Published online May 26, 2022. doi: 10.4330/wjc.v14.i5.282
Peer-review started: December 14, 2021
First decision: January 25, 2022
Revised: February 19, 2022
Accepted: April 26, 2022
Article in press: April 26, 2022
Published online: May 26, 2022
Processing time: 155 Days and 6.4 Hours
Heart failure is a health burden responsible for high morbidity and mortality worldwide, and dilated cardiomyopathy (DCM) is one of the most common causes of heart failure. DCM is a disease of the heart muscle and is characterized by enlargement and dilation of at least one ventricle alongside impaired contractility with left ventricular ejection fraction < 40%. It is also associated with abnormalities in cytoskeletal proteins, mitochondrial ATP transporter, microvasculature, and fibrosis. However, the pathogenesis and potential biomarkers of DCM remain to be investigated.
To investigate the candidate genes and pathways involved in DCM patients.
Two expression datasets (GSE3585 and GSE5406) were downloaded from the Gene Expression Omnibus database. The differentially expressed genes (DEGs) between the DCM patients and healthy individuals were identified using the R package “linear models for microarray data.” The pathways with common DEGs were analyzed via Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene set enrichment analyses. Moreover, a protein-protein interaction network (PPI) was constructed to identify the hub genes and modules. The MicroRNA Database was applied to predict the microRNAs (miRNAs) targeting the hub genes. Additionally, immune cell infiltration in DCM was analyzed using CIBERSORT.
In total, 97 DEGs (47 upregulated and 50 downregulated) were identified. GO analysis showed that the DEGs were mainly enriched in “response to growth factor,” “extracellular matrix,” and “extracellular matrix structural constituent.” KEGG pathway analysis indicated that the DEGs were mainly enriched in “protein digestion and absorption” and “interleukin 17 (IL-17) signaling pathway.” The PPI network suggested that collagen type III alpha 1 chain (COL3A1) and COL1A2 contribute to the pathogenesis of DCM. Additionally, visualization of the interactions between miRNAs and the hub genes revealed that hsa-miR-5682 and hsa-miR-4500 interacted with both COL3A1 and COL1A2, and thus these miRNAs might play roles in DCM. Immune cell infiltration analysis revealed that DCM patients had more infiltrated plasma cells and fewer infiltrated B memory cells, T follicular helper cells, and resting dendritic cells.
COL1A2 and COL3A1 and their targeting miRNAs, hsa-miR-5682 and hsa-miR-4500, may play critical roles in the pathogenesis of DCM, which are closely related to the IL-17 signaling pathway and acute inflammatory response. These results may provide useful clues for the diagnosis and treatment of DCM.
Core Tip: As the most common cause of heart failure, the diagnosis and therapy for dilated cardiomyopathy (DCM) are still unsatisfactory because of its indistinct pathogenesis and specific biomarkers. Thus, we comprehensively utilized the microarray data from the Gene Expression Omnibus database to uncover the biomarker and mechanisms underlying DCM development. Collagen type III alpha 1 chain and collagen type I alpha 2 chain, which are regulated by hsa-miR-5682 and hsa-miR-4500, may play critical roles in the pathogenesis of DCM through the acute inflammatory response and interleukin 17 signaling pathway. These biomarkers and mechanisms need to be further studied.
- Citation: Liu Z, Song YN, Chen KY, Gao WL, Chen HJ, Liang GY. Bioinformatics prediction of potential mechanisms and biomarkers underlying dilated cardiomyopathy. World J Cardiol 2022; 14(5): 282-296
- URL: https://www.wjgnet.com/1949-8462/full/v14/i5/282.htm
- DOI: https://dx.doi.org/10.4330/wjc.v14.i5.282
Dilated cardiomyopathy (DCM) is a progressive myocardial disease. It accounts for 30%–40% of heart failure cases and leads to high mortality worldwide[1]. DCM is characterized by biventricular dilatation, cardiac systolic dysfunction, and ventricular remodeling[2]. Recently, several studies have reported that mutations, myocarditis, hypertension, and ischemia are the induction factors of DCM[3,4]. Increasing evidence shows that various gene mutations and biomarkers are associated with DCM[5-7]. Mutations in cytoskeletal proteins, including dystrophin[8] and desmin[9], impair muscular force transmission and thereby contribute to the development of DCM. Mutations in lamin A/C, a nuclear membrane protein, usually cause DCM with atrioventricular block and atrial fibrillation[10]. Li et al[11] reported that mutation of aryl hydrocarbon receptor nuclear translocator-like protein 1 (known as BMAL1) plays a critical role in the development of DCM through the regulation of mitochondrial fission and mitophagy via mitochondrial protein B cell leukemia/lymphoma 2 interacting protein 3. Mutations in thin filament regulatory proteins including cardiac troponin T, cardiac troponin I, and α-tropomyosin can cause DCM with systolic dysfunction by reducing fractional shortening and systolic calcium level[12]. Moreover, some biomarkers associated with the development of DCM have been reported. For example, syndecan-1 and syndecan-4 may serve as useful biomarkers for predicting adverse cardiovascular events in DCM patients[13,14]. Carbonic anhydrase 2 and 3 are associated with heart failure and are potential risk biomarkers for DCM[15], and serum fibroblast growth factor 21 level is linked to the prognosis of DCM[16].
Several studies have sought DCM-related genes and mechanisms via bioinformatic methods and found some meaningful results. Huang et al[17] found that Fos proto-oncogene, AP-1 transcription factor subunit, tissue inhibitor of metalloprotease-1, and serpin family E member 1 may serve as therapeutic targets in DCM. Zhao et al[18] identified 89 differentially expressed genes (DEGs), mainly enriched in the extracellular matrix and biological adhesion signaling pathways, which may play significant roles in the development of DCM. However, the main cause(s) and pathogenic mechanism(s) underlying DCM are still unknown; thus, DCM is mostly diagnosed late, which causes a poor prognosis in turn. More studies are urgently needed to improve the diagnostic and therapeutic efficiency in DCM. The Gene Expression Omnibus (GEO) database includes many DCM-related microarray data, which have not been fully utilized. These data can be used to identify additional candidate biomarkers and pathways to further explore the cause(s) of DCM. To investigate the candidate genes and pathways involved in DCM patients, we analyzed the two gene expression data sets GSE3585 and GSE5406. Using the “linear models for microarray data” (limma) method, we identified 97 DEGs between healthy individuals and DCM patients. In addition, we identified the mechanisms commonly regulated by the DEGs via Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, and gene set enrichment analyses (GSEA). Moreover, a protein-protein interaction network (PPI) network was applied to identify the hub genes that may contribute to the pathogenesis of DCM and predict the microRNAs (miRNAs) targeting the hub genes. Furthermore, we investigated the pattern of immune cell infiltration in DCM.
The mRNA expression profiles GSE3585 and GSE5406 in the GEO database (http://www.ncbi.nlm.nih.gov/geo/), which is a shared platform in which researchers deposit their microarray data related to various diseases, were downloaded. The GSE3585 dataset, generated by Barth et al[19], and the GSE5406 dataset, generated by Hannenhalli et al[20], consisted of 7 DCM patients and 5 healthy individuals, and 86 DCM patients and 16 healthy individuals, respectively. In total, 114 samples of the left ventricular myocardium, consisting of 93 DCM and 21 healthy samples (control group), were included in this study.
The two datasets GSE3585 and GSE5406 were loaded onto the GPL96 platform (Affymetrix Human Genome U133A Array [HG-U133A]). Additionally, the series matrix and platform annotation for the two databases were downloaded from the GEO database. The probe identity documents were transformed into gene symbols. Then, via the R package “Surrogate Variable Analysis,” the two databases were merged, and any batch effect was removed using the “Empirical Bayes” method[21]. The R package “limma” was applied to identify the DEGs between the DCM and healthy myocardium tissues[22]. The screening criteria were set as P < 0.05 and |log fold change (FC)| > 0.589 (FC > 1.5). Volcano and heat maps were generated using R software.
The gene expression enrichment analysis in this study included GO analysis (https://www.geneontology.org)[23], KEGG (https://www.genome.jp/kegg)[24] pathway analysis, and GSEA (https://www.gsea-msigdb.org/gsea) analysis[25]. The DEGs were inputted into Metascape (https://metascape.org)[26]: The species was selected as Homo sapiens; The screening standard was set as P < 0.05; and The GO terms of biological process (BP), cellular component (CC), and molecular function (MF) were analyzed and KEGG pathway analysis was performed with the criteria of P < 0.05. GSEA interprets the biological function of the expression dataset. The expression dataset in the DCM cases vs healthy tissues was loaded into GSEA 4.0.3 software, set gene sets database as GO gene set (c5.all.v7.1.symbols.gmt), set number of permutations as 1000, set phenotype labels as control vs DCM, set collapse/remap to gene symbols as no collapse, set permutation type as phenotype, and the other parameters were set at default parameters. Then the GSEA software was used to obtain the enrichment results. The enriched terms were defined as significant with nominal P < 0.05.
The PPI network was constructed with the online website Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, https://stringdb.org/cgi/input.pl)[27] to contribute to the understanding of the interactive relationship among DEGs. The DEGs were inputted into this website, species of Homo sapiens was selected, and the identification criterion was set as combined score > 0.4 (medium confidence). Then the profile of interacting node pairs was imported into Cytoscape (https://cytoscape.org)[28] to visualize the PPI network. The top 10 hub genes were identified with the standard of connectivity degree by using the CytoHubba plugin. The plugin Molecular Complex Detection (MCODE) was applied to identify the essential module within the PPI network in Cytoscape with the default parameters (degree cutoff, 2; node score cutoff, 0.2; kcore, 2; and maximum depth, 100).
miRNAs, a class of small non-coding RNAs, regulate the expression of various genes by binding to their transcripts and play critical roles in DCM progression[29]. By using the MicroRNA Database (miRDB)[30] (http://mirdb.org/), we predicted miRNAs targeting any of the top 10 hub genes. Then, we sorted these miRNAs according to their prediction scores and selected the top 10 miRNAs. The mRNA-miRNA pairs were imported into Cytoscape to visualize the miRNA-mRNA network.
The CIBERSORT (cibersort.stanford.edu) algorithm was applied to analyze the normalized gene expression data, and the proportions of 22 types of immune cells in each sample were analyzed[31]. The gene expression data were normalized via “limma” and transformed into the 22 types of immune cell expression data through the source of CIBERSORT[32] via R. Then the results were filtered out via Perl (https://www.perl.org) with P < 0.05, and the immune cell infiltration matrix was obtained. Next, the “vioplot” package was used to draw violin diagrams to visualize the difference in immune cell infiltration between the DCM and healthy groups in detail. The “ggplot2”[33] package was applied to perform principal component analysis (PCA) and draw a PCA clustering map. The “corrplot” package was used to analyze the correlation among immune cell infiltration and draw a correlation heatmap.
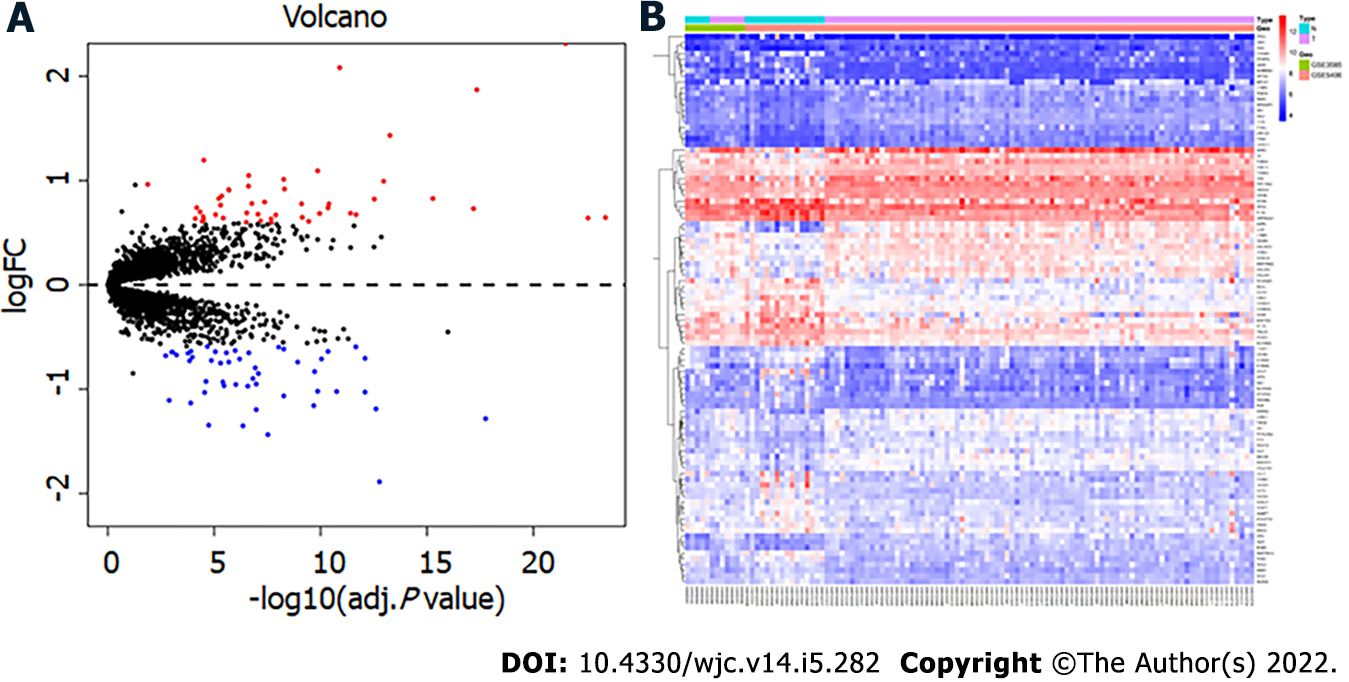
After merging the two datasets, 97 DEGs, including 47 upregulated and 50 downregulated genes, were obtained in the DCM group compared with the control group. Figure 1 shows the volcano map and heatmap of the 97 DEGs. The details of the top 10 upregulated or downregulated genes are shown in Table 1.
| Expression | Gene symbol | Description | logFC | adj. P value |
| ASPN | Asporin | 2.31 | 3.05E-22 | |
| NPPA | Natriuretic peptide A | 2.08 | 1.26E-11 | |
| LUM | Lumican | 1.87 | 4.56E-18 | |
| MXRA5 | Matrix remodeling associated 5 | 1.43 | 5.47E-14 | |
| UP | HBB | Hemoglobin subunit beta | 1.20 | 3.08E-05 |
| COL1A2 | Collagen type I alpha 2 chain | 1.09 | 1.37E-10 | |
| COL3A1 | Collagen type III alpha 1 chain | 1.05 | 2.39E-07 | |
| OGN | Osteoglycin | 1.01 | 5.38E-09 | |
| FRZB | Frizzled related protein | 0.99 | 1.08E-13 | |
| EIF1AY | Eukaryotic translation initiation factor 1A Y-linked | 0.96 | 1.33E-02 | |
| MYOT | Myotilin | -1.89 | 1.70E-13 | |
| ANKRD2 | Ankyrin repeat domain 2 | -1.44 | 3.00E-08 | |
| PLA2G2A | Phospholipase A2 group IIA | -1.35 | 4.43E-07 | |
| MYH6 | Myosin heavy chain 6 | -1.35 | 1.80E-05 | |
| SERPINA3 | Serpin family A member 3 | -1.28 | 1.75E-18 | |
| Down | CYP4B1 | Cytochrome P450 family 4 subfamily B member 1 | -1.20 | 1.06E-07 |
| FCN3 | Ficolin 3 | -1.19 | 2.48E-13 | |
| CNN1 | Calponin 1 | -1.16 | 2.09E-10 | |
| PTX3 | Pentraxin 3 | -1.13 | 1.27E-04 | |
| NRAP | Nebulin related anchoring protein | -1.11 | 1.30E-03 |
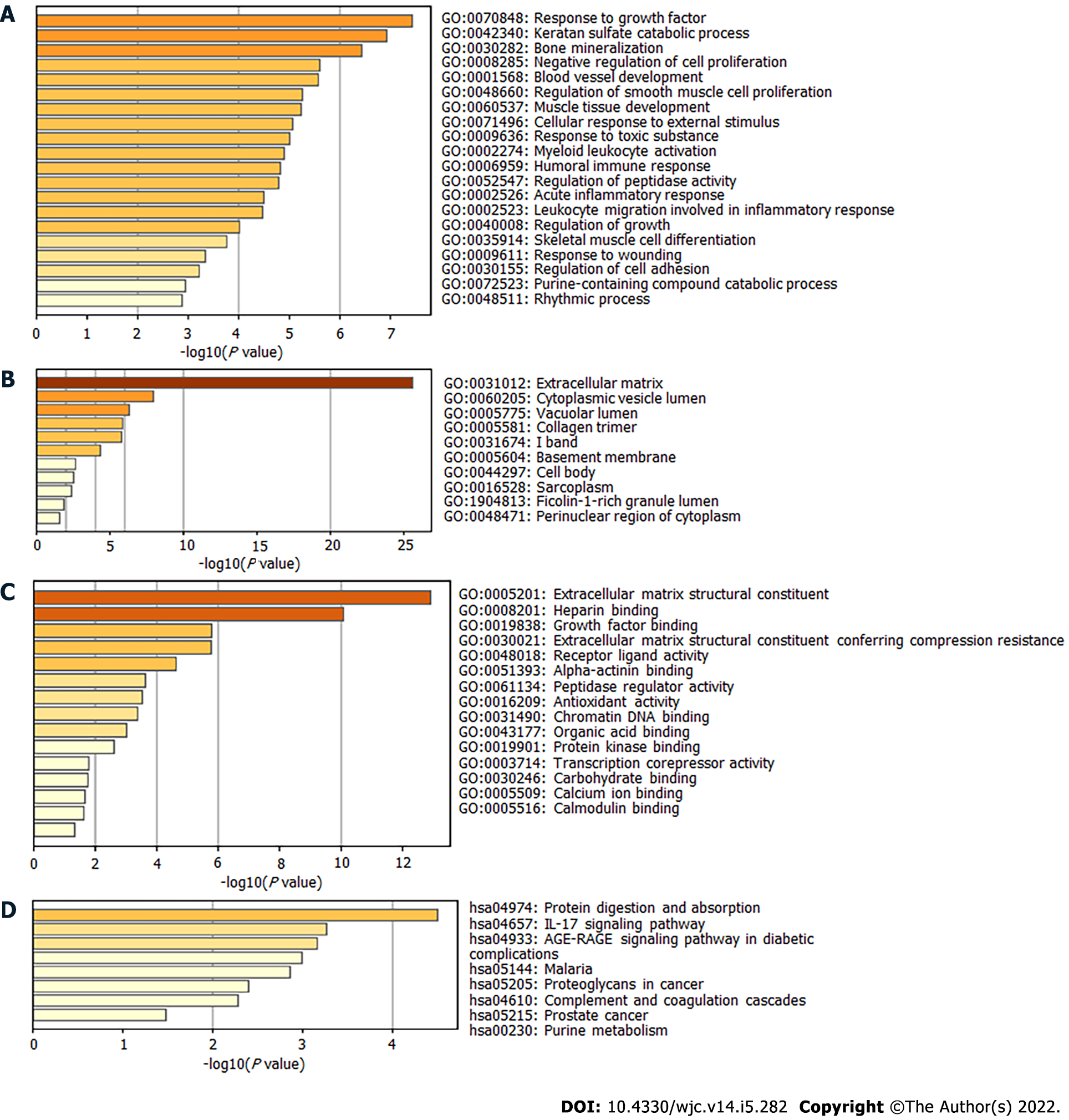
Next, the DEGs were used to perform enrichment analysis for BP, CC, MF, and KEGG pathways. By using the Metascape website, the BP of GO was found to be significantly enriched in “response to growth factor,” “blood vessel development,” “regulation of smooth muscle cell proliferation,” “muscle tissue development,” and “acute inflammatory response” (Figure 2A). The DEGs in CC were mainly enriched in “extracellular matrix,” “cytoplasmic vesicle lumen,” “collagen trimer,” and “sarcoplasm” (Figure 2B). Regarding MF, the DEGs were mainly enriched in “extracellular matrix structural constituent,” “growth factor binding,” “alpha-actinin binding,” and “calcium ion binding” (Figure 2C). KEGG pathway analysis revealed significant pathway enrichment of DEGs in “protein digestion and absorption,” “interleukin 17 (IL-17) signaling pathway,” “advanced glycation end products-receptor for advanced glycation end products signaling pathway in diabetic complications,” “complement,” and “coagulation cascades” (Figure 2D).
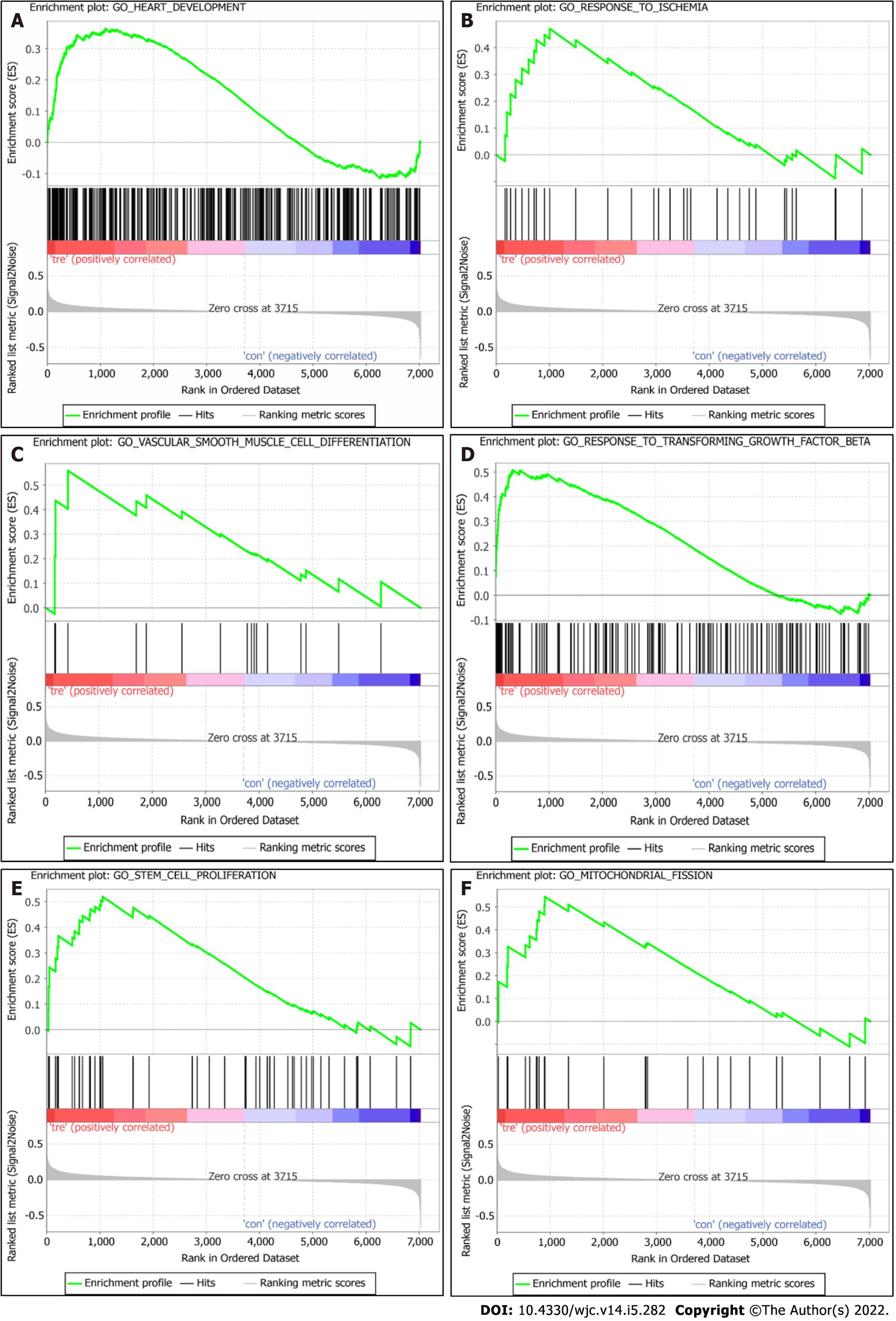
GSEA analysis results revealed that, compared with the control group, the DCM group was significantly enriched in GO terms “heart development,” “response to ischemia,” “vascular smooth muscle cell differentiation,” “response to transforming growth factor beta,” “stem cell proliferation,” and “regulation of mitochondrial fission” (Figure 3).
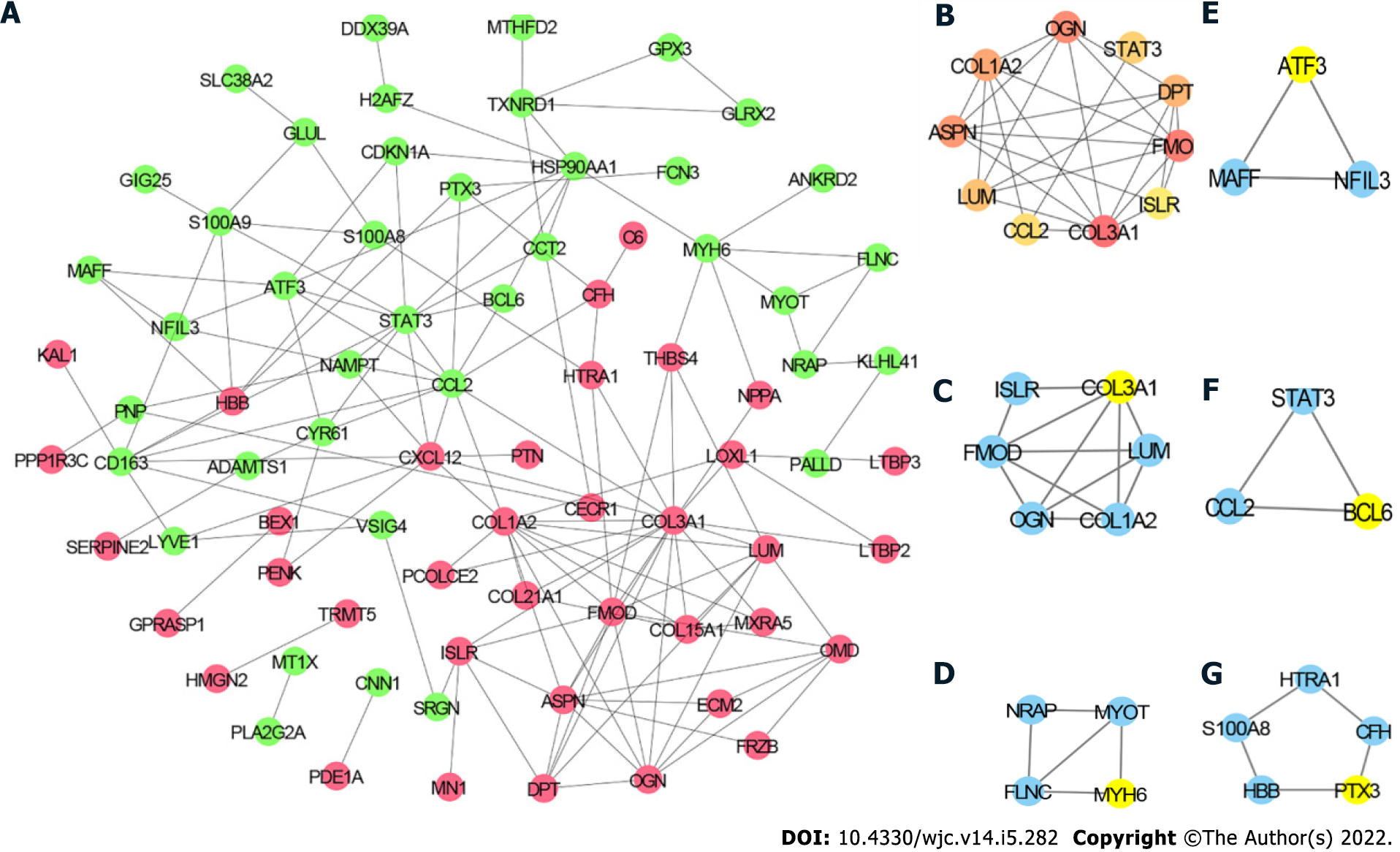
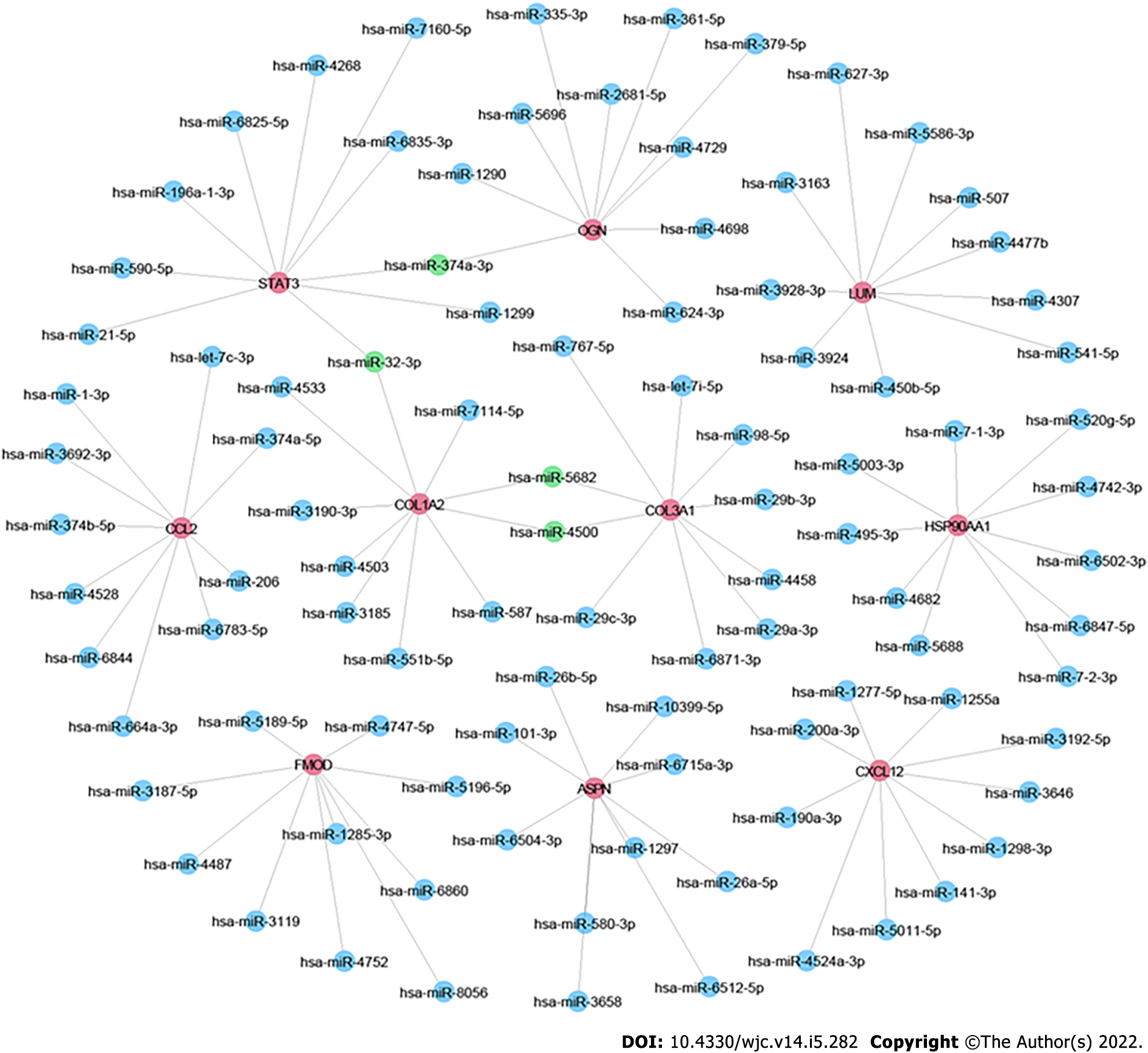
To further explore the relationship among the DEGs at the protein level, the PPI network of the 97 DEGs was constructed using STRING with the criterion of combined score > 0.4 and visualized using Cytoscape. The PPI network consisted of 77 nodes and 145 edges (Figure 4A). The top 10 hub genes included collagen type III alpha 1 chain (COL3A1), COL1A2, signal transducer and activator of transcription 3 (STAT3), C-C motif chemokine ligand 2 (CCL2), fibromodulin (FMOD), asporin (ASPN), C-X-C motif chemokine ligand 12 (CXCL12), lumican (LUM), heat shock protein 90 alpha family class A member 1 (HSP90AA1), and osteoglycin (OGN) (Figure 4B). The detailed information of these hub genes is provided in Table 2. MCODE analysis identified five essential modules, and COL3A1, myosin heavy chain 6, activating transcription factor 3, B cell leukemia/lymphoma 6 transcription repressor, and pentraxin 3 were the seeds of clusters 1, 2, 3, 4, and 5, respectively (Figure 4C–G).
| Gene symbol | Description | Rank | Degree |
| COL3A1 | Collagen type III alpha 1 chain | 1 | 18 |
| COL1A2 | Collagen type I alpha 2 chain | 2 | 12 |
| STAT3 | Signal transducer and activator of transcription 3 | 3 | 11 |
| CCL2 | C-C motif chemokine ligand 2 | 3 | 11 |
| FMOD | Fibromodulin | 5 | 10 |
| ASPN | Asporin | 6 | 9 |
| CXCL12 | C-X-C motif chemokine ligand 12 | 6 | 9 |
| LUM | Lumican | 8 | 8 |
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | 8 | 8 |
| OGN | Osteoglycin | 8 | 8 |
Increasing evidence shows that miRNAs play important roles in the development and progression of DCM. By using the miRDB database, we predicted miRNAs targeting any of the top 10 hub genes. Then we sorted these miRNAs according to their prediction scores and selected the top 10 miRNAs. Additionally, the top 100 miRNA–mRNA pairs were visualized using Cytoscape (Figure 5). Consequently, hsa-miR-5682, hsa-miR-4500, hsa-miR-32-3p, and hsa-miR-374a-3p were each found to target ≥ 2 hub genes.
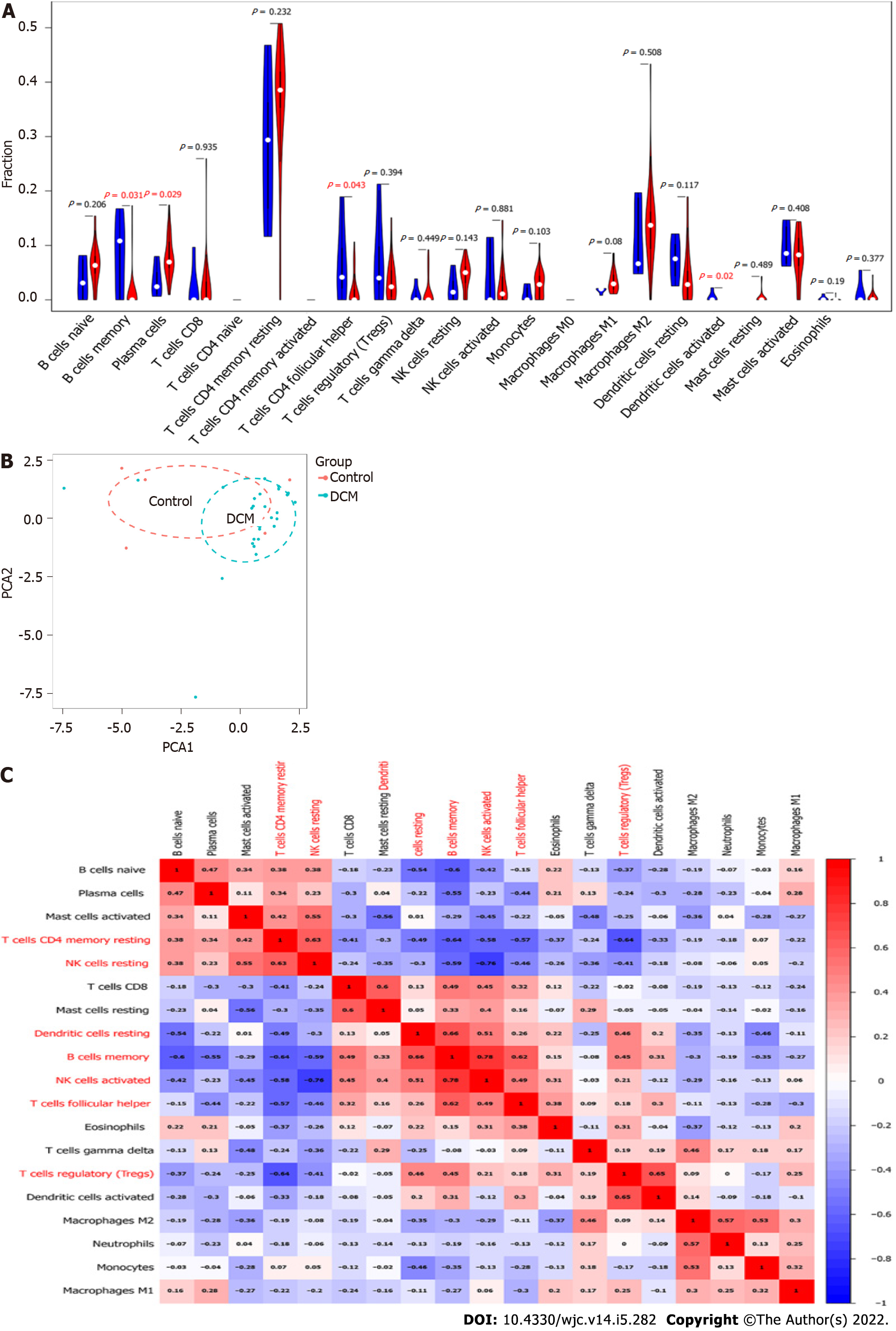
Immune cells infiltrate into the myocardium upon myocardial injury[28]. Thus, a violin plot was constructed to investigate the difference in immune-cell infiltration between the DCM and control groups (Figure 6A). Compared with the control group, the DCM group had more infiltrated plasma cells and fewer infiltrated B memory cells, T follicular helper cells, and resting dendritic cells (DCs), whereas there was no significant difference in the remaining 18 types of immune cells. However, the PCA results showed that the control and DCM groups could not be well distinguished according to the infiltration patterns of the 22 types of immune cells (Figure 6B). We generated a correlation heatmap to assess the correlation among the 19 immune cells that were found infiltrated in the DCM or control group. As shown in Figure 6C, the number of infiltrated B memory cells was positively correlated with that of the infiltrated resting DCs, activated natural killer (NK) cells and T follicular helper cells, the number of infiltrated activated NK cells was negatively correlated with that of infiltrated resting NK cells, and the number of infiltrated resting memory CD4 T cells was negatively associated with that of infiltrated B memory cells and regulatory T cells.
DCM is one of the main reasons of sudden cardiac death and heart failure. It is a heterogeneous disease caused by various types of pathogenic factors including genetic, infectious, hormonal, and environmental factors[34]. The causes of DCM should be explored in depth to improve the diagnosis, treatment, and prognosis of DCM patients. Therefore, it is of great significance to elucidate the genetic mechanisms involved in DCM.
In this study, 97 DEGs, consisting of 47 upregulated and 50 downregulated genes, were identified between the DCM and control groups. GO of BP and GSEA analysis revealed that the DEGs were not only enriched in the development of the cardiovascular system, such as in the development of the muscle tissue, heart, and blood vessels, but also in the etiology of DCM, such as in acute inflammatory response and mitochondrial fission (Figures 2 and 3). Growing evidence shows that infiltration of inflammatory cells is associated with the pathogenesis of DCM[35-37], and prelamin A accumulation[38] and myosin binding protein C3 mutation[39] can promote DCM pathogenesis via regulation of inflammation. Xia et al[40] reported that dynamin-related protein 1 (Drp1, myocardial fission protein) is significantly upregulated in DCM patients. Moreover, Sacubitril (known as LCZ696), a novel inhibitor of the angiotensin receptor neprilysin, can alleviate the cardiac dysfunction in doxorubicin-induced DCM and reduce apoptosis by inhibiting mitochondrial fission via the Drp1-mediated pathway.
Regarding CC, the DEGs were enriched in sarcoplasm. Previous studies have reported that mutations in phospholamban (related to abnormal contractility)[41] and B-cell lymphoma 2-associated athanogene 3 (alter the cardiac response)[42] are closely associated with DCM in the sarcoplasmic reticulum. GO analysis of MF indicated that the DEGs were enriched in alpha-actinin binding and calcium ion binding. Alpha-actinin and calcium ion are critical for myocardial contraction[43,44]. Other studies have demonstrated that most DCM patients exhibit abnormalities related to calcium ion and α-actinin, which cause decreased heart contractility[45-47]. Cardiac troponin contributes to myocardial contraction[48]. Mutations in cardiac troponin T, troponin C, and troponin I are mainly related to DCM pathogenesis[49,50]
KEGG pathway analysis showed that the DEGs were significantly enriched in the IL-17 signaling pathway. DCM is induced by viral myocarditis, accompanying autoimmune dysfunction, affecting the secretion of IL-17 cytokine by T helper 17 cells, and IL-17 itself promotes myocardial cell injury[51]. Wang et al[52] reported that elevated IL-17 levels are significantly associated with DCM incidence and progression. Additionally, the serum levels of other inflammatory factors such as IL-6, tumor necrosis factor-α, and IL-21 are significantly increased in DCM patients[53]. Thus, the IL-17 signaling pathway may be one of the major signaling pathways involved in the development of DCM.
Through construction of PPI and miRNA-mRNA interaction networks, we identified the hub genes and the miRNAs targeting them. The hub genes, COL1A2 and COL3A1, encode the pro-alpha2/1 chains of type I and III collagens, respectively. Collagens I and III, the main collagens of cardiac extracellular matrix, are classical biomarkers of cardiac fibrosis in DCM[54]. Mihailovici et al[55] reported that collagens I and III are upregulated in DCM patients compared with matched healthy controls. Additionally, Zhao et al[18] reported that COL1A2 may participate in DCM pathogenesis by regulating the cardiac remodeling characterized by collagen deposition in the extracellular matrix[56]. Consistent with our results, Liu et al[53] identified STAT3 as a hub gene in DCM via bioinformatic analysis. Other studies have also indicated a role of STAT3 in DCM. Podewski et al[57] showed that STAT3 protein level is significantly decreased in the cardiomyocytes of patients with end-stage DCM. Moreover, inhibition of the IL-6–mediated STAT3 signaling pathway can improve myocardial remodeling through reducing myocardial apoptosis in a mouse model of DCM[58]. Thus, the roles of COL1A2, COL3A1, and STAT3 in DCM should be further investigated. Moreover, the identified miRNAs hsa-miR-5682, hsa-miR-4500, hsa-miR-32-3p, and hsa-miR-374a-3p may participate in DCM pathogenesis through their interaction with ≥ two hub genes. Previous studies have also suggested that miRNAs play significant roles in DCM. It has been found that miR-21, miR-29a, and miR-133b are differentially expressed in DCM patients[59]. miR-133a expression is associated with fibrosis, myocyte necrosis, left ventricular function, and clinical outcome in patients with inflammatory DCM[60]. Moreover, Satoh et al[61] showed that a low let-7i level can serve as an independent predictor of cardiac death and heart failure (relative risk = 3.76).
Immune cells commonly infiltrate into the myocardium upon various types of cardiac damage[49,62]. Overactivation of immune cells could be investigated in pathological examination about cardiac biopsy specimens in DCM patients. Noutsias et al[63] reported that the upregulation of genes associated with T cells exacerbates DCM progression. Therefore, our study also assessed the correlation between DEGs and immune cell infiltration. The results indicated infiltration of 19 types of immune cells in DCM pathogenesis. Notably, compared with the control group, the DCM group had more infiltrated plasma cells and fewer infiltrated B memory cells, T follicular helper cells, and resting DCs. However, Liu et al[53] have demonstrated that, compared with healthy controls, DCM patients have more infiltrated T follicular helper cells and fewer T follicular regulatory cells, and infiltration of T follicular regulatory cells is positively correlated with left ventricular ejection fraction.
Our study had some limitations. First, the gene expression data were acquired from a public database. Moreover, we did not experimentally verify the relevance of the identified DEGs with DCM and their enriched functions or hub genes. Likewise, we did not verify the predicted miRNA-mRNA interactions and their relevance.
In summary, we first identified that COL1A2 and COL3A1 may be both presumably regulated by hsa-miR-5682 and hsa-miR-4500, and play significant roles in the pathogenesis of DCM through acute inflammatory response and IL-17 signaling pathway. These results may provide useful biomarkers for the diagnosis and treatment of DCM, but further research is needed to clarify the roles of the predicted genes and pathways.
Dilated cardiomyopathy (DCM), a disease of the heart muscle, is one of the most common causes of heart failure. However, the original cause and pathogenesis in development of DCM are still remain elusive.
The early diagnosis and prognosis of DCM patients are unsatisfactory because of DCM main cause and pathogenesis are still unclear. Increasing DCM datasets were provided online but little was been explored. Bioinformatics could further investigate the DCM mechanism and biomarkers for improving the diagnostic and therapeutic efficiency.
This study investigated the candidate genes and pathways involved in DCM patients.
Expression datasets were downloaded from the Gene Expression Omnibus database. Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and gene set enrichment analyses investigated the key pathway in differentially expressed genes (DEGs) between the DCM patients and healthy individuals. Protein-protein interaction network identified the hub genes and modules in DCM. MicroRNA Database predicted the microRNAs which targeting the hub genes. CIBERSORT analyzed the immune- ell infiltration in DCM.
Ninety-seven DEGs mainly enriched in “response to growth factor,” “extracellular matrix,” and “extracellular matrix structural constituent.” Moreover, the top two pathways were “protein digestion and absorption” and “interleukin 17 signaling pathway.” Collagen type III alpha 1 chain (COL3A1) and COL1A2, whose were regulated by hsa-miR-5682 and hsa-miR-4500, mainly contributed to the pathogenesis of DCM. Compared with the control group, DCM patients had more infiltrated plasma cells and fewer infiltrated B memory cells, T follicular helper cells, and resting dendritic cells.
DCM progression closely related to IL-17 signaling pathway and acute inflammatory response. COL1A2 and COL3A1 and their targeting miRNAs, hsa-miR-5682 and hsa-miR-4500, are the potential biomarkers of DCM.
This study may provide valuable pathways and biomarkers for the diagnosis or treatment of DCM. Further studies should investigate the functions of the predicted genes and pathways.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Engineering, biomedical
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Emran TB, Bangladesh; Tanabe S, Japan S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Ma YJ
| 1. | Papadakis M, Sharma S, Cox S, Sheppard MN, Panoulas VF, Behr ER. The magnitude of sudden cardiac death in the young: a death certificate-based review in England and Wales. Europace. 2009;11:1353-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 111] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 2. | Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, Feng Z, Müller S, Kayvanpour E, Vogel B, Sedaghat-Hamedani F, Lim WK, Zhao X, Fradkin D, Köhler D, Fischer S, Franke J, Marquart S, Barb I, Li DT, Amr A, Ehlermann P, Mereles D, Weis T, Hassel S, Kremer A, King V, Wirsz E, Isnard R, Komajda M, Serio A, Grasso M, Syrris P, Wicks E, Plagnol V, Lopes L, Gadgaard T, Eiskjær H, Jørgensen M, Garcia-Giustiniani D, Ortiz-Genga M, Crespo-Leiro MG, Deprez RH, Christiaans I, van Rijsingen IA, Wilde AA, Waldenstrom A, Bolognesi M, Bellazzi R, Mörner S, Bermejo JL, Monserrat L, Villard E, Mogensen J, Pinto YM, Charron P, Elliott P, Arbustini E, Katus HA, Meder B. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123-135a. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 367] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 3. | Bozkurt B, Colvin M, Cook J, Cooper LT, Deswal A, Fonarow GC, Francis GS, Lenihan D, Lewis EF, McNamara DM, Pahl E, Vasan RS, Ramasubbu K, Rasmusson K, Towbin JA, Yancy C; American Heart Association Committee on Heart Failure and Transplantation of the Council on Clinical Cardiology; Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; Council on Epidemiology and Prevention; and Council on Quality of Care and Outcomes Research. Current Diagnostic and Treatment Strategies for Specific Dilated Cardiomyopathies: A Scientific Statement From the American Heart Association. Circulation. 2016;134:e579-e646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 525] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 4. | Vikhorev PG, Vikhoreva NN. Cardiomyopathies and Related Changes in Contractility of Human Heart Muscle. Int J Mol Sci. 2018;19:2234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 5. | Weintraub RG, Semsarian C, Macdonald P. Dilated cardiomyopathy. Lancet. 2017;390:400-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 443] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 6. | Willott RH, Gomes AV, Chang AN, Parvatiyar MS, Pinto JR, Potter JD. Mutations in Troponin that cause HCM, DCM AND RCM: what can we learn about thin filament function? J Mol Cell Cardiol. 2010;48:882-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 163] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 7. | Pyun JH, Kim HJ, Jeong MH, Ahn BY, Vuong TA, Lee DI, Choi S, Koo SH, Cho H, Kang JS. Cardiac specific PRMT1 ablation causes heart failure through CaMKII dysregulation. Nat Commun. 2018;9:5107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 8. | Restrepo-Cordoba MA, Wahbi K, Florian AR, Jiménez-Jáimez J, Politano L, Arad M, Climent-Paya V, Garcia-Alvarez A, Hansen RB, Larrañaga-Moreira JM, Kubanek M, Lopes LR, Ros A, Jurcut R, Rasmussen TB, Ruiz-Guerrero L, Pribe-Wolferts R, Palomino-Doza J, Bilinska Z, Rodríguez-Palomares JF, Van Loon RLE, Basurte Elorz MT, Quarta G, Robledo Iñarritu M, Verdonschot JAJ, Stojkovic T, Shomanova Z, Bermudez-Jimenez F, Palladino A, Freimark D, García-Álvarez MI, Jorda P, Dominguez F, Ochoa JP, Girolami F, Brugada R, Meder B, Barriales-Villa R, Mogensen J, Laforêt P, Yilmaz A, Elliott P, Garcia-Pavia P; European Genetic Cardiomyopathies Initiative Investigators (see online supplementary Appendix S1). Prevalence and clinical outcomes of dystrophin-associated dilated cardiomyopathy without severe skeletal myopathy. Eur J Heart Fail. 2021;23:1276-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Pawlak A, Rejmak-Kozicka E, Gil KE, Ziemba A, Kaczmarek L, Gil RJ. Patterns of desmin expression in idiopathic dilated cardiomyopathy are related to the desmin mRNA and ubiquitin expression. J Investig Med. 2019;67:11-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Peterson A, Li D, Jakobs P, Litt M, Porter CB, Rahko PS, Hershberger RE. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 197] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 11. | Li E, Li X, Huang J, Xu C, Liang Q, Ren K, Bai A, Lu C, Qian R, Sun N. BMAL1 regulates mitochondrial fission and mitophagy through mitochondrial protein BNIP3 and is critical in the development of dilated cardiomyopathy. Protein Cell. 2020;11:661-679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 12. | Robinson P, Sparrow AJ, Patel S, Malinowska M, Reilly SN, Zhang YH, Casadei B, Watkins H, Redwood C. Dilated cardiomyopathy mutations in thin-filament regulatory proteins reduce contractility, suppress systolic Ca2+, and activate NFAT and Akt signaling. Am J Physiol Heart Circ Physiol. 2020;319:H306-H319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 13. | Liu W, Wang Y, Zheng J, Song D, Zheng S, Ren L, Yao Y, Liu Y, Bai R, Dong J, Liu T. Syndecan-1 as an independent risk factor for the incidence of adverse cardiovascular events in patients having stage C and D heart failure with non-ischemic dilated cardiomyopathy. Clin Chim Acta. 2019;490:63-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Bielecka-Dabrowa A, Sakowicz A, Misztal M, von Haehling S, Ahmed A, Pietrucha T, Rysz J, Banach M. Differences in biochemical and genetic biomarkers in patients with heart failure of various etiologies. Int J Cardiol. 2016;221:1073-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Su H, Hu K, Liu Z, Chen K, Xu J. Carbonic anhydrase 2 and 3 as risk biomarkers for dilated cardiomyopathy associated heart failure. Ann Palliat Med. 2021;10:12554-12565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Gu L, Jiang W, Zheng R, Yao Y, Ma G. Fibroblast Growth Factor 21 Correlates with the Prognosis of Dilated Cardiomyopathy. Cardiology. 2021;146:27-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Huang H, Luo B, Wang B, Wu Q, Liang Y, He Y. Identification of Potential Gene Interactions in Heart Failure Caused by Idiopathic Dilated Cardiomyopathy. Med Sci Monit. 2018;24:7697-7709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Zhao J, Lv T, Quan J, Zhao W, Song J, Li Z, Lei H, Huang W, Ran L. Identification of target genes in cardiomyopathy with fibrosis and cardiac remodeling. J Biomed Sci. 2018;25:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Barth AS, Kuner R, Buness A, Ruschhaupt M, Merk S, Zwermann L, Kääb S, Kreuzer E, Steinbeck G, Mansmann U, Poustka A, Nabauer M, Sültmann H. Identification of a common gene expression signature in dilated cardiomyopathy across independent microarray studies. J Am Coll Cardiol. 2006;48:1610-1617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 143] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 20. | Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 196] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 21. | Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2446] [Cited by in RCA: 3995] [Article Influence: 307.3] [Reference Citation Analysis (0)] |
| 22. | Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16184] [Cited by in RCA: 25671] [Article Influence: 2567.1] [Reference Citation Analysis (0)] |
| 23. | Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325-D334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2681] [Cited by in RCA: 2225] [Article Influence: 556.3] [Reference Citation Analysis (0)] |
| 24. | Kanehisa M, Sato Y, Kawashima M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022;31:47-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 427] [Article Influence: 142.3] [Reference Citation Analysis (0)] |
| 25. | Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-15550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27252] [Cited by in RCA: 37449] [Article Influence: 1872.5] [Reference Citation Analysis (0)] |
| 26. | Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3766] [Cited by in RCA: 8765] [Article Influence: 1460.8] [Reference Citation Analysis (0)] |
| 27. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11736] [Article Influence: 1956.0] [Reference Citation Analysis (1)] |
| 28. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33531] [Article Influence: 1596.7] [Reference Citation Analysis (0)] |
| 29. | Isserlin R, Merico D, Wang D, Vuckovic D, Bousette N, Gramolini AO, Bader GD, Emili A. Systems analysis reveals down-regulation of a network of pro-survival miRNAs drives the apoptotic response in dilated cardiomyopathy. Mol Biosyst. 2015;11:239-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127-D131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 855] [Cited by in RCA: 1992] [Article Influence: 398.4] [Reference Citation Analysis (0)] |
| 31. | Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 993] [Cited by in RCA: 2453] [Article Influence: 350.4] [Reference Citation Analysis (0)] |
| 32. | Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4763] [Cited by in RCA: 8898] [Article Influence: 889.8] [Reference Citation Analysis (0)] |
| 33. | Wickham H. ggplot2. New York: Springer, Cham; 2016. |
| 34. | Merlo M, Cannatà A, Gobbo M, Stolfo D, Elliott PM, Sinagra G. Evolving concepts in dilated cardiomyopathy. Eur J Heart Fail. 2018;20:228-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 237] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 35. | Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O'Connell J, Olsen E, Thiene G, Goodwin J, Gyarfas I, Martin I, Nordet P. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation. 1996;93:841-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2218] [Cited by in RCA: 2131] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 36. | Maisch B, Pankuweit S. Inflammatory dilated cardiomyopathy : Etiology and clinical management. Herz. 2020;45:221-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 37. | Barcena ML, Pozdniakova S, Haritonow N, Breiter P, Kühl AA, Milting H, Baczko I, Ladilov Y, Regitz-Zagrosek V. Dilated cardiomyopathy impairs mitochondrial biogenesis and promotes inflammation in an age- and sex-dependent manner. Aging (Albany NY). 2020;12:24117-24133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Brayson D, Frustaci A, Verardo R, Chimenti C, Russo MA, Hayward R, Ahmad S, Vizcay-Barrena G, Protti A, Zammit PS, dos Remedios CG, Ehler E, Shah AM, Shanahan CM. Prelamin A mediates myocardial inflammation in dilated and HIV-associated cardiomyopathies. JCI Insight. 2019;4:e126315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 39. | Lynch TL 4th, Ismahil MA, Jegga AG, Zilliox MJ, Troidl C, Prabhu SD, Sadayappan S. Cardiac inflammation in genetic dilated cardiomyopathy caused by MYBPC3 mutation. J Mol Cell Cardiol. 2017;102:83-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 40. | Xia Y, Chen Z, Chen A, Fu M, Dong Z, Hu K, Yang X, Zou Y, Sun A, Qian J, Ge J. LCZ696 improves cardiac function via alleviating Drp1-mediated mitochondrial dysfunction in mice with doxorubicin-induced dilated cardiomyopathy. J Mol Cell Cardiol. 2017;108:138-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 41. | Liu GS, Morales A, Vafiadaki E, Lam CK, Cai WF, Haghighi K, Adly G, Hershberger RE, Kranias EG. A novel human R25C-phospholamban mutation is associated with super-inhibition of calcium cycling and ventricular arrhythmia. Cardiovasc Res. 2015;107:164-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 42. | Knezevic T, Myers VD, Gordon J, Tilley DG, Sharp TE 3rd, Wang J, Khalili K, Cheung JY, Feldman AM. BAG3: a new player in the heart failure paradigm. Heart Fail Rev. 2015;20:423-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 43. | Sutanto H, Lyon A, Lumens J, Schotten U, Dobrev D, Heijman J. Cardiomyocyte calcium handling in health and disease: Insights from in vitro and in silico studies. Prog Biophys Mol Biol. 2020;157:54-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 44. | Sheng JJ, Feng HZ, Pinto JR, Wei H, Jin JP. Increases of desmin and α-actinin in mouse cardiac myofibrils as a response to diastolic dysfunction. J Mol Cell Cardiol. 2016;99:218-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 45. | Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 536] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 46. | Li W, Lu M, Banerjee S, Zhong J, Ye A, Molter J, Yu X. Ex vivo diffusion tensor MRI reflects microscopic structural remodeling associated with aging and disease progression in normal and cardiomyopathic Syrian hamsters. NMR Biomed. 2009;22:819-825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 47. | Cetinkaya A, Berge B, Sen-Hild B, Troidl K, Gajawada P, Kubin N, Valeske K, Schranz D, Akintürk H, Schönburg M, Kubin T, Choi YH, Richter M. Radixin Relocalization and Nonmuscle α-Actinin Expression Are Features of Remodeling Cardiomyocytes in Adult Patients with Dilated Cardiomyopathy. Dis Markers. 2020;2020:9356738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 48. | Park KC, Gaze DC, Collinson PO, Marber MS. Cardiac troponins: from myocardial infarction to chronic disease. Cardiovasc Res. 2017;113:1708-1718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 317] [Cited by in RCA: 333] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 49. | Schultheiss HP, Fairweather D, Caforio ALP, Escher F, Hershberger RE, Lipshultz SE, Liu PP, Matsumori A, Mazzanti A, McMurray J, Priori SG. Dilated cardiomyopathy. Nat Rev Dis Primers. 2019;5:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 434] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 50. | Chang AN, Parvatiyar MS, Potter JD. Troponin and cardiomyopathy. Biochem Biophys Res Commun. 2008;369:74-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 51. | Baldeviano GC, Barin JG, Talor MV, Srinivasan S, Bedja D, Zheng D, Gabrielson K, Iwakura Y, Rose NR, Cihakova D. Interleukin-17A is dispensable for myocarditis but essential for the progression to dilated cardiomyopathy. Circ Res. 2010;106:1646-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 260] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 52. | Wang ZH, Liao YH, Yuan J, Jin XJ, Yu M, Chen RZ, Xu DJ, Wei J, Wan J, Zhao DC, Han HY, Li B, Tian G, Hu G, Xu J. Continued Elevation of Plasma IL-4 and IL-17 Predicts the Progression from VMC to DCM. Dis Markers. 2020;2020:9385472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 53. | Liu X, Zhang W, Han Z. Decreased circulating follicular regulatory T cells in patients with dilated cardiomyopathy. Braz J Med Biol Res. 2021;54:e11232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 54. | Nagao K, Inada T, Tamura A, Kajitani K, Shimamura K, Yukawa H, Aida K, Sowa N, Nishiga M, Horie T, Makita T, Ono K, Tanaka M. Circulating markers of collagen types I, III, and IV in patients with dilated cardiomyopathy: relationships with myocardial collagen expression. ESC Heart Fail. 2018;5:1044-1051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 55. | Mihailovici AR, Deliu RC, Mărgăritescu C, Simionescu CE, Donoiu I, Istrătoaie O, Tudoraşcu DR, Târtea EA, Gheonea DI. Collagen I and III, MMP-1 and TIMP-1 immunoexpression in dilated cardiomyopathy. Rom J Morphol Embryol. 2017;58:777-781. [PubMed] |
| 56. | Tsoutsman T, Wang X, Garchow K, Riser B, Twigg S, Semsarian C. CCN2 plays a key role in extracellular matrix gene expression in severe hypertrophic cardiomyopathy and heart failure. J Mol Cell Cardiol. 2013;62:164-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 57. | Podewski EK, Hilfiker-Kleiner D, Hilfiker A, Morawietz H, Lichtenberg A, Wollert KC, Drexler H. Alterations in Janus kinase (JAK)-signal transducers and activators of transcription (STAT) signaling in patients with end-stage dilated cardiomyopathy. Circulation. 2003;107:798-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 106] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 58. | Li Q, Ye WX, Huang ZJ, Zhang Q, He YF. Effect of IL-6-mediated STAT3 signaling pathway on myocardial apoptosis in mice with dilated cardiomyopathy. Eur Rev Med Pharmacol Sci. 2019;23:3042-3050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 59. | Wang Y, Li M, Xu L, Liu J, Wang D, Li Q, Wang L, Li P, Chen S, Liu T. Expression of Bcl-2 and microRNAs in cardiac tissues of patients with dilated cardiomyopathy. Mol Med Rep. 2017;15:359-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 60. | Besler C, Urban D, Watzka S, Lang D, Rommel KP, Kandolf R, Klingel K, Thiele H, Linke A, Schuler G, Adams V, Lurz P. Endomyocardial miR-133a levels correlate with myocardial inflammation, improved left ventricular function, and clinical outcome in patients with inflammatory cardiomyopathy. Eur J Heart Fail. 2016;18:1442-1451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 61. | Satoh M, Minami Y, Takahashi Y, Tabuchi T, Nakamura M. A cellular microRNA, let-7i, is a novel biomarker for clinical outcome in patients with dilated cardiomyopathy. J Card Fail. 2011;17:923-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 62. | Carrillo-Salinas FJ, Ngwenyama N, Anastasiou M, Kaur K, Alcaide P. Heart Inflammation: Immune Cell Roles and Roads to the Heart. Am J Pathol. 2019;189:1482-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 63. | Noutsias M, Rohde M, Göldner K, Block A, Blunert K, Hemaidan L, Hummel M, Blohm JH, Lassner D, Kühl U, Schultheiss HP, Volk HD, Kotsch K. Expression of functional T-cell markers and T-cell receptor Vbeta repertoire in endomyocardial biopsies from patients presenting with acute myocarditis and dilated cardiomyopathy. Eur J Heart Fail. 2011;13:611-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |