253 codons of rat p53, that is equal to 255 codons in humans[86,101]. A conformation-specific p53 antibody, PAb240 and other p53 antibodies have been used to identify an increase in the quantities of mutant p53 protein in serum samples from workers exposed to VC[99,102].
Drinking alcohol regularly is bad for our health. It causes liver cirrhosis, which then develops into HCC. One of the major causes of HCC is alcoholic liver disease[103,104]. Alcohol usage is linked to an up to twofold increased risk of HCC, say case studies from throughout the globe[105,106]. Studies on mice on an alcohol diet have also revealed exacerbation of inflammation, epithelial-mesenchymal transition (EMT), fibrosis, as well as progression to HCC[105]. However, hazardous by-products of alcohol catabolism, such as a buildup of acetaldehyde and free radicals, might affect oxidative stress, apoptotic cell death, necrosis, and necroptosis[106]. Pure ethanol cannot directly induce inflammation and liver damage[106]. Elevated inflammatory cytokine release brought on by ongoing inflammatory pathways leads to the creation of ROS[103,106]. HCC growth is aided by ROS-induced DNA damage, hepatocyte genomic susceptibility, and T-lymphocyte suppression[103].
Additionally, alcohol catabolism affects a number of lipid metabolism processes, which results in liver steatosis and fatty acid oxidation inhibition[105,106].
An essential biological process known as epigenetic reversibility of gene expression frequently contributes to cancer[106]. Alcohol abuse alters epigenetic pathways that result in altered DNA methylation and acetylation. For instance, aberrant acetylation has been linked to alcohol-induced HCC and hepatic steatosis[106]. In a syngeneic BALB/c mouse tumour model, promoter hypomethylation in circulating tumour cells of HCC is strongly correlated with overexpression of c-Met and hepatocyte growth factor[107].
Alcohol consumption also affects metabolism, causing acetaldehyde to accumulate, hypomethylation, an absence of antioxidants and retinoic acid, as well as inflammation, oxidative stress, hypoxia, and genetic instability[106].
NASH, non-alcoholic fatty liver disease (NAFLD), fibrosis, cirrhosis and HCC are among the illnesses that make up NAFLD. Obesity, type 2 diabetes and metabolic syndrome are all linked to NAFLD[108]. NAFLD affects 20%-30% of people in the West, according to estimates[109]. Just 11.5% of individuals with NAFLD-caused cirrhosis go on to grow HCC and roughly 50% of NASH-caused HCCs happen in people who don't have cirrhosis[110,111]. These findings suggest that additional oncogenic events are necessary for NAFLD-caused HCC[86]. The cellular pathways in NAFLD caused HCC, however, remain poorly known. Its development has been linked to a number of mediators, including insulin resistance, deregulation of NF-kB signalling, the Pl3K/ATK-PTEN pathway and the activity of certain miRNAs (e.g., miR-34)[112,113].
Additionally, p53 has been linked to the development of NAFLD via a variety of pathways. In a mouse model for NAFLD, in which p53 +/+ and p53 -/- mice have nourished a diet deficient in choline and methionine, p53 +/+ mice exhibit enhancement in histologically visible steatohepatitis, fibrosis and ROS formation as well as higher protein levels of p66Shc, a protein linked to oxidative stress, in comparison to p53 Human NASH hepatocytes have increased p53 activity along with elevated p21 and p66Shc mRNA levels, that is highly linked to the degree of fibrosis[86]. Additionally, a rise in miR-34 expression and reduced SIRT1 protein content were found in human NAFLD liver tissues along with a rise in acetylation of p53, both of that were associated with the condition[114]. By promoting hepatocyte death, stimulation of the miR-34a-SIRT1-p53 axis has also been demonstrated to lead to liver fibrosis or NASH[115]. Additionally, p53 has the ability to activate miR-34, which suppresses the expression of SIRT1 mRNA and raises p53's acetylation in a positive feedback loop[116]. These findings suggest that NAFLD and high expression of miR-34 and p53 are related. However, it should be highlighted that p53-dependent and p53-independent apoptosis can be mediated by miR-34a[117]. Nevertheless, some evidence points to p53's involvement in the development of NAFLD and NASH; however, more research is needed to show if p53 actually have a critical role in the development of NAFLD-mediated HCC[86].
Iron is a crucial mineral that is involved in several metabolic processes, including heme production, the formation of the Fe-S cluster and the transportation of oxygen by haemoglobin[118].
Nevertheless, there can be serious implications when iron homeostasis is disturbed, whether as a result of hereditary or environmental factors, such as hepatic fibrosis, cardiomyopathy and arthropathy, endocrine abnormalities[119,120]. Crucially, having too much iron raises the risk of developing a variety of neoplasias, such as breast cancer, colorectal cancer and HCC[121]. Dietary iron excess, primarily in beer cooked in iron pots, is closely linked to a higher risk of HCC in several areas of sub-Saharan Africa[122]. HCC has been demonstrated experimentally in Wistar rats on iron-rich diet[86,123]. Multiple rodent models and investigations of human HCCs have connected higher iron content with increment in 8-oxo-2-deoxyguanosine adducts and oxidizing agents like malondialdehyde, suggesting that ROS-inducing DNA alterations are one pathway involved in iron overload-linked HCC genesis[86,124].
Moreover, there is proof that an excess of iron directly affects p53 activity[86]. A high-iron diet causes p53 protein levels in the liver of C57BL/6 mice to drop[125]. Additionally, male Sprague-Dawley rats exhibit a rise in MDM2 and a reduction in p53 in the liver when fed a high-iron diet over an extended length of time[126]. Another chemical mechanism for why there are fewer p53 molecules in the blood when there is too much iron is that p53 is linked to heme, transferred to the cytoplasm and then destroyed by the proteasomal pathway in HepG2 cells[125]. As a result, p53 levels are decreased by both iron overload and dysregulated heme, which aids in the development of HCC[86]. It's interesting to note that p53 also affects intracellular iron content via transactivating iron-sulfur cluster enzyme 2, which results in less efficient iron uptake[127,128]. As a result of chronic iron overload, decreased p53 function raises intracellular iron levels, which further encourages the development of HCC. It is important to emphasized that people with hereditary hemochromatosis have greater frequencies of p53 mutations (64%-71%) than those with sporadic HCC, indicating a possible involvement of p53 in the development of HCC brought on by iron overload[129,130]. 45% A>C transitions and 33% G>C transversions, involving two hotspots at codons 275 and 298, are seen in HCC tissues from hereditary hemochromatosis[86,130]. The p53 mutation spectrum in the research utilizing British families with hereditary hemochromatosis is made up of 60% A>G transitions and 40% A>T transversions, though[129]. However, it is still unclear if iron excess does in fact cause HCC in animal models in a p53-dependent manner[86].
According to estimates, 248 million people worldwide have chronic HBV infection and are hepatitis surface antigen positive[131]. HBV is the main contributor of HCC, with chronic HBV infection accounting for the majority of cases[86,132]. Repeated episodes of immune-linked hepatocyte death and consequent tissue regeneration can result in cirrhosis of the liver and HBV-mediated HCC carcinogenesis[133]. Importantly, 10%-30% of HCCs linked to HBV do not develop in the context of cirrhosis, pointing to additional oncogenic pathways involved in the development of HCCs linked to HBV[86,134].
The genome of HBV, a circular, partly double-stranded DNA virus, has four overlapping open reading frames: A core area, a surface region, a polymerase region and an X region[86]. These four regions together develop seven viral proteins: Precore, polymerase, S, M, core, HBx and L[135,136]. The HBx protein, which is crucial for viral replication and is most connected to the development of HCC, is one of these[136]. In fact, HBx causes HCC in transgenic mice models by confining p53 to the cytoplasm[137,138].
Additionally connected to hepatocyte apoptosis is HBx[86]. In many situations, HBx prevents apoptosis by adhering to and sequestering p53 in the cytoplasm in addition to raising levels of the anti-apoptotic protein surviving[69,139]. Additionally, HBx is said to suppress tumour growth factor (TGF)-mediated apoptosis[140]. On the other hand, in rare circumstances, HBx has been demonstrated to cause apoptosis without the involvement of p53[141,142]. The double functions of HBx in hepatocyte apoptosis and its connection to the development of HCC, therefore, call for additional research[86].
Although there is no concrete proof for it, HBx variants with C-terminal truncations (Ct-HBx) are typically found in HCC and may potentially have a role in the progression of HCC[143,144]. In numerous cell lines, Ct-HBx suppresses apoptosis and encourages hepatocyte growth[145]. It has also been demonstrated that Ct-transcriptional HBx's repression of USP16 (ubiquitin-specific peptidase 16) enhances the tumourigenicity and stem-like characteristics of HCC cells[86]. Additionally, like HBx, Ct-HBx links to p53 and blocks p53-linked apoptosis[135]. Likewise, few Ct-HBx variations have the capacity to inhibit GAS2, a modulator of p53-linked apoptosis, from expressing its mRNA[146]. As a result, USP16 is downregulated and p53-mediated apoptosis is inhibited, which may help explain how Ct-HBx contributes to the pathogenesis of HBV-related HCC[86]. Given that p53 mutations are uncommon in HBV-related HCC, are linked to advanced disease and are inhibited by both HBx and Ct-HBx, suppression of p53 activity might be more advantageous for HBV-linked HCC carcinogenesis than p53 mutation[147]. Crucially, HCC patients with wild-type p53 outlive those with p53 mutations in terms of both overall survival and recurrence-free survival[86].
According to estimates, 184 million people worldwide have anti-HCV positivity and those who have HCV have a 15-to-20-fold higher risk of developing HCC[148]. HCV is a single open reading, positive sense, 9600 nucleotide long single-stranded RNA virus[149]. The HCV genome produces a polyprotein that is broken into nine viral proteins, encompassing structural proteins (C, E1 and E2) and non-structural proteins (N, S and S2) (p7, NS2, NS3, NS4A, NS5A, NS5B)[86,150]. Although cirrhosis is the primary setting for the great majority of HCCs connected to HCV, there is some proof that the HCV viral proteins have the capacity to cause cancer[134]. Particularly, both p53-dependent and -independent mechanisms have been proposed to link the development of HCC to the HCV core, NS3 and NS5 proteins[86].
Without the presence of cirrhosis, HCC definitely instantaneously arise in transgenic mice that produce the HCV core protein[151]. The Wnt-β-catenin pathway is upregulated, ROS levels are increased and Fas- and tumor necrosis factor (TNF)-mediated apoptosis is inhibited by HCV core protein[86]. The core protein alters the subcellular localization of p53 to the perinuclear area and nuclear granular structures, as well as its post-translational changes, such as phosphorylation and acetylation, in HeLa and HepG2 cell lines[86]. This is significant because it reduces p53 activity[152]. Additionally, the core protein activates SIRT1, which deacetylates p53, impairing p53-dependent apoptosis in HepG2 cells. Thus, both p53-dependent and -independent mechanisms by which HCC is caused by HCV core protein are likely[86].
Another HCV protein that could alter human hepatocytes is called NS3, a non-structural HCV protein that increases cyclooxygenase-2 and activates mitogen-stimulated protein kinase[153]. In HeLa and NIH3T3 cells, NS3 also forms complexes with p53[154] and inhibits p53's transcriptional activity in NIH3T3 and Huh7 cells. Furthermore, overexpressing NS3 can cause NIH3T3 cells to convert and cause tumours in animals[155]. It is yet unknown, nevertheless, whether or not p53 is required for NS3-mediated transformation. In transgenic mice models, the non-structural HCV protein NS5A can result in steatosis and HCC[86]. It has been demonstrated that NS5A inhibits Bax-linked apoptosis, transactivates c-fos and prevents TNF-linked apoptosis without the need for p53[156]. However, p53's transcriptional activity can be inhibited when NS5A binds to and colocalizes with p53 in the perinuclear membrane. Additionally, NS5A interacts with hTAFII32 at the nucleoplasm membrane and prevents it from stabilizing p53, which prevents p53-mediated apoptosis in Hep3B cells[157]. As a result, NS5A promotes the growth and progression of HCC through both p53-dependent and -independent mecha
The structural viral protein known as the HCV core protein encases the viral genomic RNA. According to its subcellular localization and the subsequent impact on the cell cycle inhibitor p21, the core protein may also have contradictory effects on cell growth, supporting both apoptosis and cell proliferation[158,159].
The innate version of the HCV core protein is synthesized first (amino acids 1–191) and a mature form (amino acids 1–173) is created during processing[61]. The innate form and the mature form, form a heteromultimer in the cytoplasm, preventing the mature form from being transported to the nucleus. The innate form activates p53 in the cytoplasm and p53 then increases the production of p21 by acting as a transcription factor for p21. It is unclear how the innate form stimulates p53, but because it binds to the protein, it may do so by stabilizing the protein[159,160]. p21 expression can be decreased by a process unrelated to p53 if the amount of mature core protein penetrates the nucleus and surpasses the innate form's binding capacity[161,162]. A core-responsive region in the p21 promoter precisely overlaps a butyrate/TGF-RE. In this instance, core protein stimulates a butyrate route to activate p21 via the element[73].
Additionally, the HCV core protein controls p73, a tumour suppressor protein belonging to the p53 family[61]. In addition to being engaged in neurogenesis and innate immune responses, p73 also appears to have a significant role in the development and maintenance of malignancies[163]. The HCV core protein is translocated into the nucleus as a result of its interaction with p73. Additionally, in a p53-dependent way, the association with the core protein blocks p73-α, but not p73-β, dependent cell growth arrest. Consequently, the impact of the HCV core protein on p73 function may play a role in HCV pathogenesis[61].
The HCV NS3 protein may cause host cells to become hepatocarcinomas at an early stage. Additionally, it has been suggested that it might result in p53 gene mutation and hepatocyte transformation, however this is debatable[164,165]. NS3 and wild-type p53 combine to form a complex[164]. The oligomerization domain, located close to the C-terminus of wild-type p53[166] (amino acids 301-360), is crucial for the formation of this combination with NS3. In line with this discovery, NS3 protein can specifically and dose-dependently suppress the promoter activity of p21 via altering the activity of p53[61]. The impact works in concert with that of the HCV core protein and is not cell-type specific.
A phosphoprotein between 56 and 58 kDa, the HCV NS5A protein. Although NS5A is a member of the viral replicase complex, which is located on the cytoplasmic side of the endoplasmic reticulum, along with other virus-encoded proteins, its function in viral replication is yet unknown[61]. Phosphorylation and possible proteolytic processing of NS5A into lower molecular weight forms that can translocate to the nucleus are examples of post-translational changes[167]. Truncated forms of NS5A can act as transcriptional activators, but other now has discovered interrelations of NS5A with cellular proteins, such as those of the transcription activation machinery, cellular signalling apparatus and cell-cycle-regulatory kinases, suggesting that it plays a pleiotropic role in HCV-host interrelations[167,168]. Numerous of these interactions prevent the apoptotic cellular response to persistent HCV infection, that raises the possibility that NS5A plays a role in the development of chronic liver conditions and HCC linked to HCV infection[61].
By locating p53 in the perinuclear area, NS5A can prevent it from binding to certain DNA sequences. Inhibiting p21 expression and p21-mediated apoptosis in this manner[169]. Additionally, p53 and TATA-box-binding protein (TBP) connect to NS5A, generating a heteromeric complex that prevents p53 and TBP from attaching to their respective DNA binding regions. In addition, p53-TBP-excision repair cross-complementing factor 3 complex formation is inhibited by NS5A[61]. Additionally, the NS5A protein may prevent p53 from activating by securing the crucial coactivators of p53, hTAF(II)32 and hTAF(II)28. Recently, it was discovered that the protein kinase PKR, which is activated by interferon (IFN), is inhibited by NS5A. Thus, NS5A may be involved in HCC's IFN resistance[169,170].
Other potential risk factors for the development of HCC include genetic predisposition, prenatal anomalies, chemical exposures (aflatoxin or food tainted with arsenic), and autoimmune illnesses of the body’s liver[106].
Patients with a number of congenital defects are more likely to develop liver cirrhosis and HCC. These consist of hemochromatosis, alpha-1-antitrypsin deficiency, Wilson's disease, and hereditary tyrosinemia[171]. AFB1-induced HCC is caused by a number of processes, some of which involve the development of mutagenic and carcinogenic intermediates and adducts. Food contaminated by the fungus Aspergillus flavus and Aspergillus parasiticus releases aflatoxins. There are several chemical changes that happen as a result[106].
AFB1 is transformed into the following known mutagenic or carcinogenic compounds: 8,9-dihydroxy-8-(N7), aflatoxin-B1, aflatoxin B1-8,9 exo-epoxide. Aflatoxin B1 form aminopyrimidine adduct guanyl-9-hydroxy aflatoxin B1 adduct[106]. Additionally, these intermediates and adducts have the ability to directly cause a mutation at codon 249 of the p53 tumour suppressor gene[106]. This changes the amino acid arginine to serine, reversing the gene's ability to prevent tumour growth[106]. According to certain reports, AFB1 and HBV may work together to cause HCC[172]. Additionally, additive interactions have been documented. In a systematic review, Ogunwobi et al[106] showed a connection between autoimmune hepatitis (AIH) and an elevated risk of developing HCC in individuals with liver cirrhosis. The chance of developing liver cirrhosis from AIH was determined to be less than the probability of developing the condition as a result of HBV, HCV, or primary biliary cholangitis. But the risk of HCC and liver cirrhosis from AIH is clinically considerable[106].
It has been demonstrated that MDM2 inhibits p53 gene transcription and translation in addition to adversely controlling p53 functions through ubiquitination and degradation[173]. The cell fate as well as the balance between MDM2 and p53 may be impacted by factors that control the ubiquitination of MDM2 and p53[174]. The connections between MDM2 and p53 are regulated by various mechanisms[14,175]. Ones of these interactions could be.
RPs have a substantial impact on nuclear strain, cell cycle arrest and apoptosis via activating the MDM2-p53 regulatory loop[176]. Knockdown of nuclear proteins can cause nuclear/ribosomal stress by sequestering MDM2 and inhibiting its ubiquitin ligase function against p53[177], which in turn causes nuclear/ribosomal stress[35]. It has been demonstrated that ribosome biogenesis interruption leads to p53 stability. This is caused by a number of RPs that are linked to MDM2 and restrict its action for p53 being released from the nucleolus[35]. As a result of blocking E3 ubiquitin ligase, the subunit RPs rpL5, rpL23 and rpL11 have demonstrated the capacity to attach to MDM2. According to Zhang et al[178], the human protein HDM2, which is the MDM2 ortholog, interacts to RP L11[14]. The docking site is located in the middle of HDM2, which prevents HDM2-associated p53 ubiquitination and destruction and restores p53-linked transactivation and p21 protein deposition. The research confirmed the idea that L11 controls HDM2 negatively[14].
The most prevalent element found in ribosomes is ribosomal RNA, which is the RNA portion of the ribosome and is necessary for protein synthesis[14]. In human tumour cells, it has been discovered that RNA polymerase I subunit A (POLR1A) gene silencing results in a decrease of rRNA production that stabilizes p53 without altering the integrity of the nucleolus. The suppression of the MDM2-mediated p53 breakdown was what caused p53 to stabilize. However, rRNA production down-regulation that was accompanied by a decrease in protein synthesis did not result in the stability of p53[14]. The results showed that p53 functions in mammalian cells are controlled by the equilibrium of RP and rRNA production[36].
Small non-coding RNAs called microRNAs (miRs) regulate the transcription and post-transcriptional expression of many target genes. In cancer, their action is typically dysregulated[14]. Micro-RNAs generally decrease in human malignancies, suggesting that they could be involved in tumour suppression. Therefore, by focusing on oncogenes or tumour-suppressor genes, miRs can perform a variety of biological roles[14]. The first miR-34 family members to be implicated in the p53 tumour-suppressing network was identified in 2007 and are known as miR-34a, miR-34c and miR-34b[179]. The expression of members of the microRNA family (miR-34a-c) evidenced the p53 condition and the genes encoding miR-34 family members were transcriptional domains of p53 in-vivo and in-vitro, according to the authors' comparison of the microRNA expression profiles of cells with and without p-53 deficiency[14]. As a result, the p53 network inhibited the development of tumours via a variety of co-ordinated interactions and multiple transcriptional targets, such as the function of the miR-34 family members in preventing uncontrolled cell growth[14].
Bi et al[180] investigated the function of miR-125a in HCC. MiR-125a expression was found to be downregulated in HCC tissues and cell lines and it was linked to aggressive pathological characteristics[14]. According to the research, miR-125a targets vascular endothelial growth factor A and matrix metalloproteinase II, which both function in-vivo and in-vitro to prevent the progression and metastasis of HCC. As a result, miR-125a overexpression might be an effective treatment for HCC.
Experiments on miR-125b's function revealed that it negatively regulated p53 and p53-caused apoptosis in cells under stress, implying that it had a tumourigenic effect, for example[181]. miR-122, one of the several miRs found in liver tissue, has been demonstrated to function as a tumour suppressor in HCC[14]. Poorly differentiated cancer, big tumours, metastasis and a poor prognosis are usually linked to decreased miR-122 expression[182]. Simerzin and colleagues recently demonstrated that miR-122*, the passenger strand of miR-122, targeted MDM2 and had an active role in the association between MDM2 and p53. The levels of miR-122* and MDM2 were significantly inversely correlated in human HCC samples. By highlighting the tumour-suppressor properties of miR, in-vivo research demonstrated that miR-122* was capable of preventing the formation of tumours[14]. Inhibiting miR-122 using an antagomiR-122 (miR inhibitor) caused miR-122* to build up, MDM2 to be suppressed and the p53 protein to be increased. The research revealed that miR-122*, the miR-122 passenger strand, controlled the function of p53 by concentrating on MDM2. As a result, miR-122* exhibits tumour-suppressive action that was previously only associated with miR-122. Additional research is required to determine whether it is useful as a treatment for HCC as well as what key distinctions occur among miR-122* and miR-122[14].
Kim et al[183] discovered that p53 increased the expression of members of the miR-200 and miR-192 families and explained the function of p53 in controlling the EMT in HCC via the development of particular effector miRs, such as miR-141, miR-192, miR-194, miR-200b, miR-200c and miR-215[14]. These miRs specifically targeted the zinc-finger E-box-binding homebox 1 and 2 (ZEB1 and ZEB2) and EMT-activity transcription factor 2. ZIB1 and ZEB2 genes were repressed in human HCC and HCC cell lines by p53 overexpressed miRs. This reduced EMT resulted in enhanced E-cadherin and reduced vimentin expression[14].
It has been demonstrated that the protein Enigma, PDLIM 7 (Enigma LIM), which has a LIM domain, interacts with proteins included in protein kinases and that the Enigma gene has been detected in stomach and liver malignancies in humans[14]. It's also been demonstrated that even in-vitro, in human hepatoma cell lines and in mice embryonic fibroblasts, the LIM domain protein Enigma actively links with MDM2 to create a complex with p53[14]. Furthermore, it has been demonstrated that the Enigma caused p53 breakdown by reducing MDM2's ability to ubiquitinate itself and boosting its ability to bind to p53[40].
Serum growth factors were capable of rapidly stimulating the expression of HDM2 in a well-studied non-transformed human fibroblast model, as demonstrated by Phillips et al[184]. The authors noted that the increased HDM2 protein, that also accompanied growth factor activation, was mainly the consequence of phosphatidylinositol-3 kinase-dependent stabilization of the HDM2 protein along with a global rise in protein synthesis in their investigation, which investigated the mechanistic reaction for these variations[14]. According to a new study, SRF induction enhanced Enigma transcription, stabilizing HDM2, which then caused p53 to degrade[14]. The authors saw these modifications in the liver given the hepatocyte growth factor. Additionally, they discovered that MDM2 co-expressed with SRF and Enigma, but not p53, in the human stomach and liver tumours. Enigma was also discovered to decrease p53-linked apoptosis in cell lines and a mouse xenograft model, which boosted cell viability and produced chemoresistance[14].
Several malignancies, including colorectal[185], gastric and liver tumours, and especially metastatic cancers, have been discovered to overexpress the protein tyrosine phosphatase family of the PRL family. PRL-1, PRL-2 and PRL-3 displayed a higher level of structural sequence similarity within this family. According to Min et al[186], overexpression of PRL-1 prevented p-53-mediated apoptosis and decreased levels of endogenous and exogenous p53[14]. The ubiquitination of p53 and subsequent proteasomal degradation were the mechanisms by which p53 was down-regulated. The stimulation of PIRH2 (p53-induced RING-H2) transcription and the promotion of MDM2 phosphorylation via Akt signalling were the two main strategies used to ubiquitinate PRL-1[14]. These results suggested that the growth of cancer was aided by the down-regulation of p53 caused by a PRL-1-induced negative feedback mechanism. Additional research by the same team showed that PRL-3 activated PIRH2 and MDM2 in cancer cells, negatively regulating p53 in a manner akin to PRL-1[187].
The MDM2 binding protein (MTBP) was demonstrated to have metastasis-suppressing properties in earlier investigations. Additionally, it has also been proven that MTBP prevented infiltration irrespective of p53 status[172]. Currently, Bi et al[188] established the growth inhibitory action of MTBP in HCC and observed that, when compared to nearby non-tumour tissue, MTBP expression was reduced in almost 70% of human HCC cells[14]. The local invasion of lymph node metastases was also linked to this decrease. Regardless of the p53 status, the transcription rates of MTBP were found to be adversely linked with the ability of HCC cells to migrate[14]. These findings supported the findings of Vlatković et al[189], who showed that low MTBP was linked to a poor prognosis for head and neck malignancies. Moreover, there are currently a number of holes in this field. For instance, it is yet unclear how HCC cells control the expression of MTBP. Additionally, it is unclear how functionally related MTBP and MDM2 are. We have to understand why p53 is not required for these modifications in the interim[14]. The existence of a single nucleotide polymorphism (SNP309) in the MDM2 promoter is another element that might influence how p53 interacts with MDM2. It has been demonstrated that this factor raises the affinity of the transcriptional activator Sp1, leading to greater levels of MDM2 RNA and protein as well as a resulting attenuation of the p53 pathway[14]. The authors' suggestion that SNP309 serves as a rate-limiting event in carcinogenesis stems from studies showing that such mechanisms enhance the development of tumours in humans[190].
HCC typically harbours p53 mutations[191]. Despite carrying wt-p53, the majority of HCCs have abnormalities in the p53-mediated apoptotic pathway[192]. In HCC, p53 might occasionally be wild-type but inactive[193]. The expression of wt-p53 can be induced by signals to and activation of p53, which suppresses the altered phenotype of hepatocytes and boosts the actions of radiation therapy and chemotherapy[194]. In HCC, p53 can be partially or completely repressed and single tiny compounds, like the well-known antimalarial medication quinacrine, can activate p53[195]. Exogenous wt-p53 can be delivered into cancer cells by gene delivery, which effectively slows the growth of tumours that contain both mutant and wild-type p53[196]. The oncogene MDM2 generates a phosphoprotein that builds a strong complex with mutant as well as wild-type p53 protein, blocking p53-mediated transactivation and causing p53 breakdown[197]. MDM2 also has a p53-DNA binding site[195]. The MDM2-p53 auto-regulatory feedback loop[198] controls the transcription of the MDM2 gene and the activity of the p53 protein. The amount of wild-type p53 protein can control the expression of the MDM2 gene and the MDM2 protein can interact with p53 to reduce its capacity to function as a favorable transcription factor at the MDM2 gene-responsive region[195]. To boost functional p53 levels and p53-mediated therapeutic efficacy, several methods have now been used to disrupt MDM2-p53 interaction[199]. The protein-secreting pathway is significantly regulated by Adenosine diphosphate (ADP)-ribosylation factor (ARF) proteins[200]. ADP and adenosine triphosphate are converted to adenosine in a human HCC cell line and treating HCC cells with adenosine can decrease their development and activate caspase-3, showing that adenosine is a cytotoxic apoptotic agent[195]. ARF, which is regarded as a tumour suppressor[201], can start the cellular response to abnormal oncogene stimulation by attaching to and preventing MDM2 from functioning[195]. Like the murine protein, the human protein p53's capacity for transactivation can be inhibited by HDM2, which is MDM2's human counterpart[202]. Different locations on MDM2 are where ARF and p53 bind. According to MDM2's mediation, the ARF-p53 complex is created, which can improve p53 stability[203]. Successfully inducing p53-triggered tumour cell death can be achieved by blocking the routes for p53 breakdown, either through over-expression of ARF or by disrupting the connection between MDM2 and p53[195].
High levels of exogenous p53 protein might have therapeutic benefits for HCC in two ways: (1) By increasing tumour cell apoptosis by preventing cell proliferation via a number of physiological mechanisms and[123] by making HCC more susceptible to certain chemotherapeutic drugs[195]; and (2) Additionally, due to the wt-p53 protein's angiogenesis blocking properties, p53 reduces the expression of genes related to angiogenesis[204].
Due to medication resistance, treating HCC is extremely challenging. Reversing multiple drug resistance in human HCC cells and figuring out the underlying processes are both important[195]. In HCC cells expressing mt-p53, P-glycoprotein expression and function are increased, as are the products of numerous drug resistance genes (MDR)[205]. Expression of mt-p53 decreases the absorption of chemotherapeutic drugs by HCC cells and increases their treatment resistance. MDR transcription is inhibited by wt-p53 as opposed to mt-p53, which increases MDR expression[206]. Due to the reduced expression of P-glycoprotein encoded by MDR, HCC cells become more sensitive to chemotherapeutic drugs when wt-p53 function is restored in these cells[195].
Currently, the most frequently used in-vitro and in-vivo therapeutic agents for the treatment of HCC are the oncolytic virus and recombinant adenovirus p53 (rAd-p53)[195]. Both the expression of p53 and p21 proteins is observed to be up-regulated in cells following the introduction of exogenous wt-p53 generated by the recombinant adenoviral vector. It is possible to stop cell division and apoptosis. Oncolytic viruses are a group of flawed viruses[207] that cannot reproduce in healthy cells but can expand in tumour cells, ultimately lysing them[195].
Human adenovirus vector processes are typically utilized for the expression of proteins in humans or other species with a few benefits due to their wider host range and lesser pathogenicity to humans, have a binding attraction for epithelium[208], which is essential as the majority of human tumours are of epithelial origin and serotype 4 of species E[209] have a specific binding affinity for HCC cells[208] . It has been determined that the E1 region of the adenovirus, a subregion of the viral genome seen in converted cells, is what causes transformation[195]. Exogenous DNA is used to substitute the E1 region in the construction of recombinant human adenoviruses, which results in replication defects and a negligibly high level of acute toxicity[210]. Recombinant adenoviral vectors that express wt-p53 (Ad-p53) allow for the transfection of the p53 gene into HCC cell lines[195]. In-vivo tumour growth of neighbouring non-transduced cells can be inhibited by tumour cells transduced with the wt-p53 gene, according to experiments. This suggests that Ad-p53 is also anti-angiogenic[195,211], at least in part because of the bystander effect caused by the transfer of the wt-p53 gene on neighbouring tumour cells.
Oncolytic viruses that interfere with the key surveillance pathways, as those mediated by p53, might multiply selectively in them and cause lysis since HCC cells with p53 abnormalities have abandoned their cellular surveillance systems[195]. Adenoviruses have E1A and E1B genes, with E1A being an apoptosis-inducing gene and E1B being an apoptosis-inhibiting gene[195]. The p53 gene is bound to and rendered inactive by the 55kDa protein from the E1B-region of the adenovirus[212]. The 55-kDa E1B protein is not produced due to a loss in E1B and the mutant adenovirus known as ONYX-015[213] can only multiply in cells lacking wt-p53. The E1B55K-defective adenovirus ONYX-015 is a prototype for oncolytic viruses that may specifically reproduce in and kill p53-deficient HCC cells[195]. Unless cancer gene therapy is properly planned based on the biology of a particular tumour type[214], it is unlikely to be successful. Another E1B 55kDa-deficient adenovirus with a mouse endostatin gene has been developed for antitumour activities against HCC and is known as CNHK200-mE to increase the effectiveness of such oncolytic viruses[195]. A novel strategy known as a gene-viral vector or gene-viral therapeutic system has been developed using the synergic activity of a carrier virus and therapeutic gene[195]. In this method, an anti-tumour gene is inserted into the genome of a replicative virus specific for tumour cells in order to merge the benefits of gene and virus therapies. CNHK300-murine endostatin (CNHK300-mE) is created using the human telomerase reverse transcriptase promoter to activate the production of the adenovirus E1A gene and clone the therapeutic gene mouse endostatin into the adenovirus genome, indicating promising results in the treatment of HCC[195] (Table 1).
| NCT No. | Title | Drug | Present status | Population | Phase | Date | Location |
| NCT02418988 | Trans-catheter Chemo-embolization combined with rAd-p53 gene injection in treatment of advanced HCC | TACE plus rAd-p53 artery injection | Unknown status | Estimated enrolment: 120 participants. Age: 18 years to 80 years (Adult, older adult) | Advanced adult HCC: Phase 2 | Study start study: July 2014. Primary completion study: May 2017. Study completion date: December 2017 | China, Shanxi |
| NCT02509169 | Trans-catheter Chemo-embolization combined with p53 gene therapy for treatment of advanced HCC | TAE plus p53 gene | Unknown status | Estimated enrolment: 60 participants. Age: 18 years to 85 years | Advanced HCC: Phase 2 | Study start study: October 2014. Primary completion study: December 2016. Study completion date: December 2016 | China, Liaoning |
| NCT02561546 | p53 gene therapy in Treatment of diabetes concurrent with HCC | p53 gene therapy | Unknown status | Estimated enrolment: 40 participants. Age: 18 years to 85 years | HCC: Phase 2 | Study start study: December 2015. Primary completion study: October 2017. Study completion date: December 2017 | China, Liaoning |
| NCT00003147 | Gene therapy in treating patients with cancer of the liver | Ad5CMV-p53 gene | Terminated | Actual enrolment: 30 participants. Age: 18 years and older (Adult, older adult) | Liver cancer: Phase 2 | First posted: August 24, 2004. Last Update posted: February 5, 2013 | United States, New York. United States, Pennsylvania |
| NCT02432963 | Vaccine therapy and pembrolizumab in treating patients with solid tumours that have failed prior therapy | Modified Vaccinia Virus Ankara Vaccine Expressing p53 | Active and recruiting | Actual enrolment: 11 participants. Age: 18 years and older (Adult, older adult) | HCC: Phase 1 | First posted: May 4, 2015. Last Update posted: March 21, 2023 | United States, California |
| NCT03544723 | Safety and efficacy of p53 gene therapy combined with Immune checkpoint inhibitors in solid tumours | Ad-p53 | Unknown status | Estimated enrolment: 40 participants. Age: 18 years and older (Adult, older adult) | Solid tumour: Phase 2 | Study start study: October 1, 2018. Primary completion study: June 30, 2022. Study completion date: December 31, 2022 | United States, Illinois |
Through the direct inhibition of apoptotic pathways, the buildup of mutant p53 with gain of function can cause tumours to become resistant to treatment[215]. p53 has been linked to chemotherapeutic treatment resistance in HCC. Cisplatin, also known as cis-diamminedichloroplatin (CDDP), is an essential part of standard treatment plans for tumours of the gastrointestinal, pulmonary and genitourinary systems[216]. The response rate to CDDP in systemic monotherapy for HCC is 15%[217], however multidrug regimens containing this medication produce higher response rates[218]. Because arterial infusion schemes with CDDP reach larger concentrations of the drug inside the tumour and consequently exhibit a more potent anticancer effect, response rates range from 41% to 61%[219]. In most HCC cell lines, regardless of p53 status, a prior study revealed that CDDP has a synergistic effect with tumour necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis[218]. But an in-depth examination of the Bel7402 (p53 wild type), Huh 7 (p53 mutant) and Hep3B (p53 deficient) cell lines revealed that the p53 status is only partially necessary for CDDP to reverse TRAIL resistance in HCC cells[220]. Thus, p53 is linked to the effectiveness of CDDP in inducing apoptosis. Later research revealed that defective p53 contributes to CDDP resistance[218]. In HepG2 cells, the long interspersed nuclear element-1 ORF-1 protein (LINE-1 ORF-1p) enhances CDDP and epirubicin resistance by suppressing the production of the p53 gene[221]. As a result, HCC cells are more likely to be CDDP resistant when p53 expression is downregulated[218].
The cornerstone of chemotherapy for HCC is doxorubicin (Adriamycin, ADA), but doxorubicin resistance prevents these patients from receiving a complete course of treatment. Doxorubicin activates the p53 pathway to cause apoptosis in human HCC cells[222]. In contrast to wild-type p53, which enhances doxorubicin chemosensitivity in the drug-resistant human HCC cell line Bel7402/5-fluorouracil (5-FU), mutant p53 enhances doxorubicin resistance in HCC cell lines[205]. Doxorubicin sensitivity is reduced by p53 activation inhibition[223]. N-myc downstream regulated gene-1, a downstream target of p53, also contributes to doxorubicin resistance in HCC cells[224]. 5-FU and vincristine resistance in HCC cells is linked to p53 mutation and inactivation addition to doxorubicin resistance[218].
For HCC patients, doxorubicin and sorafenib combined therapy produce better results than doxorubicin alone does in terms of median time to progression, overall survival and progression-free survival[103]. These findings suggest that the use of sorafenib in combination with doxorubicin is effective in the treatment of HCC[218]. By reducing Bcl-2 expression, microRNA-34a, a specific target of p53, improves the susceptibility of human HCC cells to the anticancer impact of sorafenib, amplifying sorafenib-induced apoptosis and damage[225]. CDDP, doxorubicin, 5-FU and other treatments are ineffective against HCC cells due to p53 mutation and inactivation, among other factors. The G245D mutation in p53 reduces the chemosensitivity of HCC cells by making them resistant to histone deacetylase inhibitors[218]. Contrarily, dominant-negative p73 (Delta Np73) can suppress p53 function, leading to treatment resistance. Chemoresistance is caused by deltaNp63alpha's direct interference with the p53 family target genes' ability to activate transcription[226]. The mechanism underpinning p53-related chemoresistance may be started or controlled by both extrinsic and intrinsic factors because chemotherapy-induced apoptosis involving p53 in HCC is mediated by extrinsic and intrinsic pathways[218].
Dox is the cornerstone of chemotherapy for HCC, but Dox resistance prevents patients with HCC from receiving an effective course of treatment[227]. Dox activates the p53 pathway in human HCC cells to cause apoptosis. Ma et al[227] looked at the connection between Lgr5 and p53 expression in HCC in order to clarify the molecular mechanism of Dox resistance[227]. They discovered that Lgr5 stimulates p53 degradation in HCC cell lines and that there is an adverse connection between Lgr5 and p53 expression in HCC tissues. Target gene PDCD5 was found; PDCD5 is a protein involved in apoptosis and is thought to act as a tumour suppressor in a variety of tumour types, particularly lung cancer, breast cancer, and glioma[228,229]. In the presence of DNA damage, PDCD5 can interact with and stabilize p53 to encourage apoptosis[230]. They observed that PDCD5 serves as an independent predictive factor and serves as an antioncogene. Drug resistance caused by Lgr5 is only partially reversed by PDCD5 overexpression. They initially created transfected HCC cell lines that overexpressed Lgr5 and PDCD5, and we discovered that neither protein could enhance the expression of the other at the transcriptional and posttranscriptional stages[227]. This information allowed us to expand our understanding of the interaction between Lgr5 and PDCD5. In addition, they discovered that Lgr5 may bind with PDCD5 in the cytoplasm and stop it from translocating. As a result, PDCD5 was unable to bind to p53 and translocate to the nucleus, stabilizing p53. This suggests that Lgr5 Lowers the effective level of PDCD5 as opposed to the overall concentration of this protein. In HCC cells treated with Dox or even untreated cells, high levels of Lgr5 could lower p53 protein levels and prevent apoptosis[227]. They overexpressed PDCD5 in HCC cells that had been stably transfected with Lgr5 to examine how Lgr5 affected PDCD5. They discovered that this decreased the cells' ability to resist Dox and caused them to undergo apoptosis, which suggests that PDCD5 overexpression is antagonistic to Lgr5. These observations indicated that Lgr5 could prevent PDCD5 from translocating into the nucleus by binding directly to PDCD5 with its N-terminal extracellular domain and causing p53 to degrade. Recent studies demonstrating that PDCD5 may move from the cytoplasm to the nucleus after Dox treatment[231] reinforce this conclusion. Additionally, we discovered that Lgr5 prevents Dox-induced apoptosis in HCC cells via inhibiting the p53 pathway[227].
They offer a hypothesis for a brand-new mechanism of Dox resistance in HCC cells, one that emphasizes the crucial function of Lgr5, which is effectively triggered by this chemotherapy medication. When the N-terminal extracellular domain of the triggered Lgr5 directly binds to PDCD5, it inhibits PDCD5's nuclear translocation[227]. This is how the induced Lgr5 functions as a direct negative regulator of PDCD5. The lack of p53 stabilization caused by the decreasing PDCD5 levels within the nucleus causes an EMT-like phenotype, which inhibits apoptosis and eventually results in Dox resistance[227].
The recent literature was examined in this review to show the negative impacts of MDM2-p53 axis malfunction on the transformation of hepatoma cells. Despite numerous studies proposing tumour cell-specific signaling pathways that affect the MDM2-p53 axis, the mechanisms causing malfunction in this axis during the progression of HCC are intricate. To better diagnose and treat HCC at an early stage, future studies should concentrate on the characteristic features of MDM2-p53 axis malfunction in chronic progressive liver diseases that are closely linked to the onset and advancement of HCC. The balance of the MDM2-p53 axis in chronic liver illnesses could be modulated by a number of molecules, and it has been discovered that HCC tissues have severely altered levels of these molecules. By emphasizing on the malfunction of the MDM2-p53 axis during the conversion of healthy hepatocytes into hepatoma cells, we intended to offer some guidance for techniques to cure pre-HCC.
We would like to thank Dr. A. P. Pawar, Principal, Poona College of Pharmacy, Bharati Vidyapeeth (Deemed to be University), for providing the necessary facilities to carry out this research work.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Liao W, China; Qi XS, China; Xu X, China S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | Tian LY, Smit DJ, Jücker M. The Role of PI3K/AKT/mTOR Signaling in Hepatocellular Carcinoma Metabolism. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 124] [Reference Citation Analysis (0)] |
| 2. | Coates A, Gebski V, Bishop JF, Jeal PN, Woods RL, Snyder R, Tattersall MH, Byrne M, Harvey V, Gill G. Improving the quality of life during chemotherapy for advanced breast cancer. A comparison of intermittent and continuous treatment strategies. N Engl J Med. 1987;317:1490-1495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 3. | Guo XG, Wang ZH, Dong W, He XD, Liu FC, Liu H. Specific CYP450 Genotypes in the Chinese Population Affect Sorafenib Toxicity in HBV/HCV-associated Hepatocellular Carcinoma Patients. Biomed Environ Sci. 2018;31:586-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 4. | Zhou J, Wen Q, Li SF, Zhang YF, Gao N, Tian X, Fang Y, Gao J, Cui MZ, He XP, Jia LJ, Jin H, Qiao HL. Significant change of cytochrome P450s activities in patients with hepatocellular carcinoma. Oncotarget. 2016;7:50612-50623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 5. | Frión-Herrera Y, Gabbia D, Cuesta-Rubio O, De Martin S, Carrara M. Nemorosone inhibits the proliferation and migration of hepatocellular carcinoma cells. Life Sci. 2019;235:116817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 6. | Kurma K, Zeybek Kuyucu A, Roth GS, Sturm N, Mercey-Ressejac M, Abbadessa G, Yu Y, Lerat H, Marche PN, Decaens T, Macek Jilkova Z. Effect of Novel AKT Inhibitor Vevorisertib as Single Agent and in Combination with Sorafenib on Hepatocellular Carcinoma in a Cirrhotic Rat Model. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Lyon AR, Yousaf N, Battisti NML, Moslehi J, Larkin J. Immune checkpoint inhibitors and cardiovascular toxicity. Lancet Oncol. 2018;19:e447-e458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 418] [Article Influence: 69.7] [Reference Citation Analysis (0)] |
| 8. | Liu J, Ma Q, Zhang M, Wang X, Zhang D, Li W, Wang F, Wu E. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a systematic review and meta-analysis. Eur J Cancer. 2012;48:2328-2338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 9. | Lai PB, Chi TY, Chen GG. Different levels of p53 induced either apoptosis or cell cycle arrest in a doxycycline-regulated hepatocellular carcinoma cell line in vitro. Apoptosis. 2007;12:387-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Dowell SP, Wilson PO, Derias NW, Lane DP, Hall PA. Clinical utility of the immunocytochemical detection of p53 protein in cytological specimens. Cancer Res. 1994;54:2914-2918. [PubMed] |
| 11. | Park NH, Chung YH, Youn KH, Song BC, Yang SH, Kim JA, Lee HC, Yu E, Lee YS, Lee SG, Kim KW, Suh DJ. Close correlation of p53 mutation to microvascular invasion in hepatocellular carcinoma. J Clin Gastroenterol. 2001;33:397-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 12. | Müller M, Meyer M, Schilling T, Ulsperger E, Lehnert T, Zentgraf H, Stremmel W, Volkmann M, Galle PR. Testing for anti-p53 antibodies increases the diagnostic sensitivity of conventional tumor markers. Int J Oncol. 2006;29:973-980. [PubMed] |
| 13. | Yuan RH, Jeng YM, Chen HL, Lai PL, Pan HW, Hsieh FJ, Lin CY, Lee PH, Hsu HC. Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression, and is associated with tumour progression, early recurrence, and poor prognosis in hepatocellular carcinoma. J Pathol. 2006;209:549-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 126] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 14. | Azer SA. MDM2-p53 Interactions in Human Hepatocellular Carcinoma: What Is the Role of Nutlins and New Therapeutic Options? J Clin Med. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Miller C, Mohandas T, Wolf D, Prokocimer M, Rotter V, Koeffler HP. Human p53 gene localized to short arm of chromosome 17. Nature. 1986;319:783-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 148] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 16. | Arrowsmith CH, Morin P. New insights into p53 function from structural studies. Oncogene. 1996;12:1379-1385. [PubMed] |
| 17. | Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1625] [Cited by in RCA: 1723] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 18. | DeLeo AB, Jay G, Appella E, Dubois GC, Law LW, Old LJ. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc Natl Acad Sci U S A. 1979;76:2420-2424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 450] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 19. | Rotter V, Witte ON, Coffman R, Baltimore D. Abelson murine leukemia virus-induced tumors elicit antibodies against a host cell protein, P50. J Virol. 1980;36:547-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 140] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci U S A. 1994;91:2230-2234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 481] [Article Influence: 15.5] [Reference Citation Analysis (1)] |
| 21. | Koeffler HP, Miller C, Nicolson MA, Ranyard J, Bosselman RA. Increased expression of p53 protein in human leukemia cells. Proc Natl Acad Sci U S A. 1986;83:4035-4039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Steinmeyer K, Maacke H, Deppert W. Cell cycle control by p53 in normal (3T3) and chemically transformed (Meth A) mouse cells. I. Regulation of p53 expression. Oncogene. 1990;5:1691-1699. [PubMed] |
| 23. | Eliyahu D, Raz A, Gruss P, Givol D, Oren M. Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature. 1984;312:646-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 536] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 24. | Parada LF, Land H, Weinberg RA, Wolf D, Rotter V. Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature. 1984;312:649-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 546] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 25. | Aguilar F, Harris CC, Sun T, Hollstein M, Cerutti P. Geographic variation of p53 mutational profile in nonmalignant human liver. Science. 1994;264:1317-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 174] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 26. | Hollstein M, Rice K, Greenblatt MS, Soussi T, Fuchs R, Sørlie T, Hovig E, Smith-Sørensen B, Montesano R, Harris CC. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22:3551-3555. [PubMed] |
| 27. | Kraiss S, Quaiser A, Oren M, Montenarh M. Oligomerization of oncoprotein p53. J Virol. 1988;62:4737-4744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 113] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Koom WS, Park SY, Kim W, Kim M, Kim JS, Kim H, Choi IK, Yun CO, Seong J. Combination of radiotherapy and adenovirus-mediated p53 gene therapy for MDM2-overexpressing hepatocellular carcinoma. J Radiat Res. 2012;53:202-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Milner J. Forms and functions of p53. Semin Cancer Biol. 1994;5:211-219. [PubMed] |
| 30. | Rotter V, Aloni-Grinstein R, Schwartz D, Elkind NB, Simons A, Wolkowicz R, Lavigne M, Beserman P, Kapon A, Goldfinger N. Does wild-type p53 play a role in normal cell differentiation? Semin Cancer Biol. 1994;5:229-236. [PubMed] |
| 31. | Yang Y, Li CC, Weissman AM. Regulating the p53 system through ubiquitination. Oncogene. 2004;23:2096-2106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 136] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 32. | Kwon SK, Saindane M, Baek KH. p53 stability is regulated by diverse deubiquitinating enzymes. Biochim Biophys Acta Rev Cancer. 2017;1868:404-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 33. | Schlaeger C, Longerich T, Schiller C, Bewerunge P, Mehrabi A, Toedt G, Kleeff J, Ehemann V, Eils R, Lichter P, Schirmacher P, Radlwimmer B. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 160] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 34. | Meng X, Franklin DA, Dong J, Zhang Y. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res. 2014;74:7161-7167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 172] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 35. | Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 473] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 36. | Donati G, Bertoni S, Brighenti E, Vici M, Treré D, Volarevic S, Montanaro L, Derenzini M. The balance between rRNA and ribosomal protein synthesis up- and downregulates the tumour suppressor p53 in mammalian cells. Oncogene. 2011;30:3274-3288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 79] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 37. | Wang JH, Yun C, Kim S, Chae S, Lee YI, Kim WH, Lee JH, Kim W, Cho H. Reactivation of p53 in cells expressing hepatitis B virus X-protein involves p53 phosphorylation and a reduction of Hdm2. Cancer Sci. 2008;99:888-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 38. | Kremer-Tal S, Narla G, Chen Y, Hod E, DiFeo A, Yea S, Lee JS, Schwartz M, Thung SN, Fiel IM, Banck M, Zimran E, Thorgeirsson SS, Mazzaferro V, Bruix J, Martignetti JA, Llovet JM, Friedman SL. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J Hepatol. 2007;46:645-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 39. | Tarocchi M, Hannivoort R, Hoshida Y, Lee UE, Vetter D, Narla G, Villanueva A, Oren M, Llovet JM, Friedman SL. Carcinogen-induced hepatic tumors in KLF6+/- mice recapitulate aggressive human hepatocellular carcinoma associated with p53 pathway deregulation. Hepatology. 2011;54:522-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 40. | Jung CR, Lim JH, Choi Y, Kim DG, Kang KJ, Noh SM, Im DS. Enigma negatively regulates p53 through MDM2 and promotes tumor cell survival in mice. J Clin Invest. 2010;120:4493-4506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 41. | Park MY, Kim KR, Park HS, Park BH, Choi HN, Jang KY, Chung MJ, Kang MJ, Lee DG, Moon WS. Expression of the serum response factor in hepatocellular carcinoma: implications for epithelial-mesenchymal transition. Int J Oncol. 2007;31:1309-1315. [PubMed] |
| 42. | Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32:509-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 114] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 43. | Dongiovanni P, Fracanzani AL, Cairo G, Megazzini CP, Gatti S, Rametta R, Fargion S, Valenti L. Iron-dependent regulation of MDM2 influences p53 activity and hepatic carcinogenesis. Am J Pathol. 2010;176:1006-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 44. | Menendez JA, Joven J, Cufí S, Corominas-Faja B, Oliveras-Ferraros C, Cuyàs E, Martin-Castillo B, López-Bonet E, Alarcón T, Vazquez-Martin A. The Warburg effect version 2.0: metabolic reprogramming of cancer stem cells. Cell Cycle. 2013;12:1166-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 45. | Meng X, Leslie P, Zhang Y, Dong J. Stem cells in a three-dimensional scaffold environment. Springerplus. 2014;3:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 46. | Zhang YY, Zhou LM. Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochem Biophys Res Commun. 2012;423:26-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 47. | Dominguez-Malagón H, Gaytan-Graham S. Hepatocellular carcinoma: an update. Ultrastruct Pathol. 2001;25:497-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 59] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 48. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4464] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 49. | Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 245] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 50. | Hayes CN, Zhang P, Zhang Y, Chayama K. Molecular Mechanisms of Hepatocarcinogenesis Following Sustained Virological Response in Patients with Chronic Hepatitis C Virus Infection. Viruses. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (1)] |
| 51. | Oda T, Tsuda H, Scarpa A, Sakamoto M, Hirohashi S. p53 gene mutation spectrum in hepatocellular carcinoma. Cancer Res. 1992;52:6358-6364. [PubMed] |
| 52. | Tsuda H, Hirohashi S, Shimosato Y, Ino Y, Yoshida T, Terada M. Low incidence of point mutation of c-Ki-ras and N-ras oncogenes in human hepatocellular carcinoma. Jpn J Cancer Res. 1989;80:196-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 53. | Vogelstein B, Kinzler KW. p53 function and dysfunction. Cell. 1992;70:523-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1325] [Cited by in RCA: 1346] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 54. | Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252:1708-1711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 775] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 55. | Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1786] [Cited by in RCA: 1792] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 56. | Amundson SA, Myers TG, Fornace AJ Jr. Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene. 1998;17:3287-3299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 286] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 57. | el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 575] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 58. | Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ Jr. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2143] [Cited by in RCA: 2179] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 59. | Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 906] [Cited by in RCA: 929] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 60. | Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5482] [Cited by in RCA: 5460] [Article Influence: 195.0] [Reference Citation Analysis (0)] |
| 61. | Anzola M, Burgos JJ. Hepatocellular carcinoma: molecular interactions between hepatitis C virus and p53 in hepatocarcinogenesis. Expert Rev Mol Med. 2003;5:1-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 62. | Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2565] [Cited by in RCA: 2682] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 63. | Harris CC, Hollstein M. Clinical implications of the p53 tumor-suppressor gene. N Engl J Med. 1993;329:1318-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 943] [Cited by in RCA: 931] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 64. | Ljungman M. Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia. 2000;2:208-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 154] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 65. | Shaulsky G, Ben-Ze'ev A, Rotter V. Subcellular distribution of the p53 protein during the cell cycle of Balb/c 3T3 cells. Oncogene. 1990;5:1707-1711. [PubMed] |
| 66. | Lilling G, Nordenberg J, Rotter V, Goldfinger N, Peller S, Sidi Y. Altered subcellular localization of p53 in estrogen-dependent and estrogen-independent breast cancer cells. Cancer Invest. 2002;20:509-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 67. | Bosari S, Moneghini L, Graziani D, Lee AK, Murray JJ, Coggi G, Viale G. bcl-2 oncoprotein in colorectal hyperplastic polyps, adenomas, and adenocarcinomas. Hum Pathol. 1995;26:534-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 120] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 68. | Takada S, Kaneniwa N, Tsuchida N, Koike K. Cytoplasmic retention of the p53 tumor suppressor gene product is observed in the hepatitis B virus X gene-transfected cells. Oncogene. 1997;15:1895-1901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 69. | Truant R, Antunovic J, Greenblatt J, Prives C, Cromlish JA. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J Virol. 1995;69:1851-1859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 187] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 70. | Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1167] [Cited by in RCA: 1153] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 71. | Thomas M, Pim D, Banks L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene. 1999;18:7690-7700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 322] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 72. | Ahn JY, Chung EY, Kwun HJ, Jang KL. Transcriptional repression of p21(waf1) promoter by hepatitis B virus X protein via a p53-independent pathway. Gene. 2001;275:163-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 73. | Ahn JY, Jung EY, Kwun HJ, Lee CW, Sung YC, Jang KL. Dual effects of hepatitis B virus X protein on the regulation of cell-cycle control depending on the status of cellular p53. J Gen Virol. 2002;83:2765-2772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 74. | Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res. 2003;282:59-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 189] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 75. | O'Keefe K, Li H, Zhang Y. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol Cell Biol. 2003;23:6396-6405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 100] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 76. | Atz J, Wagner P, Roemer K. Function, oligomerization, and conformation of tumor-associated p53 proteins with mutated C-terminus. J Cell Biochem. 2000;76:572-584. [PubMed] [DOI] [Full Text] |
| 77. | Liu MC, Gelmann EP. P53 gene mutations: case study of a clinical marker for solid tumors. Semin Oncol. 2002;29:246-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 78. | Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855-4878. [PubMed] |
| 79. | Nishida N, Fukuda Y, Kokuryu H, Toguchida J, Yandell DW, Ikenega M, Imura H, Ishizaki K. Role and mutational heterogeneity of the p53 gene in hepatocellular carcinoma. Cancer Res. 1993;53:368-372. [PubMed] |
| 80. | Shen HM, Ong CN. Mutations of the p53 tumor suppressor gene and ras oncogenes in aflatoxin hepatocarcinogenesis. Mutat Res. 1996;366:23-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 110] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 81. | Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 108] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 82. | Okada F, Shiraki T, Maekawa M, Sato S. A p53 polymorphism associated with increased risk of hepatitis C virus infection. Cancer Lett. 2001;172:137-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 83. | Shank RC. Metabolic activation of mycotoxins by animals and humans: an overview. J Toxicol Environ Health. 1977;2:1229-1244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 84. | Hsieh DP, Atkinson DN. Bisfuranoid mycotoxins: their genotoxicity and carcinogenicity. Adv Exp Med Biol. 1991;283:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 85. | Liu Y, Chang CC, Marsh GM, Wu F. Population attributable risk of aflatoxin-related liver cancer: systematic review and meta-analysis. Eur J Cancer. 2012;48:2125-2136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 262] [Cited by in RCA: 217] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 86. | Link T, Iwakuma T. Roles of p53 in extrinsic factor-induced liver carcinogenesis. Hepatoma Res. 2017;3:95-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 87. | Gross-Steinmeyer K, Eaton DL. Dietary modulation of the biotransformation and genotoxicity of aflatoxin B(1). Toxicology. 2012;299:69-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 88. | Smela ME, Currier SS, Bailey EA, Essigmann JM. The chemistry and biology of aflatoxin B(1): from mutational spectrometry to carcinogenesis. Carcinogenesis. 2001;22:535-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 191] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 89. | Aguilar F, Hussain SP, Cerutti P. Aflatoxin B1 induces the transversion of G-->T in codon 249 of the p53 tumor suppressor gene in human hepatocytes. Proc Natl Acad Sci U S A. 1993;90:8586-8590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 287] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 90. | Ming L, Thorgeirsson SS, Gail MH, Lu P, Harris CC, Wang N, Shao Y, Wu Z, Liu G, Wang X, Sun Z. Dominant role of hepatitis B virus and cofactor role of aflatoxin in hepatocarcinogenesis in Qidong, China. Hepatology. 2002;36:1214-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 160] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 91. | Puisieux A, Lim S, Groopman J, Ozturk M. Selective targeting of p53 gene mutational hotspots in human cancers by etiologically defined carcinogens. Cancer Res. 1991;51:6185-6189. [PubMed] |
| 92. | Ricordy R, Gensabella G, Cacci E, Augusti-Tocco G. Impairment of cell cycle progression by aflatoxin B1 in human cell lines. Mutagenesis. 2002;17:241-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 93. | Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989-5005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 608] [Cited by in RCA: 680] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 94. | Dragani TA, Zocchetti C. Occupational exposure to vinyl chloride and risk of hepatocellular carcinoma. Cancer Causes Control. 2008;19:1193-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 95. | Dogliotti E. Molecular mechanisms of carcinogenesis by vinyl chloride. Ann Ist Super Sanita. 2006;42:163-169. [PubMed] |
| 96. | Mastrangelo G, Fedeli U, Fadda E, Valentini F, Agnesi R, Magarotto G, Marchì T, Buda A, Pinzani M, Martines D. Increased risk of hepatocellular carcinoma and liver cirrhosis in vinyl chloride workers: synergistic effect of occupational exposure with alcohol intake. Environ Health Perspect. 2004;112:1188-1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 97. | Sherman M. Vinyl chloride and the liver. J Hepatol. 2009;51:1074-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 98. | el Ghissassi F, Barbin A, Bartsch H. Metabolic activation of vinyl chloride by rat liver microsomes: low-dose kinetics and involvement of cytochrome P450 2E1. Biochem Pharmacol. 1998;55:1445-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 99. | Trivers GE, Cawley HL, DeBenedetti VM, Hollstein M, Marion MJ, Bennett WP, Hoover ML, Prives CC, Tamburro CC, Harris CC. Anti-p53 antibodies in sera of workers occupationally exposed to vinyl chloride. J Natl Cancer Inst. 1995;87:1400-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 100. | Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5532] [Cited by in RCA: 5487] [Article Influence: 161.4] [Reference Citation Analysis (0)] |
| 101. | Barbin A, Froment O, Boivin S, Marion MJ, Belpoggi F, Maltoni C, Montesano R. p53 gene mutation pattern in rat liver tumors induced by vinyl chloride. Cancer Res. 1997;57:1695-1698. [PubMed] |
| 102. | Smith SJ, Li Y, Whitley R, Marion MJ, Partilo S, Carney WP, Brandt-Rauf PW. Molecular epidemiology of p53 protein mutations in workers exposed to vinyl chloride. Am J Epidemiol. 1998;147:302-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 103. | Abou-Alfa GK, Johnson P, Knox JJ, Capanu M, Davidenko I, Lacava J, Leung T, Gansukh B, Saltz LB. Doxorubicin plus sorafenib vs doxorubicin alone in patients with advanced hepatocellular carcinoma: a randomized trial. JAMA. 2010;304:2154-2160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 340] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 104. | Yan G, Wang X, Sun C, Zheng X, Wei H, Tian Z, Sun R. Chronic Alcohol Consumption Promotes Diethylnitrosamine-Induced Hepatocarcinogenesis via Immune Disturbances. Sci Rep. 2017;7:2567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 105. | Ramadori P, Cubero FJ, Liedtke C, Trautwein C, Nevzorova YA. Alcohol and Hepatocellular Carcinoma: Adding Fuel to the Flame. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 106. | Ogunwobi OO, Harricharran T, Huaman J, Galuza A, Odumuwagun O, Tan Y, Ma GX, Nguyen MT. Mechanisms of hepatocellular carcinoma progression. World J Gastroenterol. 2019;25:2279-2293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 116] [Cited by in RCA: 168] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 107. | Ogunwobi OO, Puszyk W, Dong HJ, Liu C. Epigenetic upregulation of HGF and c-Met drives metastasis in hepatocellular carcinoma. PLoS One. 2013;8:e63765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 108. | Calzadilla Bertot L, Adams LA. The Natural Course of Non-Alcoholic Fatty Liver Disease. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 339] [Cited by in RCA: 457] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 109. | Bedogni G, Nobili V, Tiribelli C. Epidemiology of fatty liver: an update. World J Gastroenterol. 2014;20:9050-9054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 110. | Tovoli F, Ferri S, Piscaglia F. Hepatocellular Carcinoma in Non Alcoholic Fatty Liver Disease. Curr Pharm Des. 2020;26:3909-3914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 111. | Ertle J, Dechêne A, Sowa JP, Penndorf V, Herzer K, Kaiser G, Schlaak JF, Gerken G, Syn WK, Canbay A. Non-alcoholic fatty liver disease progresses to hepatocellular carcinoma in the absence of apparent cirrhosis. Int J Cancer. 2011;128:2436-2443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 382] [Article Influence: 27.3] [Reference Citation Analysis (1)] |
| 112. | Sanyal AJ, Yoon SK, Lencioni R. The etiology of hepatocellular carcinoma and consequences for treatment. Oncologist. 2010;15 Suppl 4:14-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 356] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 113. | Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10:656-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 771] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 114. | Castro RE, Ferreira DM, Afonso MB, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CM. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J Hepatol. 2013;58:119-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 288] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 115. | Tian XF, Ji FJ, Zang HL, Cao H. Activation of the miR-34a/SIRT1/p53 Signaling Pathway Contributes to the Progress of Liver Fibrosis via Inducing Apoptosis in Hepatocytes but Not in HSCs. PLoS One. 2016;11:e0158657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 116. | Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 371] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 117. | Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745-752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1580] [Cited by in RCA: 1574] [Article Influence: 87.4] [Reference Citation Analysis (0)] |
| 118. | Chifman J, Laubenbacher R, Torti SV. A systems biology approach to iron metabolism. Adv Exp Med Biol. 2014;844:201-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 119. | Siddique A, Kowdley KV. Review article: the iron overload syndromes. Aliment Pharmacol Ther. 2012;35:876-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 120. | Heeney MM. Iron clad: iron homeostasis and the diagnosis of hereditary iron overload. Hematology Am Soc Hematol Educ Program. 2014;2014:202-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 121. | Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 936] [Cited by in RCA: 1152] [Article Influence: 96.0] [Reference Citation Analysis (0)] |
| 122. | Kew MC. Hepatocellular carcinoma in African Blacks: Recent progress in etiology and pathogenesis. World J Hepatol. 2010;2:65-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 123. | Asare GA, Paterson AC, Kew MC, Khan S, Mossanda KS. Iron-free neoplastic nodules and hepatocellular carcinoma without cirrhosis in Wistar rats fed a diet high in iron. J Pathol. 2006;208:82-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 124. | Kang JO, Jones C, Brothwell B. Toxicity associated with iron overload found in hemochromatosis: possible mechanism in a rat model. Clin Lab Sci. 1998;11:350-354. [PubMed] |
| 125. | Shen J, Sheng X, Chang Z, Wu Q, Wang S, Xuan Z, Li D, Wu Y, Shang Y, Kong X, Yu L, Li L, Ruan K, Hu H, Huang Y, Hui L, Xie D, Wang F, Hu R. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Rep. 2014;7:180-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 126. | Bloomer SA, Brown KE. Tumour promotion versus tumour suppression in chronic hepatic iron overload. Cell Biochem Funct. 2015;33:241-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 127. | Zhang F, Wang W, Tsuji Y, Torti SV, Torti FM. Post-transcriptional modulation of iron homeostasis during p53-dependent growth arrest. J Biol Chem. 2008;283:33911-33918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 128. | Funauchi Y, Tanikawa C, Yi Lo PH, Mori J, Daigo Y, Takano A, Miyagi Y, Okawa A, Nakamura Y, Matsuda K. Regulation of iron homeostasis by the p53-ISCU pathway. Sci Rep. 2015;5:16497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 129. | Vautier G, Bomford AB, Portmann BC, Metivier E, Williams R, Ryder SD. p53 mutations in british patients with hepatocellular carcinoma: clustering in genetic hemochromatosis. Gastroenterology. 1999;117:154-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 50] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 130. | Marrogi AJ, Khan MA, van Gijssel HE, Welsh JA, Rahim H, Demetris AJ, Kowdley KV, Hussain SP, Nair J, Bartsch H, Okby N, Poirier MC, Ishak KG, Harris CC. Oxidative stress and p53 mutations in the carcinogenesis of iron overload-associated hepatocellular carcinoma. J Natl Cancer Inst. 2001;93:1652-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 131. | Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212-2219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1217] [Cited by in RCA: 1328] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 132. | Chan SL, Wong VW, Qin S, Chan HL. Infection and Cancer: The Case of Hepatitis B. J Clin Oncol. 2016;34:83-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 128] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 133. | Tarocchi M, Polvani S, Marroncini G, Galli A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J Gastroenterol. 2014;20:11630-11640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 132] [Cited by in RCA: 142] [Article Influence: 12.9] [Reference Citation Analysis (2)] |
| 134. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4263] [Article Influence: 236.8] [Reference Citation Analysis (2)] |
| 135. | Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26 Suppl 1:144-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 213] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 136. | Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 625] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 137. | Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 880] [Cited by in RCA: 867] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 138. | Ueda H, Ullrich SJ, Gangemi JD, Kappel CA, Ngo L, Feitelson MA, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 254] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 139. | Wang HD, Trivedi A, Johnson DL. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol Cell Biol. 1997;17:6838-6846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 140. | Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, Liu W. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10:416-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 141. | Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273:381-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 164] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 142. | Schuster R, Gerlich WH, Schaefer S. Induction of apoptosis by the transactivating domains of the hepatitis B virus X gene leads to suppression of oncogenic transformation of primary rat embryo fibroblasts. Oncogene. 2000;19:1173-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 143. | Wang Y, Lau SH, Sham JS, Wu MC, Wang T, Guan XY. Characterization of HBV integrants in 14 hepatocellular carcinomas: association of truncated X gene and hepatocellular carcinogenesis. Oncogene. 2004;23:142-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 89] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 144. | Liu LP, Hu BG, Ye C, Ho RL, Chen GG, Lai PB. HBx mutants differentially affect the activation of hypoxia-inducible factor-1α in hepatocellular carcinoma. Br J Cancer. 2014;110:1066-1073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 145. | Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M, Paterlini-Bréchot P, Bréchot C, Kremsdorf D. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803-7810. [PubMed] |
| 146. | Zhu R, Mok MT, Kang W, Lau SS, Yip WK, Chen Y, Lai PB, Wong VW, To KF, Sung JJ, Cheng AS, Chan HL. Truncated HBx-dependent silencing of GAS2 promotes hepatocarcinogenesis through deregulation of cell cycle, senescence and p53-mediated apoptosis. J Pathol. 2015;237:38-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 147. | Long J, Wang Y, Li M, Tong WM, Jia JD, Huang J. Correlation of TP53 mutations with HCV positivity in hepatocarcinogenesis: identification of a novel TP53 microindel in hepatocellular carcinoma with HCV infection. Oncol Rep. 2013;30:119-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 148. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2504] [Article Influence: 192.6] [Reference Citation Analysis (2)] |
| 149. | Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991;88:2451-2455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1133] [Cited by in RCA: 1131] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 150. | Bukh J. The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol. 2016;65:S2-S21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 176] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 151. | Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 912] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 152. | Kao CF, Chen SY, Chen JY, Wu Lee YH. Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein. Oncogene. 2004;23:2472-2483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 99] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 153. | He QQ, Cheng RX, Sun Y, Feng DY, Chen ZC, Zheng H. Hepatocyte transformation and tumor development induced by hepatitis C virus NS3 c-terminal deleted protein. World J Gastroenterol. 2003;9:474-478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 154. | Ishido S, Hotta H. Complex formation of the nonstructural protein 3 of hepatitis C virus with the p53 tumor suppressor. FEBS Lett. 1998;438:258-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 54] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 155. | Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893-3896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 210] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 156. | Ghosh AK, Majumder M, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein protects against TNF-alpha mediated apoptotic cell death. Virus Res. 2000;67:173-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 157. | Gong GZ, Jiang YF, He Y, Lai LY, Zhu YH, Su XS. HCV NS5A abrogates p53 protein function by interfering with p53-DNA binding. World J Gastroenterol. 2004;10:2223-2227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 158. | Lee MN, Jung EY, Kwun HJ, Jun HK, Yu DY, Choi YH, Jang KL. Hepatitis C virus core protein represses the p21 promoter through inhibition of a TGF-beta pathway. J Gen Virol. 2002;83:2145-2151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 159. | Yamanaka T, Kodama T, Doi T. Subcellular localization of HCV core protein regulates its ability for p53 activation and p21 suppression. Biochem Biophys Res Commun. 2002;294:528-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 160. | Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J Biol Chem. 2002;277:17713-17721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 113] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 161. | Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4008] [Cited by in RCA: 4200] [Article Influence: 131.3] [Reference Citation Analysis (0)] |
| 162. | Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 559] [Cited by in RCA: 592] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 163. | Bénard J, Douc-Rasy S, Ahomadegbe JC. TP53 family members and human cancers. Hum Mutat. 2003;21:182-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 162] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 164. | Feng DY, Chen RX, Peng Y, Zheng H, Yan YH. Effect of HCV NS(3) protein on p53 protein expression in hep atocarcinogenesis. World J Gastroenterol. 1999;5:45-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 165. | Muramatsu S, Ishido S, Fujita T, Itoh M, Hotta H. Nuclear localization of the NS3 protein of hepatitis C virus and factors affecting the localization. J Virol. 1997;71:4954-4961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 166. | Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol. 2005;25:7423-7431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 246] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 167. | Khabar KS, Polyak SJ. Hepatitis C virus-host interactions: the NS5A protein and the interferon/chemokine systems. J Interferon Cytokine Res. 2002;22:1005-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 168. | Reyes GR. The nonstructural NS5A protein of hepatitis C virus: an expanding, multifunctional role in enhancing hepatitis C virus pathogenesis. J Biomed Sci. 2002;9:187-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 169. | Ghosh AK, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol. 1999;80 ( Pt 5):1179-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 146] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 170. | Otsuka M, Kato N, Lan K, Yoshida H, Kato J, Goto T, Shiratori Y, Omata M. Hepatitis C virus core protein enhances p53 function through augmentation of DNA binding affinity and transcriptional ability. J Biol Chem. 2000;275:34122-34130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 132] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 171. | Singh AK, Kumar R, Pandey AK. Hepatocellular Carcinoma: Causes, Mechanism of Progression and Biomarkers. Curr Chem Genom Transl Med. 2018;12:9-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 172. | Agarwal N, Adhikari AS, Iyer SV, Hekmatdoost K, Welch DR, Iwakuma T. MTBP suppresses cell migration and filopodia formation by inhibiting ACTN4. Oncogene. 2013;32:462-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 173. | Shi D, Gu W. Dual Roles of MDM2 in the Regulation of p53: Ubiquitination Dependent and Ubiquitination Independent Mechanisms of MDM2 Repression of p53 Activity. Genes Cancer. 2012;3:240-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 174. | Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 584] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 175. | Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306-1316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 393] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 176. | Kapoor NR, Ahuja R, Shukla SK, Kumar V. The HBx protein of hepatitis B virus confers resistance against nucleolar stress and anti-cancer drug-induced p53 expression. FEBS Lett. 2013;587:1287-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 177. | Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 923] [Cited by in RCA: 816] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 178. | Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902-8912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 435] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 179. | He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130-1134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2135] [Cited by in RCA: 2102] [Article Influence: 116.8] [Reference Citation Analysis (0)] |
| 180. | Bi Q, Tang S, Xia L, Du R, Fan R, Gao L, Jin J, Liang S, Chen Z, Xu G, Nie Y, Wu K, Liu J, Shi Y, Ding J, Fan D. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One. 2012;7:e40169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 181. | Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 527] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 182. | Nakao K, Miyaaki H, Ichikawa T. Antitumor function of microRNA-122 against hepatocellular carcinoma. J Gastroenterol. 2014;49:589-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 183. | Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875-883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 404] [Cited by in RCA: 447] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 184. | Phillips A, Jones CJ, Blaydes JP. The mechanisms of regulation of Hdm2 protein level by serum growth factors. FEBS Lett. 2006;580:300-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 185. | Zhou C, Liu G, Wang L, Lu Y, Yuan L, Zheng L, Chen F, Peng F, Li X. MiR-339-5p regulates the growth, colony formation and metastasis of colorectal cancer cells by targeting PRL-1. PLoS One. 2013;8:e63142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 186. | Min SH, Kim DM, Heo YS, Kim YI, Kim HM, Kim J, Han YM, Kim IC, Yoo OJ. New p53 target, phosphatase of regenerating liver 1 (PRL-1) downregulates p53. Oncogene. 2009;28:545-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 187. | Min SH, Kim DM, Heo YS, Kim HM, Kim IC, Yoo OJ. Downregulation of p53 by phosphatase of regenerating liver 3 is mediated by MDM2 and PIRH2. Life Sci. 2010;86:66-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 188. | Bi Q, Ranjan A, Fan R, Agarwal N, Welch DR, Weinman SA, Ding J, Iwakuma T. MTBP inhibits migration and metastasis of hepatocellular carcinoma. Clin Exp Metastasis. 2015;32:301-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 189. | Vlatković N, El-Fert A, Devling T, Ray-Sinha A, Gore DM, Rubbi CP, Dodson A, Jones AS, Helliwell TR, Jones TM, Boyd MT. Loss of MTBP expression is associated with reduced survival in a biomarker-defined subset of patients with squamous cell carcinoma of the head and neck. Cancer. 2011;117:2939-2950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 190. | Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, Onel K, Yip L, Hwang SJ, Strong LC, Lozano G, Levine AJ. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 926] [Cited by in RCA: 980] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 191. | Hirohashi S. Pathology and molecular mechanisms of multistage human hepatocarcinogenesis. Princess Takamatsu Symp. 1991;22:87-93. [PubMed] |
| 192. | Elmore LW, Hancock AR, Chang SF, Wang XW, Chang S, Callahan CP, Geller DA, Will H, Harris CC. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc Natl Acad Sci U S A. 1997;94:14707-14712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 253] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 193. | Gurova KV, Hill JE, Guo C, Prokvolit A, Burdelya LG, Samoylova E, Khodyakova AV, Ganapathi R, Ganapathi M, Tararova ND, Bosykh D, Lvovskiy D, Webb TR, Stark GR, Gudkov AV. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc Natl Acad Sci U S A. 2005;102:17448-17453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 224] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 194. | Horowitz J. Adenovirus-mediated p53 gene therapy: overview of preclinical studies and potential clinical applications. Curr Opin Mol Ther. 1999;1:500-509. [PubMed] |
| 195. | Guan YS, La Z, Yang L, He Q, Li P. p53 gene in treatment of hepatic carcinoma: status quo. World J Gastroenterol. 2007;13:985-992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 196. | Wang W, Rastinejad F, El-Deiry WS. Restoring p53-dependent tumor suppression. Cancer Biol Ther. 2003;2:S55-S63. [PubMed] |
| 197. | Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2174] [Cited by in RCA: 2310] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 198. | Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1257] [Cited by in RCA: 1323] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 199. | Zhang, Wang H. MDM2 oncogene as a novel target for human cancer therapy. Curr Pharm Des. 2000;6:393-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 86] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 200. | Stearns T, Willingham MC, Botstein D, Kahn RA. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc Natl Acad Sci U S A. 1990;87:1238-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 290] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 201. | Rocha S, Perkins ND. ARF the integrator: linking NF-kappaB, p53 and checkpoint kinases. Cell Cycle. 2005;4:756-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 202. | Brown DR, Deb S, Muñoz RM, Subler MA, Deb SP. The tumor suppressor p53 and the oncoprotein simian virus 40 T antigen bind to overlapping domains on the MDM2 protein. Mol Cell Biol. 1993;13:6849-6857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 204. | Oliveira AM, Ross JS, Fletcher JA. Tumor suppressor genes in breast cancer: the gatekeepers and the caretakers. Am J Clin Pathol. 2005;124 Suppl:S16-S28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 205. | Chan KT, Lung ML. Mutant p53 expression enhances drug resistance in a hepatocellular carcinoma cell line. Cancer Chemother Pharmacol. 2004;53:519-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 79] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 206. | Li YX, Lin ZB, Tan HR. Wild type p53 increased chemosensitivity of drug-resistant human hepatocellular carcinoma Bel7402/5-FU cells. Acta Pharmacol Sin. 2004;25:76-82. [PubMed] |
| 207. | Cherubini G, Petouchoff T, Grossi M, Piersanti S, Cundari E, Saggio I. E1B55K-deleted adenovirus (ONYX-015) overrides G1/S and G2/M checkpoints and causes mitotic catastrophe and endoreduplication in p53-proficient normal cells. Cell Cycle. 2006;5:2244-2252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 208. | Zhang LQ, Mei YF, Wadell G. Human adenovirus serotypes 4 and 11 show higher binding affinity and infectivity for endothelial and carcinoma cell lines than serotype 5. J Gen Virol. 2003;84:687-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 209. | Ebner K, Rauch M, Preuner S, Lion T. Typing of human adenoviruses in specimens from immunosuppressed patients by PCR-fragment length analysis and real-time quantitative PCR. J Clin Microbiol. 2006;44:2808-2815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 210. | Wills KN, Maneval DC, Menzel P, Harris MP, Sutjipto S, Vaillancourt MT, Huang WM, Johnson DE, Anderson SC, Wen SF. Development and characterization of recombinant adenoviruses encoding human p53 for gene therapy of cancer. Hum Gene Ther. 1994;5:1079-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 160] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 211. | Nishizaki M, Fujiwara T, Tanida T, Hizuta A, Nishimori H, Tokino T, Nakamura Y, Bouvet M, Roth JA, Tanaka N. Recombinant adenovirus expressing wild-type p53 is antiangiogenic: a proposed mechanism for bystander effect. Clin Cancer Res. 1999;5:1015-1023. [PubMed] |
| 212. | Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff DD, Kirn DH. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 663] [Cited by in RCA: 636] [Article Influence: 22.7] [Reference Citation Analysis (1)] |
| 213. | Rothmann T, Hengstermann A, Whitaker NJ, Scheffner M, zur Hausen H. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J Virol. 1998;72:9470-9478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 243] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 214. | Stoff-Khalili MA, Dall P, Curiel DT. Gene therapy for carcinoma of the breast. Cancer Gene Ther. 2006;13:633-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 215. | Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2022] [Cited by in RCA: 2066] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 216. | Go RS, Adjei AA. Review of the comparative pharmacology and clinical activity of cisplatin and carboplatin. J Clin Oncol. 1999;17:409-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 509] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 217. | Okada S, Okazaki N, Nose H, Shimada Y, Yoshimori M, Aoki K. A phase 2 study of cisplatin in patients with hepatocellular carcinoma. Oncology. 1993;50:22-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 218. | Li S, Gao M, Li Z, Song L, Gao X, Han J, Wang F, Chen Y, Li W, Yang J. p53 and P-glycoprotein influence chemoresistance in hepatocellular carcinoma. Front Biosci (Elite Ed). 2018;10:461-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 219. | Ikeda M, Maeda S, Shibata J, Muta R, Ashihara H, Tanaka M, Fujiyama S, Tomita K. Transcatheter arterial chemotherapy with and without embolization in patients with hepatocellular carcinoma. Oncology. 2004;66:24-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 65] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 220. | Zhang B, Shan H, Li D, Li ZR, Zhu KS, Jiang ZB, Huang MS. Cisplatin sensitizes human hepatocellular carcinoma cells, but not hepatocytes and mesenchymal stem cells, to TRAIL within a therapeutic window partially depending on the upregulation of DR5. Oncol Rep. 2011;25:461-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 221. | Feng F, Lu YY, Zhang F, Gao XD, Zhang CF, Meredith A, Xu ZX, Yang YT, Chang XJ, Wang H, Qu JH, Zeng Z, Yang JL, Wang CP, Zhu YF, Cui JJ, Yang YP. Long interspersed nuclear element ORF-1 protein promotes proliferation and resistance to chemotherapy in hepatocellular carcinoma. World J Gastroenterol. 2013;19:1068-1078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 222. | Zheng T, Wang J, Song X, Meng X, Pan S, Jiang H, Liu L. Nutlin-3 cooperates with doxorubicin to induce apoptosis of human hepatocellular carcinoma cells through p53 or p73 signaling pathways. J Cancer Res Clin Oncol. 2010;136:1597-1604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 223. | Fang F, Yang L, Tao Y, Qin W. FBI-1 promotes cell proliferation and enhances resistance to chemotherapy of hepatocellular carcinoma in vitro and in vivo. Cancer. 2012;118:134-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 224. | Jung EU, Yoon JH, Lee YJ, Lee JH, Kim BH, Yu SJ, Myung SJ, Kim YJ, Lee HS. Hypoxia and retinoic acid-inducible NDRG1 expression is responsible for doxorubicin and retinoic acid resistance in hepatocellular carcinoma cells. Cancer Lett. 2010;298:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 225. | Yang F, Li QJ, Gong ZB, Zhou L, You N, Wang S, Li XL, Li JJ, An JZ, Wang DS, He Y, Dou KF. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat. 2014;13:77-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 137] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 226. | Mundt HM, Stremmel W, Melino G, Krammer PH, Schilling T, Müller M. Dominant negative (DeltaN) p63alpha induces drug resistance in hepatocellular carcinoma by interference with apoptosis signaling pathways. Biochem Biophys Res Commun. 2010;396:335-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 227. | Ma Z, Guo D, Wang Q, Liu P, Xiao Y, Wu P, Wang Y, Chen B, Liu Z, Liu Q. Lgr5-mediated p53 Repression through PDCD5 leads to doxorubicin resistance in Hepatocellular Carcinoma. Theranostics. 2019;9:2967-2983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 228. | Murshed F, Farhana L, Dawson MI, Fontana JA. NF-κB p65 recruited SHP regulates PDCD5-mediated apoptosis in cancer cells. Apoptosis. 2014;19:506-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 229. | Li H, Zhang X, Song X, Zhu F, Wang Q, Guo C, Liu C, Shi Y, Ma C, Wang X, Zhang L. PDCD5 promotes cisplatin-induced apoptosis of glioma cells via activating mitochondrial apoptotic pathway. Cancer Biol Ther. 2012;13:822-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 230. | Xu L, Hu J, Zhao Y, Xiao J, Wang Y, Ma D, Chen Y. PDCD5 interacts with p53 and functions as a positive regulator in the p53 pathway. Apoptosis. 2012;17:1235-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 231. | Yang M, Li Y, Shen X, Ruan Y, Lu Y, Jin X, Song P, Guo Y, Zhang X, Qu H, Shao Y, Quan C. CLDN6 promotes chemoresistance through GSTP1 in human breast cancer. J Exp Clin Cancer Res. 2017;36:157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |