Published online Feb 26, 2017. doi: 10.4331/wjbc.v8.i1.32
Peer-review started: November 4, 2016
First decision: December 14, 2016
Revised: December 24, 2016
Accepted: January 11, 2017
Article in press: January 14, 2017
Published online: February 26, 2017
Processing time: 116 Days and 12 Hours
Retroviral replication proceeds through the integration of a DNA copy of the viral RNA genome into the host cellular genome, a process that is mediated by the viral integrase (IN) protein. IN catalyzes two distinct chemical reactions: 3’-processing, whereby the viral DNA is recessed by a di- or trinucleotide at its 3’-ends, and strand transfer, in which the processed viral DNA ends are inserted into host chromosomal DNA. Although IN has been studied as a recombinant protein since the 1980s, detailed structural understanding of its catalytic functions awaited high resolution structures of functional IN-DNA complexes or intasomes, initially obtained in 2010 for the spumavirus prototype foamy virus (PFV). Since then, two additional retroviral intasome structures, from the α-retrovirus Rous sarcoma virus (RSV) and β-retrovirus mouse mammary tumor virus (MMTV), have emerged. Here, we briefly review the history of IN structural biology prior to the intasome era, and then compare the intasome structures of PFV, MMTV and RSV in detail. Whereas the PFV intasome is characterized by a tetrameric assembly of IN around the viral DNA ends, the newer structures harbor octameric IN assemblies. Although the higher order architectures of MMTV and RSV intasomes differ from that of the PFV intasome, they possess remarkably similar intasomal core structures. Thus, retroviral integration machineries have adapted evolutionarily to utilize disparate IN elements to construct convergent intasome core structures for catalytic function.
Core tip: This review examines the history of retroviral integrase structural biology and covers the currently available high-resolution structures of retroviral intasomes in detail. We in particular focus on the similarities and differences among the intasome structures of prototype foamy virus, Rous sarcoma virus and mouse mammary tumor virus.
- Citation: Grawenhoff J, Engelman AN. Retroviral integrase protein and intasome nucleoprotein complex structures. World J Biol Chem 2017; 8(1): 32-44
- URL: https://www.wjgnet.com/1949-8454/full/v8/i1/32.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v8.i1.32
Retroviral replication requires the incorporation of the viral genetic information into the host cellular genome, which occurs via two main steps: (1) the reverse transcription of single-stranded viral RNA into linear double-stranded DNA; and (2) the integration of this DNA into a host chromosome. These steps occur in the context of two subviral nucleoprotein complexes: The reverse transcription complex (reviewed in[1]) and the pre-integration complex (PIC)[2], each of which consists of a variety of cellular and viral proteins including reverse transcriptase (RT) and integrase (IN)[3-7]. In the cytoplasm, RT mediates the synthesis of a linear viral DNA (vDNA) molecule that harbors a copy of the viral long-terminal repeat (LTR) at each end[8-10]. In the confines of the PIC, vDNA is trafficked toward the nucleus, where its integration into host cell target DNA (tDNA) is promoted by IN. Here, we discuss the current knowledge of IN structural determinants and intasome function, highlighting both key similarities and differences among the retroviruses.
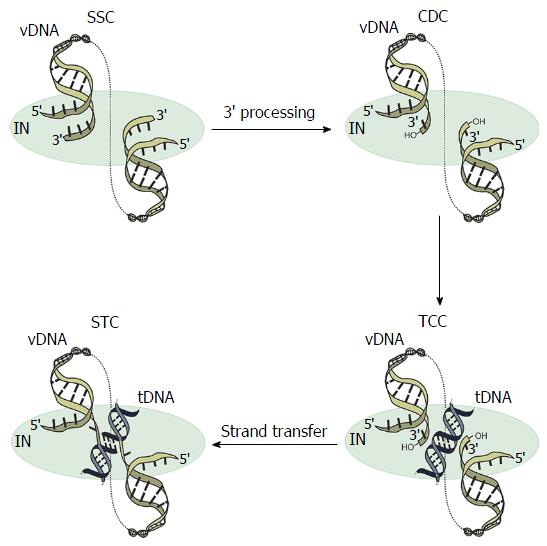
Retroviral IN performs two biochemically and temporally distinct bimolecular nucleophilic substitution (SN2) reactions[11]: 3’-processing and strand transfer (Figure 1). During 3’-processing, a di- or trinucleotide is hydrolytically cleaved from each 3’ vDNA end[12-14], exposing reactive hydroxyl groups of invariant CA dinucleotides. These groups act as nucleophiles for subsequent strand transfer whereby the newly processed 3’ vDNA ends are covalently inserted into a major groove of tDNA in a staggered fashion. The product of the second reaction is an integration intermediate that is characterized by unjoined 5’ vDNA overhangs[15,16]. Following disassembly of the associated strand transfer complex (STC, Figure 1), a DNA polymerase, 5’ flap endonuclease, and DNA ligase are required to fill in the single-strand gap regions in tDNA, excise 5’ vDNA overhangs, and join the vDNA 5’ ends to host DNA strands, respectively (reviewed in[17]). During this process, short target site duplications are generated, which flank the integrated provirus. Depending on the genus of retrovirus, the size of these target site duplications ranges from 4-6 base pairs (bp). Whereas spumavirus prototype foamy virus (PFV)[18,19] and lentivirus human immunodeficiency virus 1 (HIV-1)[20,21] integration yield 4 bp and 5 bp target site duplications, respectively, mouse mammary tumor virus (MMTV)[22] and Rous sarcoma virus (RSV)[23,24] INs cleave tDNA phosphodiester bonds that are separated by 6 bp.
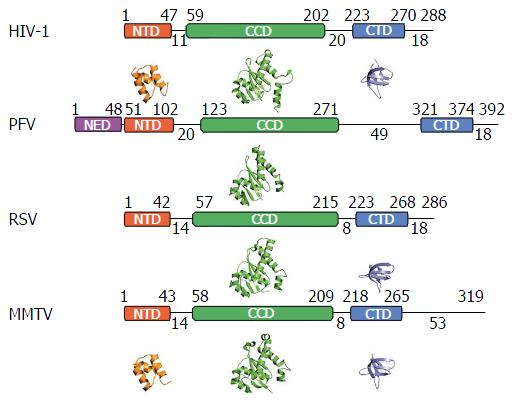
Retroviral IN proteins consist of approximately 275-470 amino acid residues. The INs to be discussed in detail in this review amount to 288 (HIV-1), 392 (PFV), 286 (RSV), and 319 (MMTV) residues[25-27]. Retroviral INs comprise three domains common to all genera: The N-terminal domain (NTD), the catalytic core domain (CCD), and the C-terminal domain (CTD)[28-32], which connect to one another via flexible linkers that vary in length across the viruses (Figure 2). The NTD adopts a helix-turn-helix fold and harbors two pairs of Zn2+-coordinating histidine and cysteine residues (HHCC motif), which are additionally conserved in retrotransposon INs and are involved in the recognition of the viral LTRs[30,31,33-35]. Accordingly, Zn2+ binding triggers HIV-1 IN multimerization and increases its catalytic activity[36,37]. The CCD adopts an RNase H fold and coordinates two Mg2+ ions via the invariant Asp and Glu amino acid residues that comprise the D, DX35E catalytic triad motif[28,29,38-40]. The coordination of Mg2+ ions chemically activates the nucleophiles for 3’-processing (water) as well as for strand transfer (3’-OH groups of vDNA) and destabilizes the respective scissile phosphodiester bonds[41-43]. The CTD is the least conserved among the shared IN domains, however, the tertiary structures of resolved CTDs show similar characteristics: They adopt a Src homology 3 fold[44,45], are involved in DNA binding[46], and promote IN multimerization[47,48]. Some retroviruses, namely spuma-, ε- and γ-retroviruses, harbor an additional NTD extension domain (NED) that precedes the NTD[26,40,49] and engages vDNA in the context of the intasome structure[40]. These IN proteins accordingly are larger than their lenti-, α-, β- and δ-retroviral cousins that lack the NED.
Numerous biochemical studies revealed that the active form of retroviral IN is a multimer that engages vDNA and tDNA in the confines of a nucleoprotein complex[50-58]. Bacteriophage Mu-mediated PCR footprinting of PICs extracted from infected cells revealed the protection of several hundred bp at the vDNA ends, and the associated complex was termed “intasome” to distinguish it from the larger PICs[59,60]. Subsequently, the intasome term was adopted by structural biologists who constructed and purified distinct, functional IN-DNA complexes capable of efficient concerted integration of two synapsed oligonucleotide vDNA ends, the structures of which were solved by X-ray crystallography[40,43,61-63] or single particle cryo-electron microscopy (EM)[26]. Although retroviral INs have been studied for decades, the 3-dimensional structures of PFV intasomes greatly aided the elucidation of the details of 3’-processing and strand transfer reaction mechanisms[40,43,62].
The “intasome” term today applies to the family of nucleoprotein complexes that are known to mediate retroviral DNA integration (Figure 1), which encompasses the stable synaptic complex (SSC)[43], the cleaved donor complex (CDC) or cleaved intasome[40,43], the target capture complex (TCC)[43,62], and the STC[62,63] (Figure 1). The SSC forms upon IN binding to the vDNA ends[5,40,55,57-60,64,65]. The hydrolytic cleavage of a di- or trinucleotide from each 3’-end marks the transition to the CDC. The TCC forms when the CDC engages tDNA, whereas the STC is formed when the vDNA ends are inserted into tDNA and thus strand transfer is completed[57,58,61-64,66]. The PFV system has importantly afforded high-resolution structures for each of these complexes[40,43,62,63].
Mechanistic studies of retroviral DNA integration began in earnest in the late 1980s and early 1990s when PICs partially purified from infected cells were shown to promote correct DNA integration in vitro[5,14,60,67-70]. Nearly parallel biochemical studies, which utilized purified avian, murine, and human retroviral proteins, importantly showed that IN alone was sufficient to catalyze both the 3’-processing and strand transfer of recombinant vDNA substrates[71-77]. The results of these experiments opened up a new field dedicated to the structural and functional analysis of retroviral INs. Initial work on HIV-1 IN quickly revealed the relatively poor solubility of the full-length enzyme in vitro, which limited further biochemical and thus structural characterization[48]. To date, there is no high-resolution structure of a full-length retroviral IN in the absence of DNA, which is likely attributable to the inherent flexibility of the interdomain linkers[78-80]. In essence, the intertwined architecture of IN with vDNA in the context of the intasome complex necessarily “locks down” the inherently flexible enzymes, which afforded platforms for their detailed structural analyses.
Initial structures of individual IN domains or two-domain constructs in the absence of DNA (reviewed in[81,82]) (Figure 2) turned out to be challenging in many cases, and were only possible for the HIV-1 IN protein with the help of solubility-enhancing mutations. By systematically replacing hydrophobic residues in the HIV-1 IN CCD, Phe185 was identified as a solubility-limiting residue[83]: Substituting either Lys or His for Phe185 dramatically improved HIV-1 IN protein solubility[48,84]. Although the F185K change enabled the determination of the HIV-1 CCD X-ray structure[85], it conferred a lethal viral phenotype due to deficiencies in virus particle assembly and reverse transcription in addition to defective vDNA integration[48]. Indeed, the vast majority of IN mutations elicit such pleotropic defects on HIV-1 replication (reviewed in[86]). In contrast, the F185H change was tolerated by the virus, and importantly enabled crystallization of the CCD[84,87]. In both cases, the positively charged substituent residue reaches across the CCD dimerization interface and makes a hydrogen bond contact with the main chain carbonyl of Ala105 of the partner IN monomer. The F185K and F185H changes are therefore likely to dramatically increase HIV-1 IN solubility by removing a surface exposed aromatic residue that may nucleate protein aggregation, as well as by enhancing multimerization by adding two hydrogen bonds per IN dimer. Even greater solubility of the HIV-1 CCD was achieved by mutating the tryptophan at position 131 to glutamic acid in the context of the F185K change[88]. Simultaneous to the work on the HIV-1 IN CCD, the X-ray structure of the avian sarcoma virus (ASV) IN CCD was solved[89]. Of note, ASV IN harbors His at the position analogous to Phe185 in HIV-1 IN, and solubility-enhancing mutations accordingly were not required to crystallize the ASV IN CCD. Succeeding structures of HIV-1[88,90-93] and ASV[94-97] IN CCDs elucidated binding sites for metal ion cofactors as well as for early inhibitors. NTD and CTD structures of HIV-1 IN and HIV-2 IN, which were solved by nuclear magnetic resonance spectroscopy[34,35,44,45,98,99], suggested functions of the NTDs in metal ion coordination and the binding of the CTD to vDNA, each of which is required for integration.
Two-domain IN constructs were initially studied as an approach to understand how individual IN domains might interact to form an active nucleoprotein complex. Three different CCD-CTD two-domain structures for HIV-1, simian immunodeficiency virus (SIV), and avian sarcoma-leukosis virus (ASLV) were reported in 2000[100-102]. Each construct harbored at least one solubility-enhancing mutation: C56S, W131D, F139D, F185K, and C280S for HIV-1, F185H for SIV, and F199K for ASLV, the latter two of which are analogous to the F185K change in HIV-1. The HIV-1 CCD-CTD structure revealed an extended α-helix for the CCD-CTD linker region[100]. Whereas the SIV IN CCD-CTD linker could not be traced[101], the RSV IN CCD-CTD linker had a rather extended, non-helical form compared to the HIV-1 IN CCD-CTD linker[102]. The appearance of three different linker configurations in three different IN CCD-CTD constructs led some to suggest that such configurations may result from crystal packing and therefore represent limited physiological relevance[82].
Crystallization of a HIV-1 IN NTD-CCD construct was achieved in 2001 by including W131D, F139D and F185K solubility-enhancing mutations[103]. The resulting X-ray structure revealed possible interactions between two of the NTDs with two CCDs of opposing NTD-CCD molecules, which was of potential physiological relevance due to the fact that it was known from prior biochemical studies that the NTD functioned in trans with the CCD[51-53]. However, the inability to trace the NTD-CCD linker regions limited the confidence of this interpretation. Importantly, the domain sharing arrangement suggested by this structure was later confirmed by additional NTD-CCD structures and mutagenesis[104], and ultimately through the elucidation of intasome structures (see below).
In the early 2000s, a host factor implicated in the nuclear retention of HIV-1 IN, lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75), was reported to increase the solubility of HIV-1 IN through its tight binding interaction[105-107]. LEDGF/p75 is a lentiviral-specific IN-binding protein[105,108,109] that tethers vDNA integration to transcriptionally active regions of the host genome (reviewed in[110,111]). LEDGF/p75 engages lentiviral IN via its C-terminally located IN-binding domain (IBD)[112]. Although the HIV-1 IN CCD was sufficient for LEDGF/p75 binding, the NTD was required for the high efficiency interaction[107]. LEDGF/p75 binding stabilizes lentiviral IN tetramerization[104,113], which is likely related to its ability to enhance the solubility of the viral proteins.
Crystal structures of lentiviral INs in complex with LEDGF/p75 include an HIV-1 IN F185K CCD construct[114] as well as HIV-2[115] and maedi-visna virus (MVV) IN NTD-CCD two-domain fragments[104]. Though HIV-2 and MVV INs harbor hydrophobic residues at the positions analogous to Phe185 in HIV-1 IN (Phe and Ile, respectively), the favorable solubility properties of lentiviral IN-LEDGF/p75 complexes likely dispelled the need for solubility-enhancing mutations for the crystallization of these constructs. The LEDGF/p75 IBD is a PHAT domain composed of two helix-hairpin-helix motifs[116], with Asp366 at the tip of the N-terminal hairpin nestling into a binding cleft at the HIV-1 IN CCD dimerization interface and contacting the main chain amides of IN residues Glu170 and His171 via hydrogen bonds[114]. A novel class of potent anti-HIV compounds, known as LEDGINs (LEDGF-IN inhibitors) or ALLINIs (allosteric IN inhibitors), structurally mimic the role of Asp366 in their binding to HIV-1 IN, which accounts for their abilities to compete for LEDGF/p75 binding to IN (reviewed in[117]). The two domain NTD-CCD constructs revealed the structural basis for the IN NTD interaction with the LEDGF/p75 IBD, which was ionic in nature. Interestingly, the polarities of the participating salt bridges were functionally reversible, such that HIV-1 particles carrying NTD reverse charge substitutions that were otherwise dead regained partial activity in the presence of the complementary reverse charge LEDGF/p75 partner protein[115,118].
Although the aforementioned individual and two-domain constructs provided insight into retroviral IN function, the field sorely required the structural determination of a functional intasome. The sole class of clinically approved HIV-1 IN inhibitors, known as IN strand transfer inhibitors (INSTIs), displays little if any binding affinity for free IN protein; their clinical target is the IN-vDNA complex[119]. Fortuitously, INSTIs are active against most types of retroviruses[120-122], so intasome structures derived from basically any retroviral genus would have in theory provided a backdrop for understanding the structural basis for INSTI action and the clinical emergence of drug resistance.
Due to the poor solubility of HIV-1 and other early studied retroviral INs, the search for an enzyme with more favorable biochemical properties for in vitro experimentation and crystallography was initiated. Though early work had revealed that relatively short oligonucleotide substrates, which modeled the vDNA ends, supported IN 3’ processing and strand transfer activities[72-77], not all enzymes behaved similarly. Most critical for intasome structural biology was the ability for the IN multimer to coordinate the binding of two vDNA ends, and insert these in concerted fashion into opposing strands of tDNA. Critically, PFV IN was discovered to promote efficient concerted integration of oligonucleotide vDNA ends[121]. By contrast, HIV-1 IN had revealed the tendency to insert just one vDNA end at a time[77]. Subsequent modifications of HIV-1 IN expression systems, including protein purification under relatively dilute conditions to prevent IN aggregation[123], or by fusing the small Sso7d DNA binding domain from Sulfolobus solfataricus to the IN N-terminus to mimic the NED that naturally exists in PFV IN[124], yielded proteins that supported efficient concerted integration activity. Such modifications might eventually prove useful to characterize HIV-1 intasomes structurally[123,124].
Functional PFV-vDNA complexes assembled by differential salt dialysis migrated as a distinct species on gel filtration columns, and remained intact and active following challenge with high salt concentrations[40]. The initial X-ray crystal structure of the PFV intasome, representing the CDC, was reported in 2010[40] (Figure 3). To date, 37 PFV intasome structures composed of wild-type IN or mutant variants that contain clinically relevant amino acid substitutions have been solved by X-ray crystallography or cryo-EM, representing complexes in the presence of divalent metal ion cofactors, tDNA/nucleosomes, and INSTIs[40,43,62,63,125-130]. The INSTI-bound structures elucidated the mechanism of drug action: The halo-benzyl chemical group common to these compounds assumes the position of the invariant 3’ deoxyadenylate in vDNA with its critical hydroxyl group, thus ejecting the strand transfer nucleophile from the enzyme active site and disarming the nucleoprotein complex[40].
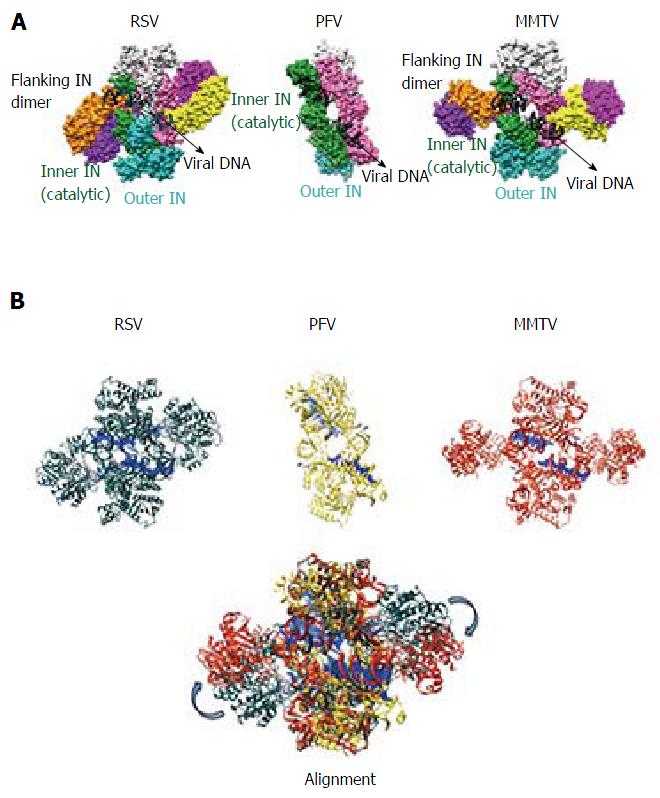
The PFV intasome consists of a tetrameric assembly of IN arranged around a dimer-of-dimers architecture[40] (Figure 3). The inner dimer is composed of two inner monomers (green and pink in Figure 3A), whereas each outer dimer is composed of an inner IN monomer and an outer IN monomer (cyan-green and pink-light grey in Figures 3 and 4). The inner IN monomers make all contacts with vDNA and thus are the catalytic subunits, with each of their constitutive domains mediating vDNA in addition to IN-IN contacts. As previously alluded to, the catalytic subunits are established via a domain sharing mechanism whereby the NTD of each inner IN monomer interacts intimately with the CCD of the opposing IN monomer. The outer IN dimers center around the extensive CCD dimeric interface observed in prior retroviral IN CCD crystal structures[85,87-97]. The CTDs, NTDs, and NEDs of the outer IN monomers are not resolved in the electron density maps, and it is currently unclear what precise role(s) they may play in the catalysis of vDNA integration[131]. It is generally thought that the outer IN monomers mainly play a supportive architectural role to truss the inner IN monomers and vDNA together. As the outer CTDs can contribute to nucleosome binding in vitro[130], it seems possible they might play a role during vDNA integration into chromatinized templates, as occurs during virus infection. The NTD-CCD and CCD-CTD linkers, which are only visible for the inner IN monomers in the crystal structures, adopt extended conformations and contact the vDNA[40].
MMTV and RSV intasome structures were recently solved using single-particle cryo-EM[26] and X-ray crystallography[61], respectively. The MMTV intasome was assembled using pre-processed 22 bp vDNA, and thus represents the CDC[26]. The RSV intasome structure by contrast is the STC, which was assembled using a so-called X-mer disintegration substrate[132] where three oligonucleotide strands were annealed together to yield a synapsed complex composed of two 22 bp vDNA branches covalently linked through a central 6 bp stagger to a 38 bp palindromic tDNA[61]. The crystal structure of the PFV STC assembled with its analogous X-mer DNA substrate[63] was virtually identical to the structure that was solved when the CDC integrated into tDNA during crystallogenesis[62], validating the X-mer substrate design approach for RSV STC crystallography.
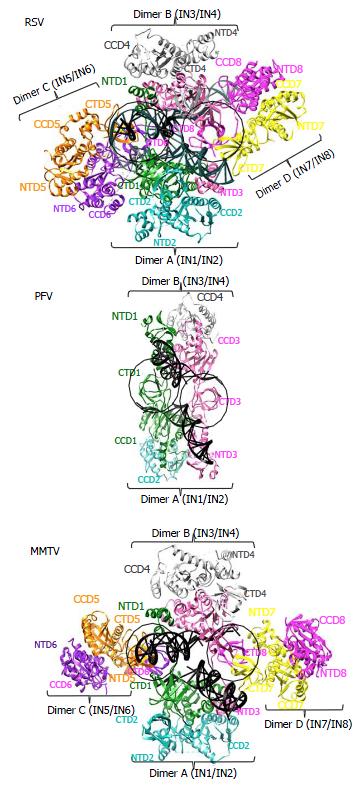
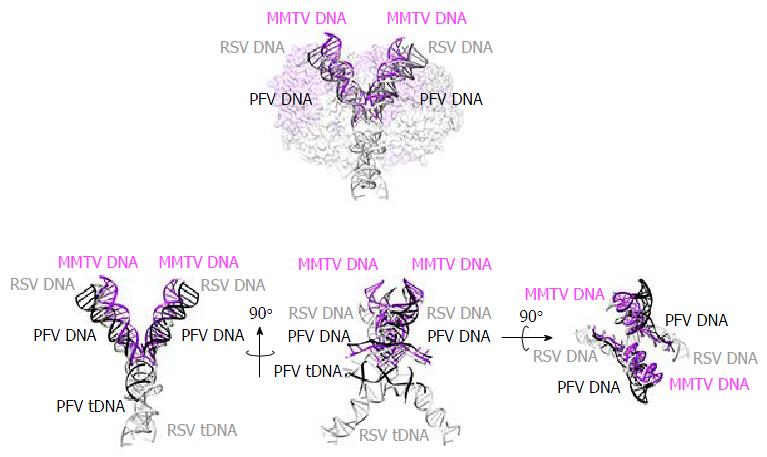
Although the tetrameric IN4-to-vDNA2 stoichiometry represented by the PFV intasome was generally thought to be evolutionarily conserved across the retroviruses[54,55,57,58], the intasome structures of MMTV and RSV strikingly revealed octameric IN assemblies[26,61] (Figures 3 and 4). The MMTV and RSV intasomes comprise a core density region consisting of IN dimers A and B, as well as flanking density regions that consist of IN dimers C and D (Figure 3). Analogous to the PFV structure, core inner IN monomers IN1 and IN3 intimately contact the vDNA ends and are catalytically active, with their NTDs reaching out and contacting the CCDs of the opposing inner monomer (Figure 4). The core structure moreover is primarily supported through CCD dimerization interfaces with outer IN monomers IN2 and IN4. In contrast to PFV, flanking structures in the MMTV and RSV intasomes constitute additional IN dimers, which on their own multimerize primarily through the familiar CCD dimer interface[26,61]. The NTD-CCD linker that is extended in IN1 and IN3 to contact the opposing CCD in trans is contracted in the other IN monomers[26]. This observation highlights the necessity for NTD-CCD linker flexibility: Though principally contracted, it must also possess the ability to extend when situated at the IN1 and IN3 positions to support IN catalytic function. In hindsight, it is not surprising that the linker regions in the original HIV-1 IN NTD-CCD structure, which lacked LEDGF/p75 or DNA binding partners, were untraceable[103]. Of note, whereas the crystallographic PFV intasome structure is rather rigid, the flanking dimer regions in the MMTV and RSV intasome structures reveal significant flexibility (Figure 3B). As small angle X-ray scattering analysis of the PFV intasome revealed significant conformational flexibility for the outer subunit NEDs, NTDs and CTDs of the IN tetramer[79], it is tempting to describe retroviral intasomes as common rigid core structures surrounded by extraneous elements that, although likely to play physiologically relevant roles during virus infection, display marked movement as purified biochemical entities.
The most striking difference between the tetrameric and octameric IN assemblies is the unique function attributed to the CTDs of the flanking dimers in the MMTV and RSV intasome structures. While contacts with vDNA in the PFV structure are restricted to the inner IN1 and IN3 subunits, MMTV and RSV INs donate their CTDs in trans to the core region of the intasome[26,61]. The locations of six CTDs, including those of the flanking IN dimers, are conserved in the MMTV and RSV intasomes (Figure 4). The exclusive conformation of the CTDs allows them to tightly associate near the vDNA and assume positions resembling those of the inner PFV CTDs (Figure 4). Biochemical complementation assays revealed that the flanking MMTV IN dimers are crucial for IN catalytic function[26]. Intriguingly, the length of the CCD-CTD linker is quite variable among retroviral INs, amounting to about 50 residues for PFV, but only to 8 residues for MMTV and RSV IN[26] (Figure 2). The extended conformation of the PFV IN 49-mer CTD-CCD linker affords the positioning of IN1 and IN3 CTDs to enable critical contacts with vDNA and tDNA during integration[40,62]. The analogous HIV-1 IN linker, composed of 20 residues (Figure 2), could be stretched to similarly position inner IN monomer CTDs in a molecular model of the HIV-1 intasome based on the PFV structure[133]. However, it is physically impossible for an 8 amino acid region to span the required distance. MMTV and RSV accordingly solve this conundrum by employing additional IN molecules to donate their CTDs to the required positions in the intasome core structure (Figure 4). Hence, retroviral IN CTD-CCD linker length is suggested to be a key determinant for the higher-order architecture of the respective intasome structures[26,61].
The integration of vDNA into tDNA does not occur randomly in the host genome, with integration site selection preferences varying among retroviruses (recently reviewed in[134]). Whereas HIV-1 and other lentiviruses favor integration into highly expressed genes that are rich in introns[135-139], the spumavirus PFV avoids active gene regions[130,140,141]. Interestingly, α- and β-retroviruses such as RSV and MMTV are the least selective in integration site selection, displaying patterns that much more closely approach random[138,142,143].
The co-crystallization of the PFV intasome with an oligonucleotide tDNA[62] derived from the PFV consensus integration sequence[121,140] elucidated the mechanism of strand transfer. Whereas crystallization in the absence of Mg2+ or the presence of a dideoxy viral 3’ end led to high-resolution TCC structures, the addition of Mg2+ with the normal vDNA end afforded integration during crystallogenesis, yielding the first high-resolution structure of a retroviral or bacterial transposon STC[62] (Figure 5). The tDNA adopted a highly bent conformation at the PFV DNA insertion sites, with the major groove widened to 26.3 Å and the minor groove compressed to 9.6 Å[62]. This conformation enables the accommodation of the inner IN1 and IN3 D,DX35E catalytic triads to the scissile phosphodiester bonds of tDNA, thus promoting integration[62]. As SN2 transesterification reactions are iso-energetic, they have the potential to reverse direction if chemical leaving groups remain associated with the catalytic active site. Following strand transfer, the newly formed phosphodiester bonds are displaced by 2.3 Å from the IN active sites, effectively suppressing the probability for strand transfer reversal[62]. A similar displacement is described for the tDNA in the RSV intasome[61].
Early studies revealed that retroviral INs prefer chromatinized tDNA templates over naked DNA for integration in vitro[144-147], and subsequent work revealed the propensity to similarly target nucleosomes during virus infection[135,148]. PFV IN prefers relatively flexible tDNA sequences for integration[62], and a cryo-EM structure of an intasome-nucleosome complex revealed the same degree of local tDNA distortion can occur on the nucleosome surface during PFV integration[130]. Since spumaviral INs cleave tDNA with a 4 bp stagger, it has been suggested that the degree of tDNA kink has to be greater for spumaviral integration than for viruses that cleave target DNA with a 6 bp stagger, such as the α- and β-retroviruses[149]. Although the IN1 and IN3 catalytic triads of the PFV and MMTV intasomes are superimposable, modeling of tDNA into the MMTV CDC revealed a relatively unbent conformation to accommodate a 6 bp staggered cut as compared to the highly bent tDNA conformation for the 4 bp stagger in the PFV TCC[26] (Figure 5).
The RSV STC harbors a highly bent tDNA conformation despite the fact that RSV IN cleaves the DNA with a 6 bp stagger (Figure 5). Kinks located at the vDNA/tDNA junctions of the RSV intasome provoke a 20 Å shift in the helical axis, leading to an overall tDNA twist[61]. Hence, the tDNA conformation in the RSV STC differs significantly from the tDNA conformation in the PFV structure (Figure 5) and also from the relatively unbent tDNA confirmation in the MMTV TCC model[26]. These observations suggest potentially different modes of integration into nucleosomal DNA among the studied viruses. Whereas PFV and RSV are predicted to target chromatinized DNA during virus infection, MMTV was unique among a study of 10 exogenous retroviruses for its apparent avoidance of nucleosomal DNA in vivo[149]. Elucidation of high-resolution MMTV STC/TCC structures will help to illuminate the degree of tDNA bending that occurs during MMTV integration.
Based on the variety of tDNA structural properties that influence target site selectivity, including bendability[62,149-152], major groove widening, and nucleosomal packaging[144-147,153,154], retroviral INs can be classified as shape-readout DNA binding proteins[155]. DNA minicircles, which mimic nucleosome-induced tDNA circularization in the absence of histones, represent a relatively new tool to tease out physiologically-relevant influences of tDNA structure on integration site selectivity in vitro, and the roles of IN-binding cofactors such as LEDGF/p75[156].
The study of retroviral integration has come a long way since its beginnings in the late 1970s. The relatively large repertoire of individual and two-domain retroviral IN structures that were solved initially has since expanded to a set of high-resolution intasome structures, including those from the spumavirus PFV, β-retrovirus MMTV, and α-retrovirus RSV. To date, the plethora of PFV intasome structures represents a remarkable advance for the field of retroviral integration. Not only have they elucidated the mechanism of INSTI action, they provide high-resolution structures of the entire set of complexes (SSC, CDC, TCC and STC) that mediate retroviral DNA integration[40,43,62,63].
The recently emerged octameric intasome structures of MMTV[26] and RSV[61] reveal an unexpected evolutionary diversity among retroviruses. As the intasomal core is conserved among the three studied retroviruses, the utilization of flanking dimers to complete the functional MMTV and RSV intasome structures represents a remarkable example of convergent evolution of the DNA integration apparatus[26,61]. Considering CCD-CTD linker length as a predictor of the state of IN multimerization within functional intasomes[26], it remains to be investigated whether retroviral INs with intermediary linker lengths, including those of HIV-1 and the δ-retrovirus human T-cell lymphotropic virus, which harbor 20 and 19 residues, respectively[26], will reveal tetrameric, octameric, or perhaps even higher-order IN assemblies.
Motivated by the recent advances in the intasome field, new IN-DNA complexes are currently being investigated in various laboratories, including those derived from lentiviruses. The emergence of new three-dimensional intasome structures will help to model novel interactions between HIV-1 IN and DNA, and thus should reveal new insights into the mechanisms of emergence of drug resistance to clinical INSTIs.
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): A, A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Arriagada GL, Kodama EN, Pandey KK S- Editor: Ji FF L- Editor: A E- Editor: Li D
| 1. | Iordanskiy S, Bukrinsky M. Reverse transcription complex: the key player of the early phase of HIV replication. Future Virol. 2007;2:49-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 2. | Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 300] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 3. | Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910-1915. [PubMed] |
| 4. | Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125-6129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 357] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 5. | Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382-5390. [PubMed] |
| 6. | Raghavendra NK, Shkriabai N, Graham RLj, Hess S, Kvaratskhelia M, Wu L. Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology. 2010;7:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 7. | Schweitzer CJ, Jagadish T, Haverland N, Ciborowski P, Belshan M. Proteomic analysis of early HIV-1 nucleoprotein complexes. J Proteome Res. 2013;12:559-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Vande Woude GF, Oskarsson M, Enquist LW, Nomura S, Sullivan M, Fischinger PJ. Cloning of integrated Moloney sarcoma proviral DNA sequences in bacteriophage lambda. Proc Natl Acad Sci USA. 1979;76:4464-4468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Van Beveren C, Goddard JG, Berns A, Verma IM. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5’ long terminal repeat and adjacent cellular sequences. Proc Natl Acad Sci USA. 1980;77:3307-3311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 99] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Ju G, Skalka AM. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980;22:379-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 150] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 511] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 12. | Pauza CD. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 13. | Roth MJ, Schwartzberg PL, Goff SP. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 221] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 14. | Lee YM, Coffin JM. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol Cell Biol. 1991;11:1419-1430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 33] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 305] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 16. | Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525-2529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 293] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 17. | Lesbats P, Engelman AN, Cherepanov P. Retroviral DNA Integration. Chem Rev. 2016;116:12730-12757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 162] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 18. | Schweizer M, Fleps U, Jäckle A, Renne R, Turek R, Neumann-Haefelin D. Simian foamy virus type 3 (SFV-3) in latently infected Vero cells: reactivation by demethylation of proviral DNA. Virology. 1993;192:663-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Neves M, Périès J, Saïb A. Study of human foamy virus proviral integration in chronically infected murine cells. Res Virol. 1998;149:393-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Vincent KA, York-Higgins D, Quiroga M, Brown PO. Host sequences flanking the HIV provirus. Nucleic Acids Res. 1990;18:6045-6047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 21. | Vink C, Groenink M, Elgersma Y, Fouchier RA, Tersmette M, Plasterk RH. Analysis of the junctions between human immunodeficiency virus type 1 proviral DNA and human DNA. J Virol. 1990;64:5626-5627. [PubMed] |
| 22. | Majors JE, Varmus HE. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981;289:253-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 214] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 23. | Hishinuma F, DeBona PJ, Astrin S, Skalka AM. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981;23:155-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 119] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 24. | Hughes SH, Mutschler A, Bishop JM, Varmus HE. A Rous sarcoma virus provirus is flanked by short direct repeats of a cellular DNA sequence present in only one copy prior to integration. Proc Natl Acad Sci USA. 1981;78:4299-4303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Grandgenett DP, Pandey KK, Bera S, Aihara H. Multifunctional facets of retrovirus integrase. World J Biol Chem. 2015;6:83-94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (1)] |
| 26. | Ballandras-Colas A, Brown M, Cook NJ, Dewdney TG, Demeler B, Cherepanov P, Lyumkis D, Engelman AN. Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature. 2016;530:358-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 27. | Andrake MD, Skalka AM. Retroviral Integrase: Then and Now. Annu Rev Virol. 2015;2:241-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331-2338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 281] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 29. | Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361-6369. [PubMed] |
| 30. | Burke CJ, Sanyal G, Bruner MW, Ryan JA, LaFemina RL, Robbins HL, Zeft AS, Middaugh CR, Cordingley MG. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992;267:9639-9644. [PubMed] |
| 31. | Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428-3432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 289] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 32. | Woerner AM, Klutch M, Levin JG, Marcus-Sekura CJ. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses. 1992;8:297-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 33. | Khan E, Mack JP, Katz RA, Kulkosky J, Skalka AM. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 34. | Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 255] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 35. | Eijkelenboom AP, van den Ent FM, Vos A, Doreleijers JF, Hård K, Tullius TD, Plasterk RH, Kaptein R, Boelens R. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr Biol. 1997;7:739-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 88] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 36. | Zheng R, Jenkins TM, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659-13664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 261] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 37. | Lee SP, Han MK. Zinc stimulates Mg2+-dependent 3’-processing activity of human immunodeficiency virus type 1 integrase in vitro. Biochemistry. 1996;35:3837-3844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 52] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 38. | Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 170] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 39. | Leavitt AD, Shiue L, Varmus HE. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113-2119. [PubMed] |
| 40. | Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 552] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 41. | Mizuuchi K. Polynucleotidyl transfer reactions in transpositional DNA recombination. J Biol Chem. 1992;267:21273-21276. [PubMed] |
| 42. | Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 455] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 43. | Hare S, Maertens GN, Cherepanov P. 3’-processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 2012;31:3020-3028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 44. | Eijkelenboom AP, Lutzke RA, Boelens R, Plasterk RH, Kaptein R, Hård K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 189] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 45. | Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826-9833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 222] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 46. | Engelman A, Hickman AB, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911-5917. [PubMed] |
| 47. | Andrake MD, Skalka AM. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299-29306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 48. | Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271:7712-7718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 225] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 49. | Krishnan L, Engelman A. Retroviral integrase proteins and HIV-1 DNA integration. J Biol Chem. 2012;287:40858-40866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 50. | Jones KS, Coleman J, Merkel GW, Laue TM, Skalka AM. Retroviral integrase functions as a multimer and can turn over catalytically. J Biol Chem. 1992;267:16037-16040. [PubMed] |
| 51. | Engelman A, Bushman FD, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269-3275. [PubMed] |
| 52. | van Gent DC, Vink C, Groeneger AA, Plasterk RH. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261-3267. [PubMed] |
| 53. | Jonsson CB, Donzella GA, Gaucan E, Smith CM, Roth MJ. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J Virol. 1996;70:4585-4597. [PubMed] |
| 54. | Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. Functional oligomeric state of avian sarcoma virus integrase. J Biol Chem. 2003;278:1323-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977-986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 153] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 56. | Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J Biol Chem. 2006;281:22707-22719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 123] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 57. | Li M, Mizuuchi M, Burke TR, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295-1304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 202] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 58. | Bera S, Pandey KK, Vora AC, Grandgenett DP. Molecular Interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J Mol Biol. 2009;389:183-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Wei SQ, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511-7520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 81] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 60. | Chen H, Wei SQ, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J Biol Chem. 1999;274:17358-17364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 82] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 61. | Yin Z, Shi K, Banerjee S, Pandey KK, Bera S, Grandgenett DP, Aihara H. Crystal structure of the Rous sarcoma virus intasome. Nature. 2016;530:362-366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 62. | Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326-329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 267] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 63. | Yin Z, Lapkouski M, Yang W, Craigie R. Assembly of prototype foamy virus strand transfer complexes on product DNA bypassing catalysis of integration. Protein Sci. 2012;21:1849-1857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 64. | Kotova S, Li M, Dimitriadis EK, Craigie R. Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J Mol Biol. 2010;399:491-500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 65. | Kessl JJ, Li M, Ignatov M, Shkriabai N, Eidahl JO, Feng L, Musier-Forsyth K, Craigie R, Kvaratskhelia M. FRET analysis reveals distinct conformations of IN tetramers in the presence of viral DNA or LEDGF/p75. Nucleic Acids Res. 2011;39:9009-9022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 66. | Pandey KK, Bera S, Zahm J, Vora A, Stillmock K, Hazuda D, Grandgenett DP. Inhibition of human immunodeficiency virus type 1 concerted integration by strand transfer inhibitors which recognize a transient structural intermediate. J Virol. 2007;81:12189-12199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 67. | Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 362] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 68. | Farnet CM, Haseltine WA. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164-4168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 206] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 69. | Ellison V, Abrams H, Roe T, Lifson J, Brown P. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711-2715. [PubMed] |
| 70. | Wei SQ, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535-10540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 41] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 71. | Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 254] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 72. | Katzman M, Katz RA, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319-5327. [PubMed] |
| 73. | Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 292] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 74. | Pahl A, Flügel RM. Endonucleolytic cleavages and DNA-joining activities of the integration protein of human foamy virus. J Virol. 1993;67:5426-5434. [PubMed] |
| 75. | Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119-5123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 281] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 76. | Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 339] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 77. | Bushman FD, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 340] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 78. | Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A. HIV-1 Integrase. Neamati N, editor. Hoboken (NJ): John Wiley & Sons, Inc 2011; 35-49. [DOI] [Full Text] |
| 79. | Gupta K, Curtis JE, Krueger S, Hwang Y, Cherepanov P, Bushman FD, Van Duyne GD. Solution conformations of prototype foamy virus integrase and its stable synaptic complex with U5 viral DNA. Structure. 2012;20:1918-1928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 80. | Zhao Z, McKee CJ, Kessl JJ, Santos WL, Daigle JE, Engelman A, Verdine G, Kvaratskhelia M. Subunit-specific protein footprinting reveals significant structural rearrangements and a role for N-terminal Lys-14 of HIV-1 Integrase during viral DNA binding. J Biol Chem. 2008;283:5632-5641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 81. | Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009;276:2926-2946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 82. | Li X, Krishnan L, Cherepanov P, Engelman A. Structural biology of retroviral DNA integration. Virology. 2011;411:194-205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 83. | Jenkins TM, Hickman AB, Dyda F, Ghirlando R, Davies DR, Craigie R. Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc Natl Acad Sci USA. 1995;92:6057-6061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 117] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 84. | Engelman A, Liu Y, Chen H, Farzan M, Dyda F. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J Virol. 1997;71:3507-3514. [PubMed] |
| 85. | Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 623] [Cited by in RCA: 620] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 86. | Engelman A. In vivo analysis of retroviral integrase structure and function. Adv Virus Res. 1999;52:411-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 141] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 87. | Bujacz G, Alexandratos J, Qing ZL, Clément-Mella C, Wlodawer A. The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant. FEBS Lett. 1996;398:175-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 90] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 88. | Goldgur Y, Craigie R, Cohen GH, Fujiwara T, Yoshinaga T, Fujishita T, Sugimoto H, Endo T, Murai H, Davies DR. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc Natl Acad Sci USA. 1999;96:13040-13043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 396] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 89. | Bujacz G, Jaskólski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka AM. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J Mol Biol. 1995;253:333-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 172] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 90. | Goldgur Y, Dyda F, Hickman AB, Jenkins TM, Craigie R, Davies DR. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc Natl Acad Sci USA. 1998;95:9150-9154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 300] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 91. | Greenwald J, Le V, Butler SL, Bushman FD, Choe S. The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochemistry. 1999;38:8892-8898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 118] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 92. | Maignan S, Guilloteau JP, Zhou-Liu Q, Clément-Mella C, Mikol V. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J Mol Biol. 1998;282:359-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 218] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 93. | Molteni V, Greenwald J, Rhodes D, Hwang Y, Kwiatkowski W, Bushman FD, Siegel JS, Choe S. Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain. Acta Crystallogr D Biol Crystallogr. 2001;57:536-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 94. | Bujacz G, Alexandratos J, Wlodawer A, Merkel G, Andrake M, Katz RA, Skalka AM. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J Biol Chem. 1997;272:18161-18168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 113] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 95. | Bujacz G, Jaskólski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka AM. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure. 1996;4:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 141] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 96. | Lubkowski J, Yang F, Alexandratos J, Merkel G, Katz RA, Gravuer K, Skalka AM, Wlodawer A. Structural basis for inactivating mutations and pH-dependent activity of avian sarcoma virus integrase. J Biol Chem. 1998;273:32685-32689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 97. | Lubkowski J, Dauter Z, Yang F, Alexandratos J, Merkel G, Skalka AM, Wlodawer A. Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant. Biochemistry. 1999;38:13512-13522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 98. | Eijkelenboom AP, Sprangers R, Hård K, Puras Lutzke RA, Plasterk RH, Boelens R, Kaptein R. Refined solution structure of the C-terminal DNA-binding domain of human immunovirus-1 integrase. Proteins. 1999;36:556-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 99. | Eijkelenboom AP, van den Ent FM, Wechselberger R, Plasterk RH, Kaptein R, Boelens R. Refined solution structure of the dimeric N-terminal HHCC domain of HIV-2 integrase. J Biomol NMR. 2000;18:119-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 100. | Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci USA. 2000;97:8233-8238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 330] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 101. | Chen Z, Yan Y, Munshi S, Li Y, Zugay-Murphy J, Xu B, Witmer M, Felock P, Wolfe A, Sardana V. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform. J Mol Biol. 2000;296:521-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 85] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 102. | Yang ZN, Mueser TC, Bushman FD, Hyde CC. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J Mol Biol. 2000;296:535-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 95] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 103. | Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333-7343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 272] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 104. | Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009;5:e1000515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 105. | Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J Biol Chem. 2005;280:17841-17847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 169] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 106. | Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 551] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 107. | Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528-33539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 394] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 108. | Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 141] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 109. | Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78:9524-9537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 243] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 110. | Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4:e1000046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 184] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 111. | Poeschla EM. Integrase, LEDGF/p75 and HIV replication. Cell Mol Life Sci. 2008;65:1403-1424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 112. | Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem. 2004;279:48883-48892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 231] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 113. | McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J Biol Chem. 2008;283:31802-31812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 114. | Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA. 2005;102:17308-17313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 337] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 115. | Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 128] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 116. | Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol. 2005;12:526-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 192] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 117. | Engelman A, Kessl JJ, Kvaratskhelia M. Allosteric inhibition of HIV-1 integrase activity. Curr Opin Chem Biol. 2013;17:339-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 118. | Wang H, Shun MC, Li X, Di Nunzio F, Hare S, Cherepanov P, Engelman A. Efficient Transduction of LEDGF/p75 Mutant Cells by Gain-of-Function HIV-1 Integrase Mutant Viruses. Mol Ther Methods Clin Dev. 2014;1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 119. | Espeseth AS, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed JY, Young S, Hamill T. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci USA. 2000;97:11244-11249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 255] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 120. | Beck-Engeser GB, Eilat D, Harrer T, Jäck HM, Wabl M. Early onset of autoimmune disease by the retroviral integrase inhibitor raltegravir. Proc Natl Acad Sci USA. 2009;106:20865-20870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 121. | Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 118] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 122. | Koh Y, Matreyek KA, Engelman A. Differential sensitivities of retroviruses to integrase strand transfer inhibitors. J Virol. 2011;85:3677-3682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 123. | Pandey KK, Bera S, Grandgenett DP. The HIV-1 integrase monomer induces a specific interaction with LTR DNA for concerted integration. Biochemistry. 2011;50:9788-9796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 124. | Li M, Jurado KA, Lin S, Engelman A, Craigie R. Engineered hyperactive integrase for concerted HIV-1 DNA integration. PLoS One. 2014;9:e105078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 125. | Zhao XZ, Smith SJ, Maskell DP, Metifiot M, Pye VE, Fesen K, Marchand C, Pommier Y, Cherepanov P, Hughes SH. HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases. ACS Chem Biol. 2016;11:1074-1081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 126. | Raheem IT, Walji AM, Klein D, Sanders JM, Powell DA, Abeywickrema P, Barbe G, Bennet A, Childers K, Christensen M. Discovery of 2-Pyridinone Aminals: A Prodrug Strategy to Advance a Second Generation of HIV-1 Integrase Strand Transfer Inhibitors. J Med Chem. 2015;58:8154-8165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 127. | Métifiot M, Maddali K, Johnson BC, Hare S, Smith SJ, Zhao XZ, Marchand C, Burke TR, Hughes SH, Cherepanov P. Activities, crystal structures, and molecular dynamics of dihydro-1H-isoindole derivatives, inhibitors of HIV-1 integrase. ACS Chem Biol. 2013;8:209-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 128. | Hare S, Smith SJ, Métifiot M, Jaxa-Chamiec A, Pommier Y, Hughes SH, Cherepanov P. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol Pharmacol. 2011;80:565-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 215] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 129. | Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci USA. 2010;107:20057-20062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 252] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 130. | Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 118] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 131. | Li M, Lin S, Craigie R. Outer domains of integrase within retroviral intasomes are dispensible for catalysis of DNA integration. Protein Sci. 2016;25:472-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 132. | Mazumder A, Engelman A, Craigie R, Fesen M, Pommier Y. Intermolecular disintegration and intramolecular strand transfer activities of wild-type and mutant HIV-1 integrase. Nucleic Acids Res. 1994;22:1037-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 133. | Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci USA. 2010;107:15910-15915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 134. | Serrao E, Engelman AN. Sites of retroviral DNA integration: From basic research to clinical applications. Crit Rev Biochem Mol Biol. 2016;51:26-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 135. | Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 362] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 136. | Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1324] [Cited by in RCA: 1350] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 137. | Singh PK, Plumb MR, Ferris AL, Iben JR, Wu X, Fadel HJ, Luke BT, Esnault C, Poeschla EM, Hughes SH. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 2015;29:2287-2297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 138. | Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 695] [Cited by in RCA: 730] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 139. | Sowd GA, Serrao E, Wang H, Wang W, Fadel HJ, Poeschla EM, Engelman AN. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc Natl Acad Sci USA. 2016;113:E1054-E1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 192] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 140. | Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem HP, Kaul R, Russell DW. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci USA. 2006;103:1498-1503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 191] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 141. | Nowrouzi A, Dittrich M, Klanke C, Heinkelein M, Rammling M, Dandekar T, von Kalle C, Rethwilm A. Genome-wide mapping of foamy virus vector integrations into a human cell line. J Gen Virol. 2006;87:1339-1347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 142. | Faschinger A, Rouault F, Sollner J, Lukas A, Salmons B, Günzburg WH, Indik S. Mouse mammary tumor virus integration site selection in human and mouse genomes. J Virol. 2008;82:1360-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 143. | Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78:11656-11663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 131] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 144. | Pryciak PM, Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 223] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 145. | Pryciak PM, Sil A, Varmus HE. Retroviral integration into minichromosomes in vitro. EMBO J. 1992;11:291-303. [PubMed] |
| 146. | Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci USA. 1994;91:5913-5917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 187] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 147. | Pruss D, Reeves R, Bushman FD, Wolffe AP. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J Biol Chem. 1994;269:25031-25041. [PubMed] |
| 148. | Roth SL, Malani N, Bushman FD. Gammaretroviral integration into nucleosomal target DNA in vivo. J Virol. 2011;85:7393-7401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 149. | Serrao E, Ballandras-Colas A, Cherepanov P, Maertens GN, Engelman AN. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology. 2015;12:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 150. | Müller HP, Varmus HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704-4714. [PubMed] |
| 151. | Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol. 2005;79:5211-5214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 123] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 152. | Serrao E, Krishnan L, Shun MC, Li X, Cherepanov P, Engelman A, Maertens GN. Integrase residues that determine nucleotide preferences at sites of HIV-1 integration: implications for the mechanism of target DNA binding. Nucleic Acids Res. 2014;42:5164-5176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 153. | Taganov KD, Cuesta I, Daniel R, Cirillo LA, Katz RA, Zaret KS, Skalka AM. Integrase-specific enhancement and suppression of retroviral DNA integration by compacted chromatin structure in vitro. J Virol. 2004;78:5848-5855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 154. | Benleulmi MS, Matysiak J, Henriquez DR, Vaillant C, Lesbats P, Calmels C, Naughtin M, Leon O, Skalka AM, Ruff M. Intasome architecture and chromatin density modulate retroviral integration into nucleosome. Retrovirology. 2015;12:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 155. | Naughtin M, Haftek-Terreau Z, Xavier J, Meyer S, Silvain M, Jaszczyszyn Y, Levy N, Miele V, Benleulmi MS, Ruff M. DNA Physical Properties and Nucleosome Positions Are Major Determinants of HIV-1 Integrase Selectivity. PLoS One. 2015;10:e0129427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 156. | Pasi M, Mornico D, Volant S, Juchet A, Batisse J, Bouchier C, Parissi V, Ruff M, Lavery R, Lavigne M. DNA minicircles clarify the specific role of DNA structure on retroviral integration. Nucleic Acids Res. 2016;44:7830-7847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |