Published online Jun 26, 2014. doi: 10.4330/wjc.v6.i6.376
Revised: January 16, 2014
Accepted: May 8, 2014
Published online: June 26, 2014
Processing time: 195 Days and 16.6 Hours
PTPN22 has been previously found associated with coronary artery disease (CAD). In the present note we have studied the effect of p53 codon 72, acid phosphatse locus 1 (ACP1) and adenosine deaminase (ADA) genetic polymorphism on the strength of association between PTPN22 and CAD. We have studied 133 non diabetic subjects with CAD, 122 non diabetic cardiovascular patients without CAD and 269 healthy blood donors. Informed written consent was obtained from all subjects and the study was approved by the Ethical Committee. A high significant association between PTPN22 and CAD is observed in carriers of *A allele of ACP1 with a higher proportion of *T allele carriers in non diabetic subjects with CAD as compared to controls and to non diabetic subjects with cardiovascular disease without CAD. A similar pattern is observed in carriers of *Pro allele of p53 codon 72 with a higher proportion of *T allele carriers in non diabetic subjects with CAD as compared to other groups. A highly significant association between PTPN22 and CAD is observed in carriers of ADA2 *2 allele with higher proportion of *T allele carriers in non diabetic subjects with CAD as compared to other group. There is a high significant correlation between the number of factors that contributes to increase the strength of association between PTPN22 *T and CAD and the proportion of *T carriers in CAD. ACP1, p53 codon 72 and ADA are involved in immune reaction and give an important additive contribution to the strength of association between PTPN22 and CAD. This study stresses the importance of the simultaneous analysis of multiple genes functionally related to a specific disease: the approach may give important hints to understand multifactorial disorders.
Core tip: Acid phosphatse locus 1, p53 codon 72 and adenosine deaminase have an important role in immune reactions and influence the strength of association between coronary artery disease (CAD) and PTPN22 an enzyme involved in autoimmunity. These results agree with multifactorial origin of CAD.
- Citation: Gloria-Bottini F, Saccucci P, Banci M, Nardi P, Scognamiglio M, Pellegrino A, Bottini E, Chiariello L. Effect of genetic factors on the association between coronary artery disease and PTPN22 polymorphism. World J Cardiol 2014; 6(6): 376-380
- URL: https://www.wjgnet.com/1949-8462/full/v6/i6/376.htm
- DOI: https://dx.doi.org/10.4330/wjc.v6.i6.376
PTPN22 gene encodes a protein tyrosine phosphatase expressed principally in lymphoid tissue and it is also named Lyp. PTPN22 protein is involved in the control of immune system activity. The gene shows a single nucleotide polymorphism C/T at +1858 resulting in the W620 variant that is associated to autoimmune diseases. We have previously found in non diabetic subjects an association of PTPN22 with coronary artery diseases (CAD)[1] confirming the relationship observed by Pertovaara et al[2] between PTPN22 and atherosclerosis.
p53 codon 72 shows a single nucleotide substitution resulting in the presence of either arginine or proline in the aminoacid sequence. Proline variant is a stronger transcriptional activator, while the arginine variant is a stronger apoptosis inducer. The impact of this polymorphism within the context of a living organism is poorly understood but several data indicate that it is involved in immunity and inflammation by regulating STAT 1 and pro-inflammatory cytokines[3,4]. We have recently reported a statistically significant effect of this polymorphism on the association between PTPN22 and CAD in non diabetic subjects[5].
Acid phosphatase locus 1 shows a genetic polymorphism that controls the synthesis of a low molecular weight protein tyrosine phosphatase. The protein is composed by two isoforms called F (fast) and S (slow). The polymorphism is due to the presence of three codominant alleles *A, *B and *C at an autosomic locus. The corresponding six genotypes show an increasing enzymatic activity in the order *A/*A < *A/*B < *B/*B ≤ *A/*C < *B/*C < *C/*C[6]. The enzyme dephosphorylate a negative regulatory phosphorylation site of the ZAP70 tyrosine kinase in T cells that leads to increased activation of the kinase resulting in enhanced signaling from T-cell antigen receptor[7]. This suggests that acid phosphatse locus 1 (ACP1) could have an important role in immune functions. An association between ACP1 and CAD has been reported[8].
Adenosine deaminase (ADA) structural gene consists of 12 exons distributed in approximately 32 kb of DNA on chromosome 20[9]. A number of differences among normal sequences have been found within exonic and intronic regions of the gene[10]. The enzyme contributes to control the concentration of adenosine that in turn regulates T cell activation with important effects on immune reactions. As ectoenzyme ADA acts as a costimulatory molecule that facilitates specific signaling events in various cell types[11].
We have studied three intragenic ADA polymorphisms (PCRPs). The three PCRPs spanning over about 28 kb have a knows molecular basis and include the presence/absence of a Taq I site (ADA1) (nt 4050-4053-exons 1), of Pst I site (ADA2) (nt 19465-19470, intron 2) and a Mlu NI site (ADA6) (nt 31230-31235, exon 6)[10]. In non diabetic subjects with CAD a preliminary analysis of association of PTPN22 with the three ADA locus has revealed a statistically significant association with ADA2 locus.
In the present note we have examined the cooperative effects of ACP1, p53 codon 72 and ADA2 genetic polymorphisms on the association of PTPN22 and CAD in non diabetic subjects.
PTPN22 and ACP1 genotype were determined in 133 non diabetic subjects admitted to hospital for CAD, in 122 non diabetic cardiovascular patients without CAD and in 269 healthy blood donors. PTPN22 and p53 codon 72 genotype were determined in 129 non diabetic subjects with CAD, in 117 non diabetic admitted for cardiovascular disease without CAD and in 256 healthy blood donors. PTPN22 and ADA2 genotypes were determined in 132 non diabetic subjects with CAD, 121 non diabetic subjects with cardiovascular diseases without CAD and in 147 healthy blood donors. All the four polymorphisms, PTPN22, ACP1, p53 codon 72 and ADA2 were determined in 128 non diabetic subjects with CAD and in 117 non diabetic subjects admitted for cardiovascular diseases without CAD.
Informed written consent was obtained from all subjects to participate to this study that was approved by the Ethical Committee of the Hospital.
ACP1, p53 codon 72, PTPN22 and ADA2 genotypes was determined by DNA analysis. Technical details about the determination of the four polymorphisms have been described in previous papers[12,13].
Statistical analysis was performed by using SPSS programs.
Table 1 shows the proportion of *T allele of PTPN22 polymorphism in relation to the presence of *A allele of ACP1 polymorphism in non diabetic subjects with CAD, in non diabetic cardiovascular patients with no CAD and in healthy subjects. A high significant association is observed in carriers of *A allele with a very high proportion of *T allele carriers in non diabetic subjects with CAD as compared to controls and to non diabetic subjects with cardiovascular diseases without CAD. Such association is not observed in subjects who do not carry the *A allele of ACP1.
| Proportion of carriers of *T allele of PTPN22 | Total of subjects, n | ||
| Non diabetic subjects with CAD | |||
| Subjects carrying the *A allele | 19.3% | 62 | |
| Other ACP1 genotypes | 7.0% | 71 | |
| Non diabetic subjects with cardiovascular diseases without CAD | |||
| Subjects carrying the *A allele | 3.4% | 59 | |
| Other ACP1 genotypes | 6.3% | 63 | |
| Blood donors | |||
| Subjects carrying the *A allele | 7.2% | 138 | |
| Other ACP1 genotypes | 4.6% | 131 | |
| Statistical analysis | χ2 test of independence | ||
| χ2 | df | P | |
| Carriers of *A allele | 10.598 | 2 | 0.005 |
| Other ACP1 genotypes | 0.998 | 2 | 0.742 |
Table 2 shows the proportion of PTPN22 *T allele carriers in relation to the presence of *Pro allele of p53 codon 72 polymorphism in the three groups of subjects. A high significant association is observed in carriers of *Pro allele with a very high proportion of *T allele carriers in non diabetic subjects with CAD as compared to controls and to non diabetic subjects with cardiovascular diseases without CAD. Such association is not observed in subjects carrying the *Arg/*Arg genotype.
| Proportion of carriers of *T allele of PTPN22 | Total of subjects, n | ||
| Non diabetic subjects with CAD | |||
| *Arg/*Arg genotype | 7.6% | 66 | |
| Carriers of *Pro allele | 17.5% | 63 | |
| Non diabetic subjects with cardiovascular diseases without CAD | |||
| *Arg/*Arg genotype | 9.2% | 65 | |
| Carriers of *Pro allele | 0.0% | 52 | |
| Blood donors | |||
| *Arg/*Arg genotype | 7.2% | 139 | |
| Carriers of *Pro allele | 5.1% | 117 | |
| Statistical analysis | χ2 test of independence | ||
| χ2 | df | P | |
| *Arg/*Arg genotype | 1.212 | 2 | 0.545 |
| Carriers of *Pro allele | 11.248 | 2 | 0.004 |
Table 3 shows the proportion of *T allele carriers in relation to the presence of the ADA2 *2 allele of ADA2 polymorphism in non diabetic subjects with CAD, in non diabetic subjects with cardiovascular diseases without CAD and in healthy blood donors. A high significant association is observed in carriers of ADA2 *2 allele with a very high proportion of *T allele carriers in non diabetic subjects with CAD as compared to controls and to non diabetic subjects with cardiovascular diseases without CAD. Such association is not observed in subjects who do not carry the ADA2 *2 allele.
| Proportion of carriers of *T allele of PTPN22 | Total of subjects, n | ||
| Non diabetic subjects with CAD | |||
| ADA2 *1/*1 genotype | 8.3% | 84 | |
| Carriers of ADA2 *2 allele | 20.8% | 48 | |
| Non diabetic subjects with cardiovascular diseases without CAD | |||
| ADA2 *1/*1 genotype | 6.7% | 75 | |
| Carriers of ADA2 *2 allele | 2.2% | 46 | |
| Blood donors | |||
| ADA2 *1/*1 genotype | 4.5% | 88 | |
| Carriers of ADA2 *2 allele | 5.1% | 59 | |
| Statistical analysis | χ2 test of independence | ||
| χ2 | df | P | |
| ADA2 *1/*1 genotype | 1.024 | 2 | 0.599 |
| Carriers of ADA2 *2 allele | 11.747 | 2 | 0.003 |
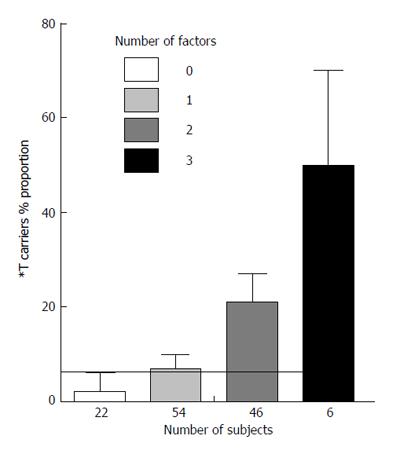
Figure 1 shows in non diabetic subjects with CAD the relationship between the number of factors (i.e., *A allele of ACP1, *Pro allele of p53 and ADA2 *2 allele) which contributes to the increase of PTPN22 *T allele carriers, and the proportion of *T carriers. There is a highly significant linear correlation between the number of factors and the proportion of *T carriers (0.0004). The relationship is compatible with an exponential function y = 5x/100 in which y = *T carriers proportion and x = the number of factors that influence the proportion of *T allele carriers.
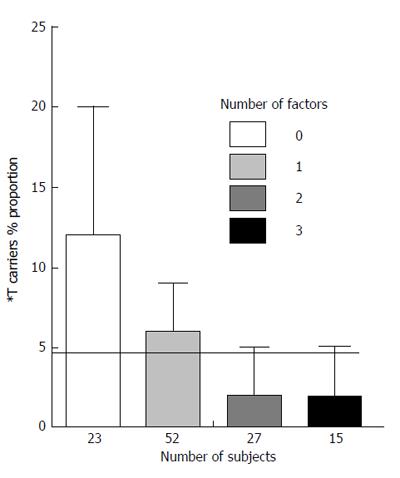
Figure 2 shows a similar analysis in non diabetic subjects with cardiovascular disease without CAD. The relationship appears opposite to that observed in non diabetic subjects with CAD.
In non diabetic subjects with CAD we have examined the relationship of PTPN22 with sex, hypertension, magnetic resonance imaging, age and total cholesterol level. No statistical significant association has been observed.
The strength of association between PTPN22 and CAD in non diabetic subjects is dependent on other genetic variables. A similar phenomenon has been recently reported also in endometriosis, a disease in which immunological factors could have a important role[14]. The data point to a multifactorial origin of CAD with a contribution of several genes involved in immune reactions.
It has been suggested that the increased susceptibility to autoimmune disorders observed in carriers of W620 variant of PTPN22 is due to failure to delete autoreactive T cells during intrathymic selection[15,16]. The Proline variant with its stronger transcriptional activity could increase the production of autoreactive T cells enhancing the effect of W620 variant of PTPN22.
Low ACP1 activity decreasing ZAP70 activity, results in a weakening of T cell receptor signaling that may contribute with W620 variant to the failure to delete autoreactive T cells during intrathymic induction.
ADA2 polymorphism could influence ADA activity and in turn the concentration of adenosine and T cell activity. The polymorphism also may have a role on ADA activity as ectoenzyme. The strength of the signal on lymphocyte would depend on the concentration of ecto-ADA available. Modulation of ecto-ADA function could influence the development and functionality of lymphoid tissue.
The simultaneous analysis of multiple genes functionally related to a specific disease would provide a productive approach to the analysis of multifactorial diseases. The mechanisms of the observed associations presented in this paper, however, remain to be elucidated.
P- Reviewers: Tagarakis G, Taguchi I S- Editor: Ma YJ L- Editor: A E- Editor: Liu SQ
| 1. | Saccucci P, Banci M, Cozzoli E, Neri A, Magrini A, Bottini E, Gloria-Bottini F. Atherosclerosis and PTPN22: a study in coronary artery disease. Cardiology. 2011;119:54-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Pertovaara M, Kähönen M, Juonala M, Laitinen T, Taittonen L, Lehtimäki T, Viikari JS, Raitakari OT, Hurme M. Autoimmunity and atherosclerosis: the presence of antinuclear antibodies is associated with decreased carotid elasticity in young women. The Cardiovascular Risk in Young Finns Study. Rheumatology (Oxford). 2009;48:1553-1556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 3. | Zheng SJ, Lamhamedi-Cherradi SE, Wang P, Xu L, Chen YH. Tumor suppressor p53 inhibits autoimmune inflammation and macrophage function. Diabetes. 2005;54:1423-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 4. | Jailwala P, Waukau J, Glisic S, Jana S, Ehlenbach S, Hessner M, Alemzadeh R, Matsuyama S, Laud P, Wang X. Apoptosis of CD4+ CD25(high) T cells in type 1 diabetes may be partially mediated by IL-2 deprivation. PLoS One. 2009;4:e6527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 67] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 5. | Saccucci P, Banci M, Amante A, Bottini E, Gloria-Bottini F. Coronary artery disease: evidence of interaction between PTPN22 and p53 genetic polymorphisms. Cardiology. 2011;120:166-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 6. | Bottini N, Meloni GF, Borgiani P, Giorgini A, Buzzetti R, Pozzilli P, Lucarelli P, Gloria-Bottini F. Genotypes of cytosolic low-molecular-weight protein-tyrosine-phosphatase correlate with age at onset of type 1 diabetes in a sex-specific manner. Metabolism. 2002;51:419-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | Bottini N, Stefanini L, Williams S, Alonso A, Jascur T, Abraham RT, Couture C, Mustelin T. Activation of ZAP-70 through specific dephosphorylation at the inhibitory Tyr-292 by the low molecular weight phosphotyrosine phosphatase (LMPTP). J Biol Chem. 2002;277:24220-24224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Banci M, Saccucci P, D’Annibale F, Dofcaci A, Trionfera G, Magrini A, Bottini N, Bottini E, Gloria-Bottini F. ACP1 genetic polymorphism and coronary artery disease: an association study. Cardiology. 2009;113:236-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | Wiginton DA, Kaplan DJ, States JC, Akeson AL, Perme CM, Bilyk IJ, Vaughn AJ, Lattier DL, Hutton JJ. Complete sequence and structure of the gene for human adenosine deaminase. Biochemistry. 1986;25:8234-8244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 123] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Tzall S, Ellenbogen A, Eng F, Hirschhorn R. Identification and characterization of nine RFLPs at the adenosine deaminase (ADA) locus. Am J Hum Genet. 1989;44:864-875. [PubMed] |
| 11. | Franco R, Casadó V, Ciruela F, Saura C, Mallol J, Canela EI, Lluis C. Cell surface adenosine deaminase: much more than an ectoenzyme. Prog Neurobiol. 1997;52:283-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 197] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 12. | Sebastiani GD, Bottini N, Greco E, Saccucci P, Canu G, Lucarelli P, Gloria-Bottini F, Fontana L. A study of Adenosine-Deaminase genetic polymorphism in rheumatoid arthritis. Int J Immunopathol Pharmacol. 2010;23:791-795. [PubMed] |
| 13. | Gloria-Bottini F, Saccucci P, ML Manca-Bitti, Rapini N, Neri A, Coppeta L, Renzetti G, Bottini E, Magrini A. Evidence of Interaction between PTPN22 and p53 codon 72 Polymorphisms on Susceptibility to Immune Related Diseases. Brit J Med and Med Res. 2013;3:1240-1247. [RCA] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 14. | Gloria-Bottini F, Ammendola M, Saccucci P, Pietropolli A, Magrini A, Bottini E. The association of PTPN22 polymorphism with endometriosis: effect of genetic and clinical factors. Eur J Obstet Gynecol Reprod Biol. 2013;169:60-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 981] [Cited by in RCA: 991] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 16. | Vang T, Congia M, Macis MD, Musumeci L, Orrú V, Zavattari P, Nika K, Tautz L, Taskén K, Cucca F. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 541] [Article Influence: 27.1] [Reference Citation Analysis (0)] |