Published online Aug 26, 2015. doi: 10.4331/wjbc.v6.i3.139
Peer-review started: April 29, 2015
First decision: May 14, 2015
Revised: May 31, 2015
Accepted: June 30, 2015
Article in press: July 2, 2015
Published online: August 26, 2015
Processing time: 120 Days and 14 Hours
Deregulated c-Myc expression is a hallmark of many human cancers. We have recently identified a role of mammalian homolog of yeast SPT-ADA-GCN5-acetyltransferas (SAGA) complex component, SAGA-associated factor 29 (SGF29), in regulating the c-Myc overexpression. Here, we discuss the molecular nature of SFG29 in SPT3-TAF9-GCN5-acetyltransferase complex, a counterpart of yeast SAGA complex, and the mechanism through which the elevated SGF29 expression contribute to oncogenic potential of c-Myc in hepatocellularcarcinoma (HCC). We propose that the upstream regulation of SGF29 elicited by sex-determining region Y (Sry) is also augmented in HCC. We hypothesize that c-Myc elevation driven by the deregulated Sry and SGF29 pathway is implicated in the male specific acquisition of human HCCs.
Core tip: Deregulated c-Myc expression is a hallmark of many human cancers. We have recently identified a role of mammalian homolog of yeast SPT-ADA-GCN5-acetyltransferas (SAGA) complex component, SAGA-associated factor 29 (SGF29), in regulating the c-Myc overexpression. We propose that the upstream regulation of SGF29 elicited by sex-determining region Y (Sry) is also augmented in hepatocellularcarcinoma (HCC). We hypothesize that c-Myc elevation driven by the deregulated Sry and SGF29 pathway is implicated in the male specific acquisition of human HCCs.
- Citation: Kurabe N, Murakami S, Tashiro F. SGF29 and Sry pathway in hepatocarcinogenesis. World J Biol Chem 2015; 6(3): 139-147
- URL: https://www.wjgnet.com/1949-8454/full/v6/i3/139.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i3.139
c-myc is a protooncogene of the viral homolog of v-myc which causes myelocytomatosis[1-4]. The expression of c-Myc is tightly regulated by the many ligand-stimulated receptor signaling under normal condition[5]. The deregulated expression level of c-Myc contributes many aspects of cancer development including proliferation, growth, DNA replication, protein synthesis, metabolism, cell adhesion, angiogenesis, metastasis and immune responses by regulating the transcription of its target genes[6-11]. Only two-fold increase in the expression level of c-Myc can affect cell cycle progression[12]. In over 50% of human cancers, c-Myc is deregulated and/or activated[13-18]. In Burkitt lymphoma, the translocation of c-myc gene to the immunoglobulin gene causes the overexpression of c-Myc[19,20]. c-Myc is also highly expressed by the gene amplification in many human and rodent cancers including hepatocellularcacinoma (HCC)[21-23]. The 8q22-24 region including c-myc locus is involved in the early onset of HCC and represent the frequent amplification in early HCC[24,25]. In mice models, the expression level of c-Myc is also associated with the development of HCC[26-28]. In colon cancer and T cell leukemia, WNT and Notch signaling pathways are involved in the upregulation of c-Myc, respectively[29-32]. Conversely, suppression of c-Myc expression can cause tumor regression by inducing cell cycle arrest, differentiation, or senescence depending on the cell contexts in mice models. The effect of c-Myc downregulation on tumor regression is permanent in some tumors such as lymphoma and osteosarcoma, whereas in hepatocellular or breast cancers this effect is reversible[33-37]. Thus, inhibiting the c-Myc expression could be a promising therapeutic strategy[38-44].
c-Myc has N-terminal transcriptional regulatory domain consisting of Myc boxes (Mb) I, II, III, IV and nuclear localization signal. The basic Helix-Loop-Helix leucine-Zipper (bHLHZ) domains encompasses C-terminal region of c-Myc[45-50]. In the nucleus, c-Myc heterodimerizes with Max through their bHLHZ domains to bind to the E-box sequence (5’-CACGTG-3’) in the gene regulatory region[51-54]. The E-box sequences mainly exist in the promoter or intron 1 of the target genes. Although E-boxes are occupied with other E-box-binding transcription factors including ChREBP, SREBP, HIF-1, NRF1, USF, TFE3, Clock, and Bmal to regulate cellular homeostasis in quiescent state, c-Myc is replaced with these factors upon its induction by ligand stimulation[11]. Genome-wide screening for c-Myc-bound target genes using chromatin immunoprecipitation (ChIP)-sequencing reveal that around 6000 genes can be bound with c-Myc in human genome[55]. Of these genes, a set of 300 genes are actually inducible by c-Myc and are involved in nucleotide metabolism, ribosome biogenesis, RNA processing, and DNA replication[56]. These genes are also suggested to be induced in c-Myc-overexpressing transformed cells. c-Myc can recruit some cofactor complexes including histone acetyltransferases, ubiquitin ligases, and kinases, and the N-terminal region of MbI and MbII domains are required for the transcriptional activation and malignant transformation activities[57-59]. These domains can recruit three classes of histone acetyltransferase (HAT) complexes including SPT3-TAF9-GCN5-acetyltransferase (STAGA) complex[60], p300/CBP-associated factor (PCAF) complex[61,62] and the TIP60-containing complex[63] to activate the adjacent gene transcription. Of these three classes of HAT complexes, STAGA complex is thought to be responsible for the transactivation of c-Myc, because the only direct interaction between STAGA and c-Myc is demonstrated[64]. The Saccharomyces cerevisiae SPT-ADA-GCN5-acetyltransferase (SAGA) complex is the 1.8 MDa transcriptional coactivator that is highly conserved counterpart of STAGA complex consisting of 18 to 20 proteins[65-71]. The detailed information about these complexes is derived from the studies of yeast SAGA complex. SAGA is essential for about 10% of whole gene transcription[65,66,70,72]. The SAGA complex subunits are classified into four functional groups. The first group includes GCN5 acetyltransferase, ADA2B, ADA3 and SAGA-associated factor 29 (SGF29) regulating the acetylation of multiple lysine residues of histone H3 including H3K9, H3K14, H3K18 and H3K23[73-76]. The acetylation of histones is significantly implicated in the activation state of transcription[77-79]. Moreover, GCN5 can bind to the acetylation mark of histone H3 by its bromo domain[80]. The second group constitutes the ubiquitin-specific protease Ubp8, SGF11, SGF73 and SUS1 to perform the deubiquitinaion of H2B[81,82]. Deubiquitination of Lys123 of histone H2B eventually induce the phosphorylation of the C-terminal domain of RNA polymerase II (Pol II) to facilitate release of Pol II into transcription elongation[83,84]. The third group contains TATA-binding protein (TBP)-associated factor (TAF) proteins that are also incorporated in the general transcription factor TFIID[67,68]. Since a complex consisting of the histone-fold domains of TAF6, TAF9, TAF12 and ADA1 have structural resemblance to the histone octamer, TAF proteins make the structural backbone of the SAGA complex[85]. The last group consists of Tra1 and SPT proteins to tether the SAGA complex to the specific transcriptional activators[67,86-91]. SPT proteins are also reported to be required for the recruitment of TBP to the target gene promoters[69,92-94]. These subunits cooperatively recruit the Pol II and assemble the preinitiation complex to induce the target gene transcription. Of these components, Tra1 and TAF proteins are essential for cellular viability in yeast[95,96]. Most of these components are highly conserved between yeast and human. The human orthologue of Tra1, Transactivation/transformation-associated protein (TRRAP), can interact with the MbI and MbII domains of c-Myc, which is essential for malignant transformation and for the hyperacetylation of histone H3 and H4[64,97-102]. In addition, TRRAP is implicated in the transactivation and transformation activity of E2F1, E2F4, p53, and E1A[98,103-105]. The acetyltransferase activity of human GCN5 is also reported to be involved in the malignant transformation potential of c-Myc[99]. Thus, STAGA complex has significant role in the c-Myc-mediated onset of many human malignancies.
The critical factor in transformation activity of c-Myc other than TRRAP and GCN5 is SGF29 which is also conserved between yeast and mammals. SGF29 is originally identified using the mass spectrometric analysis of yeast SAGA[81]. By directly interacting with ADA3, SGF29 is incorporated into the STAGA complex[106]. The structural domains of SGF29 are N-terminal coiled-coil domain and C-terminal double Tudor domains[107,108]. Tudor domain is originally cloned in Drosophila, and to date, about 30 Tudor-containing proteins have been identified in mammals such as SGF29, 53BP1, Spindlin1 and UHRF1 most of which can bind to a methylated lysine[75,109-115]. The Tudor domain-containing proteins belong to so-called histone reader proteins that can recognize the various sites for post-translational histone modifications including methylation, acetylation, ubiquitination, and phosphorylation[116]. The histone readers recruit specific protein complexes at the site of histone modifications. These modifications are referred to as histone code to regulate chromatin organization and gene transcription. Many human disorders are associated with the misreading of histone codes[117-119]. The Tudor domains of SGF29 specifically recognize the histone H3K4me3/2 sites in a manner of preference to tri-methylation in yeast and human cells[75,110]. Because tri-methylation of histone H3K4 is prerequisite for subsequent acetylation of histone H3[120] and no other subunits of SAGA complex harbor the Tudor domain, only SGF29 mediates a direct connection between histone H3K4me3/2 and acetylation of histone H3. Knocking down the yeast SGF29 or its mutated form of it decrease the acetylation levels of histone H3 without disturbing the integrity of SAGA complex because of the loss of its ability to bind histone H3K4me3/2 and to load SAGA complex at target gene promoters[67,75,121]. The histone H3K4me3/2 is frequently observed in the active promoter region of genes, and the histone H3 in the E-box sequences are also reported to be highly methylated[122-126]. These methylation of histone H3K4 in E-box occur prior to the induction of acetylation of H3 by c-Myc expression[127]. Because c-Myc is not thought to directly recognize these histone markers, SGF29 in the STAGA complex may help to recruit c-Myc to the E-box in cooperation with TRRAP[106]. The loaded c-Myc induces the local histone acetylation at the site of E-box leading to histone unwinding and significant induction of downstream target gene transcription[128]. In normal organs, SGF29 is robustly expressed in testis and modestly in thymus, spleen and lung. In rat hepatomas, three out of five cell lines overexpress SGF29 together with c-Myc[106]. In the presence of c-Myc, SGF29 enhance the transcriptional activating activity of the promoter of a c-Myc target gene, ornithine decarboxylase (ODC)[129,130], in vitro. Conversely, the deletion of SGF29 expression causes the decrease in this promoter activity in rat hepatoma K2 cells. The downregulation of SGF29 also abrogates the colony forming ability of K2 cells in soft agar and tumorigenicity in nude mice concomitant with the decreased expression of c-Myc target genes such as ODC, lactate dehydrogenase-A (LDH-A)[131,132] and metastasis-associated protein 1 (MTA1)[133], whereas growth rate in monolayer culture is not affected. Moreover, the lowered expression level of SGF29 suppresses the metastatic ability of K2 cells to lung[106]. Taken together, the increased SGF29 expression is closely associated with the oncogenic potential of c-Myc by controlling its target gene expression.
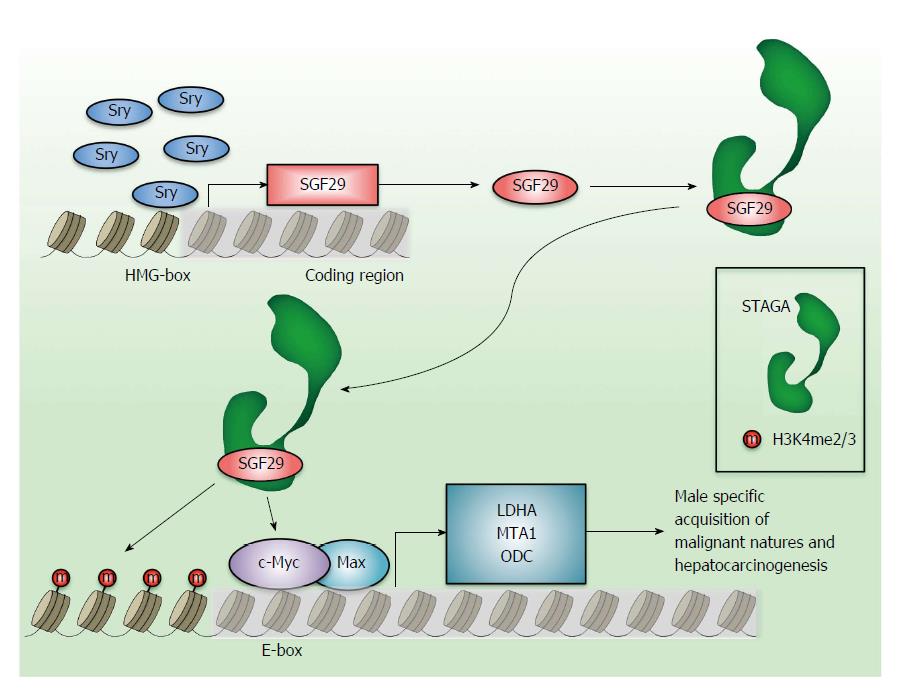
How could the factor(s) cause the deregulated expression of SGF29? The in silico search for the transcription factor binding site in the promoter region of rat SGF29 gene predicts that ten transcriptional factors could bind this region[134]. These factors contain c-Myc, sex-determining region Y (Sry), AML-1a and GATA-1. Given that there are seven high-mobility group (HMG)-boxes (5’-AACAAAG-3’), Sry-binding DNA element, and high-expression level of SGF29 in testis, Sry may be the most potent regulator of SGF29 overexpression. Indeed, SGF29 is shown to be a novel Sry target gene because Sry can directly bind the proximal promoter region of rat SGF29 gene and increase SGF29 transcript[134]. Sry is originally identified as the male sex-determining gene and is located in the male-specific region of the Y chromosome[135-137]. Induction of Sry expression in the genital ridges drives the testis differentiation and causes the activation of Sox9 to induce the downstream target genes required for male development[138-140]. Sry is not so highly conserved in its N-terminal region among mammals, but these proteins harbor the same ability to control the sex determination in developmental process in a spatiotemporally regulated manner[141-143]. The highly conserved HMG DNA-binding domain of Sry elicits the crucial function in developmental process[139,144,145]. In general, transcription factor have the DNA-binding and transactivation domains, but most of Sry proteins from different species except for mouse and rat do not have an obvious transactivation domain[145]. Although many Sry-binding partner proteins are reported to bind to the non-conserved region of Sry[146-149], physiological relevance between these interactions and testis differentiation remains obscure[150,151]. Although Sry expression is limited in brain, thymus and testis in adulthood, some HCC cell lines such as K2 cells overexpress this gene[134]. Moreover, the copy number gain or amplification of this gene locus occur in 11.8% (8/68) of human male HCC cases, which may reflect the fact that Sry is deregulated in some human HCC[152]. The deletion of Sry expression causes the lowered SGF29 expression in K2 cells together with the decreased level of LDHA and MTA1[134]. Like SGF29, the reduced Sry expression leads to the diminished colony forming ability in soft agar and tumorigenicity in vivo. The Sry expression solely does not bypath the malignant phenotype when SGF29 is deleted, which reveal that the induction of SGF29 brought by Sry is necessary for the acquisition of c-Myc-dependent transformation activity. Taken together, these findings suggest a hypothetical model of Sry and SGF29 pathway in male specific malignancy of HCC (Figure 1). The aberrant expression of Sry causes the elevation of SGF29, which is integrated into STAGA complex to augment the c-Myc target genes’ expression. Because Sry is expressed only in male, this scheme may be the explanation of male-specific acquisition of malignancy and hepatocarcinogenesis.
STAGA is one of the histone acetyltransferase complex and crucial for malignant transformation activity of c-Myc. STAGA complex can bind to c-Myc, and the deletion of key components of STAGA suppress the c-Myc target genes’ transcription. Thus, uncovering the regulatory mechanism of STAGA complex can sheds light on the oncogenesis driven by c-Myc. As a component of STAGA, SGF29 physically interact with transcriptionally active histone marker H3K4me2/3. Whether SGF29 simultaneously associates with both STAGA complex and histone H3K4me2/3 is unclear, SGF29 might facilitate the efficient recruitment of Pol II to the promoter region of c-Myc target genes. We have shown that SGF29 expression is deregulated in some rodent HCCs and is important for tumorigenic activity of c-Myc. Furthermore, because Sry is shown to be directly upregulate the SGF29 transcription and the amplification of Sry gene is observed in human male HCCs, Sry may be an attractive target for c-Myc-involved HCCs as well as SGF29. Given that c-Myc is deregulated in many human cancer types, the Sry-SGF29-c-Myc axis might be implicated in the onset of these cancers. However, not having any information about the expression level of SGF29 in cancers other than HCC, more detailed survey for this point will be required. On the other hand, in addition to Sry, c-Myc, AML-1a and GATA-1 may also bind the promoter region of SGF29 gene in in silico data analysis. Whether these factors will be the real upstream regulator(s) of SGF29 gene or not should be revealed. Although there are several protein complexes which are recruited to c-Myc, implications of these complexes in malignant transformations are unclear. Moreover, because the relationships between c-Myc and STAGA components other than SGF29, GCN5, and TRRAP are unknown, deubiquitination or TBP recruiting activities of STAGA toward c-Myc should be also clarified.
P- Reviewer: Bartova E, Wang P, Zhao Y S- Editor: Ji FF L- Editor: A E- Editor: Wang CH
| 1. | Duesberg PH, Vogt PK. Avian acute leukemia viruses MC29 and MH2 share specific RNA sequences: evidence for a second class of transforming genes. Proc Natl Acad Sci USA. 1979;76:1633-1637. [PubMed] |
| 2. | Hu SS, Lai MM, Vogt PK. Genome of avian myelocytomatosis virus MC29: analysis by heteroduplex mapping. Proc Natl Acad Sci USA. 1979;76:1265-1268. [PubMed] |
| 3. | Sheiness D, Bishop JM. DNA and RNA from uninfected vertebrate cells contain nucleotide sequences related to the putative transforming gene of avian myelocytomatosis virus. J Virol. 1979;31:514-521. [PubMed] |
| 4. | Vennstrom B, Sheiness D, Zabielski J, Bishop JM. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol. 1982;42:773-779. [PubMed] |
| 5. | Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546-5553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 618] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 6. | Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1-11. [PubMed] |
| 7. | Felsher DW. Cancer revoked: oncogenes as therapeutic targets. Nat Rev Cancer. 2003;3:375-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 350] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 8. | Shachaf CM, Felsher DW. Tumor dormancy and MYC inactivation: pushing cancer to the brink of normalcy. Cancer Res. 2005;65:4471-4474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 719] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 10. | Bachireddy P, Rakhra K, Felsher DW. Immunology in the clinic review series; focus on cancer: multiple roles for the immune system in oncogene addiction. Clin Exp Immunol. 2012;167:188-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Dang CV. MYC on the path to cancer. Cell. 2012;149:22-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1984] [Cited by in RCA: 2576] [Article Influence: 198.2] [Reference Citation Analysis (0)] |
| 12. | Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039-1048. [PubMed] |
| 13. | Escot C, Theillet C, Lidereau R, Spyratos F, Champeme MH, Gest J, Callahan R. Genetic alteration of the c-myc protooncogene (MYC) in human primary breast carcinomas. Proc Natl Acad Sci USA. 1986;83:4834-4838. [PubMed] |
| 14. | Ladanyi M, Park CK, Lewis R, Jhanwar SC, Healey JH, Huvos AG. Sporadic amplification of the MYC gene in human osteosarcomas. Diagn Mol Pathol. 1993;2:163-167. [PubMed] |
| 15. | Gamberi G, Benassi MS, Bohling T, Ragazzini P, Molendini L, Sollazzo MR, Pompetti F, Merli M, Magagnoli G, Balladelli A. C-myc and c-fos in human osteosarcoma: prognostic value of mRNA and protein expression. Oncology. 1998;55:556-563. [PubMed] |
| 16. | Kawate S, Fukusato T, Ohwada S, Watanuki A, Morishita Y. Amplification of c-myc in hepatocellular carcinoma: correlation with clinicopathologic features, proliferative activity and p53 overexpression. Oncology. 1999;57:157-163. [PubMed] |
| 17. | Stock C, Kager L, Fink FM, Gadner H, Ambros PF. Chromosomal regions involved in the pathogenesis of osteosarcomas. Genes Chromosomes Cancer. 2000;28:329-336. [PubMed] |
| 18. | Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595-5610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 342] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 19. | Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824-7827. [PubMed] |
| 20. | Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA. 1982;79:7837-7841. [PubMed] |
| 21. | Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899-905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3271] [Cited by in RCA: 3052] [Article Influence: 203.5] [Reference Citation Analysis (0)] |
| 22. | Sugiyama A, Miyagi Y, Komiya Y, Kurabe N, Kitanaka C, Kato N, Nagashima Y, Kuchino Y, Tashiro F. Forced expression of antisense 14-3-3beta RNA suppresses tumor cell growth in vitro and in vivo. Carcinogenesis. 2003;24:1549-1559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 23. | Komiya Y, Kurabe N, Katagiri K, Ogawa M, Sugiyama A, Kawasaki Y, Tashiro F. A novel binding factor of 14-3-3beta functions as a transcriptional repressor and promotes anchorage-independent growth, tumorigenicity, and metastasis. J Biol Chem. 2008;283:18753-18764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Zondervan PE, Wink J, Alers JC, IJzermans JN, Schalm SW, de Man RA, van Dekken H. Molecular cytogenetic evaluation of virus-associated and non-viral hepatocellular carcinoma: analysis of 26 carcinomas and 12 concurrent dysplasias. J Pathol. 2000;192:207-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 25. | Poon TC, Wong N, Lai PB, Rattray M, Johnson PJ, Sung JJ. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2006;131:1262-1270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 26. | Sandgren EP, Quaife CJ, Pinkert CA, Palmiter RD, Brinster RL. Oncogene-induced liver neoplasia in transgenic mice. Oncogene. 1989;4:715-724. [PubMed] |
| 27. | Murakami H, Sanderson ND, Nagy P, Marino PA, Merlino G, Thorgeirsson SS. Transgenic mouse model for synergistic effects of nuclear oncogenes and growth factors in tumorigenesis: interaction of c-myc and transforming growth factor alpha in hepatic oncogenesis. Cancer Res. 1993;53:1719-1723. [PubMed] |
| 28. | Wu Y, Renard CA, Apiou F, Huerre M, Tiollais P, Dutrillaux B, Buendia MA. Recurrent allelic deletions at mouse chromosomes 4 and 14 in Myc-induced liver tumors. Oncogene. 2002;21:1518-1526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 29. | He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509-1512. [PubMed] |
| 30. | Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261-18266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 679] [Cited by in RCA: 661] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 31. | Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022-8031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 214] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 32. | Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096-2109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 734] [Cited by in RCA: 688] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 33. | Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199-207. [PubMed] |
| 34. | Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 507] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 35. | Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 699] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 36. | Marinkovic D, Marinkovic T, Mahr B, Hess J, Wirth T. Reversible lymphomagenesis in conditionally c-MYC expressing mice. Int J Cancer. 2004;110:336-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | D’Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, Cox JD, Ha SI, Belka GK, Golant A. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 315] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 38. | Hermeking H. The MYC oncogene as a cancer drug target. Curr Cancer Drug Targets. 2003;3:163-175. [PubMed] |
| 39. | Pastorino F, Brignole C, Marimpietri D, Di Paolo D, Zancolli M, Pagnan G, Ponzoni M. Targeted delivery of oncogene-selective antisense oligonucleotides in neuroectodermal tumors: therapeutic implications. Ann N Y Acad Sci. 2004;1028:90-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 40. | Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 413] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 41. | Shachaf CM, Perez OD, Youssef S, Fan AC, Elchuri S, Goldstein MJ, Shirer AE, Sharpe O, Chen J, Mitchell DJ. Inhibition of HMGcoA reductase by atorvastatin prevents and reverses MYC-induced lymphomagenesis. Blood. 2007;110:2674-2684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 42. | Cao Z, Fan-Minogue H, Bellovin DI, Yevtodiyenko A, Arzeno J, Yang Q, Gambhir SS, Felsher DW. MYC phosphorylation, activation, and tumorigenic potential in hepatocellular carcinoma are regulated by HMG-CoA reductase. Cancer Res. 2011;71:2286-2297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 151] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 43. | Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2297] [Cited by in RCA: 2328] [Article Influence: 166.3] [Reference Citation Analysis (0)] |
| 44. | McKeown MR, Bradner JE. Therapeutic strategies to inhibit MYC. Cold Spring Harb Perspect Med. 2014;4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 179] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 45. | Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 977] [Cited by in RCA: 1003] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 46. | Amati B, Frank SR, Donjerkovic D, Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim Biophys Acta. 2001;1471:M135-M145. [PubMed] |
| 47. | Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 804] [Cited by in RCA: 868] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 48. | Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1115] [Cited by in RCA: 1246] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 49. | Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211-1217. [PubMed] |
| 50. | Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8:4048-4054. [PubMed] |
| 51. | Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211-222. [PubMed] |
| 52. | Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 352] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 53. | Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233-245. [PubMed] |
| 54. | Kato GJ, Lee WM, Chen LL, Dang CV. Max: functional domains and interaction with c-Myc. Genes Dev. 1992;6:81-92. [PubMed] |
| 55. | Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834-17839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 418] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 56. | Perna D, Fagà G, Verrecchia A, Gorski MM, Barozzi I, Narang V, Khng J, Lim KC, Sung WK, Sanges R. Genome-wide mapping of Myc binding and gene regulation in serum-stimulated fibroblasts. Oncogene. 2012;31:1695-1709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 57. | Oster SK, Mao DY, Kennedy J, Penn LZ. Functional analysis of the N-terminal domain of the Myc oncoprotein. Oncogene. 2003;22:1998-2010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16:242-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 129] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 59. | Lüscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494:145-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 113] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 60. | Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781-23785. [PubMed] |
| 61. | Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati ML, Qin J, Nakatani Y. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2:869-875. [PubMed] |
| 62. | Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35-44. [PubMed] |
| 63. | Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463-473. [PubMed] |
| 64. | Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem. 2003;278:20405-20412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 65. | Koutelou E, Hirsch CL, Dent SY. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22:374-382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 194] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 66. | Rodríguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 67. | Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, Washburn MP. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol. 2011;7:503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 68. | Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 128] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 69. | Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86-98. [PubMed] |
| 70. | Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329-5340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 149] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 71. | Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640-1650. [PubMed] |
| 72. | Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 289] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 73. | Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895-5900. [PubMed] |
| 74. | Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989-7995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 181] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 75. | Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30:2829-2842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 194] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 76. | Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155-3167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 290] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 77. | Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2582] [Cited by in RCA: 2185] [Article Influence: 156.1] [Reference Citation Analysis (0)] |
| 78. | Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 230] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 79. | Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 388] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 80. | Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369-379. [PubMed] |
| 81. | Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723-4738. [PubMed] |
| 82. | Rodríguez-Navarro S, Fischer T, Luo MJ, Antúnez O, Brettschneider S, Lechner J, Pérez-Ortín JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75-86. [PubMed] |
| 83. | Atanassov BS, Koutelou E, Dent SY. The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett. 2011;585:2016-2023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 84. | Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 509] [Cited by in RCA: 509] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 85. | Selleck W, Howley R, Fang Q, Podolny V, Fried MG, Buratowski S, Tan S. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat Struct Biol. 2001;8:695-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 70] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 86. | Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333-2337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 281] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 87. | Helmlinger D, Marguerat S, Villén J, Swaney DL, Gygi SP, Bähler J, Winston F. Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J. 2011;30:2843-2852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 88. | Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 156] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 89. | Fishburn J, Mohibullah N, Hahn S. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell. 2005;18:369-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 90. | Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092-9102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 91. | Grant PA, Schieltz D, Pray-Grant MG, Yates JR, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863-867. [PubMed] |
| 92. | Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946-1956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 256] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 93. | Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994-3006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 94. | Laprade L, Rose D, Winston F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007-2017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 95. | Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR, Lees-Miller SP, Cole MD, Brandl CJ. Tra1p is a component of the yeast Ada.Spt transcriptional regulatory complexes. J Biol Chem. 1998;273:26559-26565. [PubMed] |
| 96. | Klebanow ER, Poon D, Zhou S, Weil PA. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J Biol Chem. 1996;271:13706-13715. [PubMed] |
| 97. | Kato GJ, Barrett J, Villa-Garcia M, Dang CV. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990;10:5914-5920. [PubMed] |
| 98. | McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363-374. [PubMed] |
| 99. | McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556-562. [PubMed] |
| 100. | Park J, Kunjibettu S, McMahon SB, Cole MD. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 2001;15:1619-1624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 104] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 101. | Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069-2082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 415] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 102. | Stone J, de Lange T, Ramsay G, Jakobovits E, Bishop JM, Varmus H, Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697-1709. [PubMed] |
| 103. | Lang SE, McMahon SB, Cole MD, Hearing P. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J Biol Chem. 2001;276:32627-32634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 94] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 104. | Ard PG, Chatterjee C, Kunjibettu S, Adside LR, Gralinski LE, McMahon SB. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol Cell Biol. 2002;22:5650-5661. [PubMed] |
| 105. | Deleu L, Shellard S, Alevizopoulos K, Amati B, Land H. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene. 2001;20:8270-8275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 64] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 106. | Kurabe N, Katagiri K, Komiya Y, Ito R, Sugiyama A, Kawasaki Y, Tashiro F. Deregulated expression of a novel component of TFTC/STAGA histone acetyltransferase complexes, rat SGF29, in hepatocellular carcinoma: possible implication for the oncogenic potential of c-Myc. Oncogene. 2007;26:5626-5634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 107. | Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 805] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 108. | Li J, Xue X, Ruan J, Wu M, Zhu Z, Zang J. Cloning, purification, crystallization and preliminary crystallographic analysis of the tandem tudor domain of Sgf29 from Saccharomyces cerevisiae. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:902-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 109. | Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97-104. [PubMed] |
| 110. | Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 633] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 111. | Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 784] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 112. | Yang N, Wang W, Wang Y, Wang M, Zhao Q, Rao Z, Zhu B, Xu RM. Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1. Proc Natl Acad Sci USA. 2012;109:17954-17959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 113. | Wang W, Chen Z, Mao Z, Zhang H, Ding X, Chen S, Zhang X, Xu R, Zhu B. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep. 2011;12:1160-1166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 114. | Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155-1160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 279] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 115. | Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci USA. 2012;109:12950-12955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 161] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 116. | Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6156] [Cited by in RCA: 6144] [Article Influence: 245.8] [Reference Citation Analysis (0)] |
| 117. | Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 925] [Cited by in RCA: 844] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 118. | Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847-851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 369] [Cited by in RCA: 344] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 119. | Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 273] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 120. | Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL. Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail. J Biol Chem. 2007;282:27923-27934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 121. | Shukla A, Lahudkar S, Durairaj G, Bhaumik SR. Sgf29p facilitates the recruitment of TATA box binding protein but does not alter SAGA’s global structural integrity in vivo. Biochemistry. 2012;51:706-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 122. | Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 296] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 123. | Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 1601] [Article Influence: 69.6] [Reference Citation Analysis (0)] |
| 124. | Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553-560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3473] [Cited by in RCA: 3198] [Article Influence: 177.7] [Reference Citation Analysis (0)] |
| 125. | Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1140] [Cited by in RCA: 1169] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 126. | Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 599] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 127. | Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 227] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 128. | Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 758] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 129. | Iyengar RV, Pawlik CA, Krull EJ, Phelps DA, Burger RA, Harris LC, Potter PM, Danks MK. Use of a modified ornithine decarboxylase promoter to achieve efficient c-MYC- or N-MYC-regulated protein expression. Cancer Res. 2001;61:3045-3052. [PubMed] |
| 130. | Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 172] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 131. | Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1141] [Cited by in RCA: 1226] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 132. | Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107:2037-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 893] [Cited by in RCA: 1135] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 133. | Zhang XY, DeSalle LM, Patel JH, Capobianco AJ, Yu D, Thomas-Tikhonenko A, McMahon SB. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. Proc Natl Acad Sci USA. 2005;102:13968-13973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 102] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 134. | Murakami S, Chishima S, Uemoto H, Sakamoto E, Sato T, Kurabe N, Kawasaki Y, Shibata T, Akiyama H, Tashiro F. The male-specific factor Sry harbors an oncogenic function. Oncogene. 2014;33:2978-2986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 135. | Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2257] [Cited by in RCA: 2104] [Article Influence: 60.1] [Reference Citation Analysis (0)] |
| 136. | Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1169] [Cited by in RCA: 1173] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 137. | Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1577] [Cited by in RCA: 1451] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 138. | Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 660] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 139. | Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet. 2009;25:19-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 214] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 140. | Kashimada K, Koopman P. Sry: the master switch in mammalian sex determination. Development. 2010;137:3921-3930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 246] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 141. | Koopman P, Münsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 573] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 142. | Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603-1614. [PubMed] |
| 143. | Bullejos M, Koopman P. Spatially dynamic expression of Sry in mouse genital ridges. Dev Dyn. 2001;221:201-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 176] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 144. | Canning CA, Lovell-Badge R. Sry and sex determination: how lazy can it be? Trends Genet. 2002;18:111-113. [PubMed] |
| 145. | Zhao L, Koopman P. SRY protein function in sex determination: thinking outside the box. Chromosome Res. 2012;20:153-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 146. | Bernard P, Harley VR. Acquisition of SOX transcription factor specificity through protein-protein interaction, modulation of Wnt signalling and post-translational modification. Int J Biochem Cell Biol. 2010;42:400-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 147. | Poulat F, de Santa Barbara P, Desclozeaux M, Soullier S, Moniot B, Bonneaud N, Boizet B, Berta P. The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J Biol Chem. 1997;272:7167-7172. [PubMed] |
| 148. | Oh HJ, Li Y, Lau YF. Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol Reprod. 2005;72:407-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 149. | Thevenet L, Albrecht KH, Malki S, Berta P, Boizet-Bonhoure B, Poulat F. NHERF2/SIP-1 interacts with mouse SRY via a different mechanism than human SRY. J Biol Chem. 2005;280:38625-38630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 150. | Broere N, Hillesheim J, Tuo B, Jorna H, Houtsmuller AB, Shenolikar S, Weinman EJ, Donowitz M, Seidler U, de Jonge HR. Cystic fibrosis transmembrane conductance regulator activation is reduced in the small intestine of Na+/H+ exchanger 3 regulatory factor 1 (NHERF-1)- but Not NHERF-2-deficient mice. J Biol Chem. 2007;282:37575-37584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 151. | Polanco JC, Wilhelm D, Mizusaki H, Jackson A, Browne C, Davidson T, Harley V, Sinclair A, Koopman P. Functional analysis of the SRY-KRAB interaction in mouse sex determination. Biol Cell. 2009;101:55-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 152. | Katoh H, Ojima H, Kokubu A, Saito S, Kondo T, Kosuge T, Hosoda F, Imoto I, Inazawa J, Hirohashi S. Genetically distinct and clinically relevant classification of hepatocellular carcinoma: putative therapeutic targets. Gastroenterology. 2007;133:1475-1486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |