Published online Aug 26, 2014. doi: 10.4331/wjbc.v5.i3.301
Revised: March 19, 2014
Accepted: May 31, 2014
Published online: August 26, 2014
Processing time: 292 Days and 9.6 Hours
Colorectal cancer is the third most common cancer worldwide. Metastasis is a major cause of colorectal cancer-related death. Mechanisms of metastasis remain largely obscure. MicroRNA is one of the most important epigenetic regulators by targeting mRNAs post-transcriptionally. Accumulated evidence has supported its significant role in the metastasis of colorectal cancer, including epithelial-mesenchymal transition and angiogenesis. Dissecting microRNAome potentially identifies specific microRNAs as biomarkers of colorectal cancer metastasis. Better understanding of the complex network of microRNAs in colorectal cancer metastasis provide new insights in the biological process of metastasis and in the potential targets for colorectal cancer therapies and for diagnosis of recurrent and metastatic colorectal cancer.
Core tip: MicroRNA is one of the most important epigenetic regulators by targeting mRNAs post-transcriptionally. This article has reviewed the new evidence that has supported the significant role of microRNAs in the metastasis of colorectal cancer. Better understanding of the complex network of microRNAs in colorectal cancer metastasis provide new insights in the biological process of metastasis and in the potential targets for colorectal cancer therapies and for diagnosis of recurrent and metastatic colorectal cancer.
- Citation: Zhou JJ, Zheng S, Sun LF, Zheng L. MicroRNA regulation network in colorectal cancer metastasis. World J Biol Chem 2014; 5(3): 301-307
- URL: https://www.wjgnet.com/1949-8454/full/v5/i3/301.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v5.i3.301
Colorectal cancer is the third most common cancer in both males and females worldwide. The major cause of death from colorectal cancer is the development of metastatic diseases. For those patients who have metastatic diseases at the time of diagnosis, five-year survival rate is only 12%[1,2]. However, the molecular mechanism of metastasis still remains obscure. Multiple cellular characteristics of neoplastic cells including epithelial-mesenchymal transition (EMT), invasion and migration, and changes in the tumor microenvironment such as angiogenesis are believed to be important for the metastatic process. Genetic abnormalities accounting for metastasis have not been identified while accumulated evidence suggests that microRNAs may play a crucial role in the epigenetic regulation of the multi-step process of metastasis.
MicroRNAs are a family of small non-coding 18-22nt RNAs, which function in the post-transcriptional regulation of gene expression, by targeting mRNA for cleavage or translational repression[3]. In the nucleus, the DNA coding sequences of miRNA are firstly transcribed into pri-miRNA by RNA polymerase II. After transcription, the enzyme named Drosha cuts pri-miRNA into pre-microRNA, which is then transported into cytoplasm and further cleaved into mature microRNA by Dicer. Only one strand of the mature microRNA will be incorporated into RNA-induced silencing complex (RISC), where microRNA play its silencing functions by interacting its target mRNA[4].
Accumulated evidence has suggested that microRNAs directly participate in the tumorigenesis process of colorectal cancer, especially through the posttranscriptional regulation. Some microRNAs play roles in colorectal tumorigenesis by regulating the functional pathways of tumor suppressor genes. For example, microRNAs including miR-135a/b and miR-122a were able to inactivate the adenomatous polyposis coli (APC) tumor suppressor gene and the APC-mediated pathways, providing an alternative, epigenetic mechanism for the inactivation of this tumor suppressor gene. In the APC-mutated mouse model, microRNAs including miR-31, miR-137 and miR-215 were found to be differentially regulated in association with colorectal adenoma formation, suggesting that these microRNAs may be regulated by the APC pathway and are involved in the early stage of colorectal cancer development[5-7]. Similarly, microRNAs including miR-34, miR-145 and miR-107 are regulated by p53 to mediate the function of p53 in cell survival, proliferation and angiogenesis, respectively[8,9]. Therefore, it has been well established that microRNA is involved in the initial stage of colorectal tumorigenesis through the APC pathway and in the advanced stage of colorectal tumorigenesis through the p53 pathway. More recently, a large body of evidence has also supported the role of microRNAs in colorectal cancer metastasis. Here we will review the function of microRNAs in the regulation of EMT and angiogenesis- two biological processes that are important for colorectal cancer metastasis.
To discover aberrantly expressed microRNAs in colorectal cancer, microarray analysis was commonly used for dissecting microRNAome[10]. A microRNA microarray analysis in a discovery cohort of 84 colorectal cancer patients comparing microRNA expression profiles between colorectal tumors and paired non-tumorous tissues identified 37 microRNAs differentially expressed in colorectal cancer. Five microRNAs (miR-20a, miR-21, miR-106a, miR-181b, and miR-203) that were most differentially expressed between tumors and non-tumorous tissues and whose expression levels were also associated with patient survivals were selected for further validation. In the validation cohort, it was found that miR-21 was preferentially expressed in colorectal tumors at more advanced TNM stages. In both discovery and validation cohorts, higher miR-21 expression level was significantly associated with poorer survival[11]. Similarly, in another study, 49 microRNAs were identified to be significantly differentially expressed comparing between rectal cancer and adjacent non-tumorous mucosa. Among them, miR-135b was shown to be significantly correlated to disease-free and cancer-specific survival in the validation cohort[12].
Although these microRNAome studies included patients across different stages, there is still a lack of identification of microRNAs associated specifically with metastatic colorectal cancer. Shen et al[13] compared the miRNA expression profiles between colorectal cancers with liver metastasis and those without metastasis and found 28 differentially expressed microRNAs. Among them, four microRNAs (miR-150*, miR-125b-2*, miR-1179 and miR-139-3p) are up-regulated in colorectal cancer with metastasis. Although this study provided microRNAome data for colorectal cancer with metastasis, the sample size was small, including only 3 paired samples; and the clinical survival data was not available.
While the microRNAome specific for colorectal cancer metastasis is still highly desired, the research on microRNAome continues to strive ahead with microarray analysis on both tumoral and stromal tissues of colorectal cancer and with microarray analysis on colorectal tumors at different locations, of different subtypes, and with different mismatch repair status. Single nucleotide polymorphism (SNP) variations in microRNA coding sequences and 3’-UTR have been studied[14-18]. These new researches have shed lights on the future research direction of mciroRNA in metastatic colorectal cancer.
Despite genetic mutation is still considered as one of the key characteristics in the primary occurrence of colorectal cancer, the metastasis of colorectal cancer appears to be closely associated with epigenetic regulation, such as DNA promoter methylation, histone modification and microRNAs[19,20]. microRNAs have been demonstrated to be involved in two major pathways of colorectal cancer metastasis, EMT and angiogenesis.
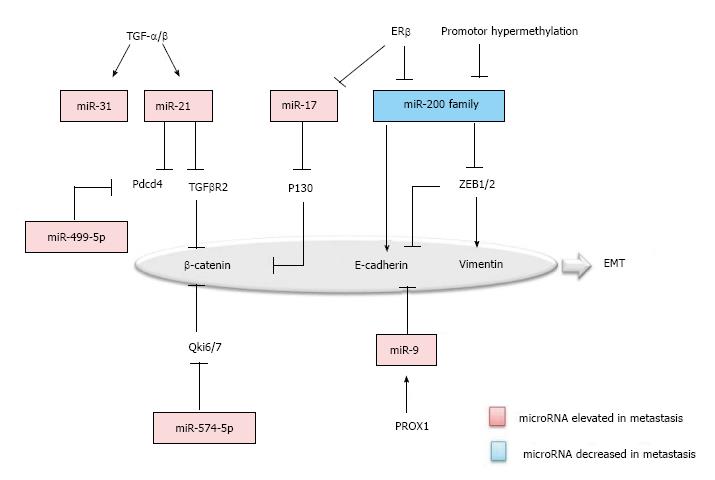
EMT is the biologic process that the polarized epithelial cells transit to a mesenchymal cell phenotype. During the course of metastasis, neoplastic cells undergo EMT, suggesting a mechanistic role of EMT in metastasis. In EMT, proteins such as Vimentin, β-Catenin, TCF8-ZEB1, E-Cadherin, Snail and Slug, are known to be specifically up or down-regulated and have been established as the markers of EMT[21]. Similarly, microRNAs associated with colorectal cancer, were also found to be specifically regulated in EMT (Figure 1).
First, TGF-β/Wnt signaling pathway is one of the prominent pathways in EMT. miR-21 and miR-31 were uncovered as the downstream effectors of TGF-β in colon carcinoma cells[22]. miR-21 is significantly elevated in the colorectal cancer with metastasis and negatively regulates the tumor suppressor programmed cell death 4 (Pdcd4) and TGF-β receptor 2, accompanied by the decrease of the downstreaming β-catenin[23,24]. Second, β-catenin was inversely correlated with Qki6/7 and P130, which are down-regulated by miR-574-5p and miR-17, respectively in colorectal carcinoma cells[25,26]. Third, the miR-200 Family was recognized as a master regulator of the epithelial phenotype, which is decreased in colorectal cancer with metastasis. The miR-200 family repressed the EMT by targeting the ZEB1/2, which down-regulate E-cadherin and up-regulate Vimentin. Furthermore, in the upstream of the miR-200 family, promoter methylation and ERβ can be the cause of decrease in colorectal metastasis[25,27,28]. Fourth, miR-9 has been suggested to be another regulator for E-cadherin. Prospero Homeobox 1 (PROX1) was shown to promote EMT by inhibiting E-cadherin via miR-9 in colon carcinoma cells[29,30]. Last, but not the least, microRNAs like miR-499-5p and miR-212 also function in regulating EMT, by targeting Pdcd4 and manganese superoxide dismutase (MnSOD)[31,32].
The mesenchymal to epithelial transition (MET) is a reverse biological process of EMT and has recently been suggested to be important for the metastatic cancer cells, by regaining epithelial properties, to establish their colonization in distant organs. MiR-147 was recently found to be able to induce MET in colon cancer cell lines by targeting the TGF-β signaling pathway[33]. Further exploring the roles of microRNAs in MET may provide a deep insight into how the MET is executed in the cancer metastasis.
Thus, many characteristic markers of EMT such as Vimentin, β-Catenin, TCF8-ZEB1, E-Cadherin, Snail and Slug, are targeted by microRNAs. Moreover, many microRNAs were found to have their targets in different types of cancer. For one example, miR-200, by targeting ZEB-1/2, is a tumor suppressor in both breast cancer and colorectal cancer. For another example, ectopic expression of miR-17 plays a role in regulating cancer cell invasion and migration of other malignancy types including colorectal cancer, breast cancer, head and neck cancer by targeting the TGF-β signaling pathway[34]. Therefore, understanding the roles of microRNAs in colorectal cancer metastasis may identify potential therapeutic targets for the treatment of many different types of malignancies.
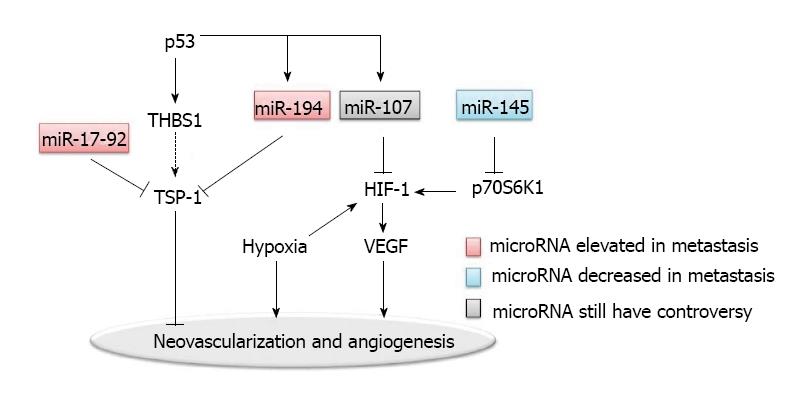
Angiogenesis is considered to be essential for the establishment of blood supply for metastatic lesions in the distant organs. Many pro-angiogenic and anti-angiogenic factors are involved (Figure 2). Tumor angiogenesis is associated with intravasation and extravasation of metastatic cancer cells, suggesting that many crucial molecules in angiogenesis and their corresponding receptors participate in the metastatic process[35]. One of the best studied molecular mechanisms involves the vascular endothelial growth factors (VEGF) and the VEGF receptors. In addition, intratumoral blood vessel is the main route of chemotherapy or targeted drug delivery; but the effectiveness of drug delivery is unsatisfactory due to the disorganized neovascularization in tumors. Clinically, VEGF targeted therapy is used for treating metastatic colorectal cancer, supporting the role of angiogenesis in colorectal cancer metastasis[36,37].
Hypoxia is one of the dominant characteristics of tumor microenvironment; and hypoxia inducible factor (HIF)-1 is one of the key regulators in hypoxia induced angiogenesis[38]. Hypoxic microenvironment drives tumor cells to undergo an angiogenic switch which leads to the production of pro-angiogenesis proteins such as VEGF. The VEGF pathway-mediated neovascularization is also driven partly by additional hypoxia responsive signals including basic FGF (bFGF) and placental growth factor (PlGF).
Epigenetic regulation via microRNAs may be one of the major regulatory mechanisms for the disorganization of neovascularization in tumor[39]. MiR-107 was shown to function as a suppresser of HIF-1 and VEGF expression[40]. In addition to miR-107, miR-145 was found to be a regulator of HIF-1 in colon cancer, by targeting p70S6K1 post-transcriptionally[42].
Another well characterized regulator for tumor angiogenesis is thrombospondin-1 (TSP-1), which belongs to the thrombospondins family[43]. TSP-1 acts as a barrier to neovascularization in tumors. MiR-17-92 and miR-194 were both found to repress TSP-1, thereby promoting the angiogenesis in colon cancers[40,44]. Interestingly, MiR-17-92 is a polycistronic microRNA cluster. The precursor transcript derived from this cluster gene produces six mature microRNAs: miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1[45]. Overexpression of the miR-17-92 cluster was observed in multiple tumor types including colon cancer. As a polycistronic cluster, it can coordinate multiple functions in tumorigenesis including increasing angiogenesis, promoting proliferation and inhibiting differentiation[45,46]. The miR-17-92 microRNA cluster is known to be regulated by the Myc oncogene. The Myc-activated miR-17-92 can stimulate tumor angiogenesis by attenuating the TGF-β signaling pathway, which provides an alternative target for miR-17-92 in addition to TSP-1[47].
Both miR-17-92 and miR-194 are upregulated by the p53 tumor suppressor. miR-194 also negatively regulates the thrombospondin (THBS1) mRNA. On another hand, it is known that p53 induces the transcription of THBS1, which would have further induced the expression of TSP-1, since enhanced expression of THBS1 leads to the induction of TSP-1. Interestingly, p53 does not induce the expression of TSP-1. Thus, it is possible that TSP-1 is suppressed by miR-194 and miR-17-92, which both are regulated by p53[40]. Such a complex regulatory network, by providing multiple feedback mechanisms, allows a more precise regulation in angiogenesis.
MicroRNA is studied as a potential diagnostic marker, considering its commonly conserved existence and remarkable stability[48]. Although plasma CEA has been used as a diagnostic marker for colorectal cancer for decades, one study had shown that only 59% of 417 monitored patients with recurrence had a preceding elevation of CEA concentration[49]. Several recent studies thus have been conducted on the identification of plasma circulating microRNAs, attempting to develop more sensitive and specific detection methods[50-53]. Especially, methods for detecting the circulating microRNAs are attracting because of their noninvasive nature. Recently, a panel of 8 plasma microRNAs (miR-532-3p, miR-331, miR-195, miR-17, miR-142-3p, miR-15b, miR-532, and miR-652) was found to be able to distinguish colonic polyps from healthy controls. In addition, this study pointed out that a panel of 3 plasma miRNAs (miR-431,miR-15b, and miR-139-3p) distinguished Stage IV CRC from controls[54].
However, there is still a lack of circulating microRNAs as an ideal biomarker for recurrence and metastasis. Plasma miR-92 and miR-29 were significantly elevated in colorectal cancer patients, but demonstrated no significant difference between different stages of colorectal cancer[50,52]. Nevertheless, in a more recent study by Wang et al[55] serum level of the miR-29a was found to be significantly higher in colorectal cancer patients with metastasis comparing to those without metastasis, with a sensitivity of 75% and a specificity of 75%. Similarly, circulating miR-221 was demonstrated to be a significant prognostic factor associated with poor overall survival. Whether it can be used for early detection of recurrence is not studied[53]. Another two more recent studies found several promising microRNAs for the detection of recurrence. Kanaan et al[51] demonstrated that plasma miR-31, miR-135b, miR-1 and miR-133a have a 100% sensitivity and a 80% specificity in detecting colorectal cancer, and also found that miR-31 was significantly more upregulated in stage III and IV than stage I and II. Hofsli et al[56] reported that in their “training” study conducted with with serum samples from 30 patients with stage IV colon cancer and from 10 healthy controls, 375 miRNAs were found to be more abundant in the sera from colon cancer patients than those from healthy controls, including miR-103, miR-107, and miR-221. These miRNAs were also found in sera from patients with stage I-II colon cancer; however, their roles in stage IV colon cancer remain interesting to be explored.
microRNAs play a substantial role in the epigenetic regulation of colorectal cancer metastasis. Differential expression of microRNAs is reported in the metastasis of colorectal cancer comparing to non-metastatic colorectal cancer (Table 1). More importantly, these microRNAs appear to form a network to coordinate the regulation of the metastatic process. Studies investigating the association of microRNAs with the metastasis process are highly desired. Plasma microRNA remains to be identified as a noninvasive biomarker for early diagnosis of colorectal cancer metastasis. Enlightened by the function of microRNAs in the EMT process and angiogenesis, mechanisms of colorectal cancer metastasis may be revealed by dissecting the regulatory network of microRNAs.
| microRNA | Description | Ref. | |
| microRNAs differentially expressed in colorectal cancers | miR-21 | Associated to more advanced TNM stages and poorer survival | [11] |
| miR-135b | Associated to disease-free and cancer-specific survival | [12] | |
| miR-150*, miR-125b-2*, miR-1179 and miR-139-3p | Up-regulated in colorectal cancer with metastasis. | [13] | |
| microRNAs as diagnostic markers | Serum miR-29a | Up-regulated in patients with metastasis | [55] |
| Circulating miR-221 | Associated to poor survival | [53] | |
| Plasma miR-31 | Up-regulated in stage III and IV | [51] | |
| microRNAs involved in epithelial-mesenchymal transition | miR-21 | Pdcd4,TGFβ receptor 2 | [23,24] |
| miR-574-5p,miR-17 | Qki6/7,P130 | [25,26] | |
| miR-200 | ZEB1/2 | [27,28] | |
| miR-9 | E-cadherin | [29,30] | |
| miR-499-5p | Pdcd4 | [31] | |
| miR-212 | MnSOD | [32] | |
| microRNAs involved in Angiogenesis | miR-17-92 | TSP-1 | [44] |
| miR-194 | TSP-1,THBS1 | [40] | |
| miR-107 | HIF-1,VEGF,DAPK,KLF4 | [41] | |
| miR-145 | p70S6K1 | [42] |
P- Reviewer: Chen JI, Guo HB, Nakajima N S- Editor: Wen LL L- Editor: A E- Editor: Lu YJ
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25546] [Article Influence: 1824.7] [Reference Citation Analysis (7)] |
| 2. | American Cancer Society. Cancer Facts & Figures 2013. Atlanta: American Cancer Society 2013; . |
| 3. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 4. | Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1217] [Cited by in RCA: 1219] [Article Influence: 76.2] [Reference Citation Analysis (0)] |
| 5. | Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795-5802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 376] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 6. | Necela BM, Carr JM, Asmann YW, Thompson EA. Differential expression of microRNAs in tumors from chronically inflamed or genetic (APC(Min/+)) models of colon cancer. PLoS One. 2011;6:e18501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 7. | Wang X, Lam EK, Zhang J, Jin H, Sung JJ. MicroRNA-122a functions as a novel tumor suppressor downstream of adenomatous polyposis coli in gastrointestinal cancers. Biochem Biophys Res Commun. 2009;387:376-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Feng Z, Zhang C, Wu R, Hu W. Tumor suppressor p53 meets microRNAs. J Mol Cell Biol. 2011;3:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 181] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 9. | He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130-1134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2135] [Cited by in RCA: 2107] [Article Influence: 117.1] [Reference Citation Analysis (0)] |
| 10. | Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687-3692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 735] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 11. | Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1037] [Cited by in RCA: 1196] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 12. | Gaedcke J, Grade M, Camps J, Søkilde R, Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi BM, Møller S. The rectal cancer microRNAome--microRNA expression in rectal cancer and matched normal mucosa. Clin Cancer Res. 2012;18:4919-4930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 13. | Lin M, Chen W, Huang J, Gao H, Ye Y, Song Z, Shen X. MicroRNA expression profiles in human colorectal cancers with liver metastases. Oncol Rep. 2011;25:739-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Balaguer F, Moreira L, Lozano JJ, Link A, Ramirez G, Shen Y, Cuatrecasas M, Arnold M, Meltzer SJ, Syngal S. Colorectal cancers with microsatellite instability display unique miRNA profiles. Clin Cancer Res. 2011;17:6239-6249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 15. | Landi D, Gemignani F, Landi S. Role of variations within microRNA-binding sites in cancer. Mutagenesis. 2012;27:205-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 16. | Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Sugihara K, Doki Y. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res. 2012;18:3054-3070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 17. | Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA, Morlan BW, Riska SM, Boardman LA, Cunningham JM. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 261] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 18. | Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011;50:196-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 117] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 19. | Kerr D. Clinical development of gene therapy for colorectal cancer. Nat Rev Cancer. 2003;3:615-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 20. | Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442-1460.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 201] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 21. | Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6575] [Cited by in RCA: 7918] [Article Influence: 494.9] [Reference Citation Analysis (0)] |
| 22. | Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem. 2010;285:35293-35302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 224] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 23. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1350] [Cited by in RCA: 1458] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 24. | Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH, Majumdar AP. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis. 2012;33:68-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 25. | Edvardsson K, Nguyen-Vu T, Kalasekar SM, Pontén F, Gustafsson JÅ, Williams C. Estrogen receptor β expression induces changes in the microRNA pool in human colon cancer cells. Carcinogenesis. 2013;34:1431-1441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Ji S, Ye G, Zhang J, Wang L, Wang T, Wang Z, Zhang T, Wang G, Guo Z, Luo Y. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62:716-726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 27. | Chen ML, Liang LS, Wang XK. miR-200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1. Clin Exp Metastasis. 2012;29:457-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, Esteller M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062-2074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 288] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 29. | Lu MH, Huang CC, Pan MR, Chen HH, Hung WC. Prospero homeobox 1 promotes epithelial-mesenchymal transition in colon cancer cells by inhibiting E-cadherin via miR-9. Clin Cancer Res. 2012;18:6416-6425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 93] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 30. | Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng S, Ge W. MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Med Oncol. 2012;29:1037-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 31. | Liu X, Zhang Z, Sun L, Chai N, Tang S, Jin J, Hu H, Nie Y, Wang X, Wu K. MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis. 2011;32:1798-1805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 32. | Meng X, Wu J, Pan C, Wang H, Ying X, Zhou Y, Yu H, Zuo Y, Pan Z, Liu RY. Genetic and epigenetic down-regulation of microRNA-212 promotes colorectal tumor metastasis via dysregulation of MnSOD. Gastroenterology. 2013;145:426-436.e1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 33. | Lee CG, McCarthy S, Gruidl M, Timme C, J . YT. MicroRNA-147 Induces a Mesenchymal-To-Epithelial Transition (MET) and Reverses EGFR Inhibitor Resistance. PloS One. 2014;9:e84597. [DOI] [Full Text] |
| 34. | Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603-1614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 570] [Cited by in RCA: 693] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 35. | Weis S, Cui J, Barnes L, Cheresh D. Endothelial barrier disruption by VEGF-mediated Src activity potentiates tumor cell extravasation and metastasis. J Cell Biol. 2004;167:223-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 361] [Cited by in RCA: 388] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 36. | Rmali KA, Puntis MC, Jiang WG. Tumour-associated angiogenesis in human colorectal cancer. Colorectal Dis. 2007;9:3-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 64] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 37. | Sun W. Angiogenesis in metastatic colorectal cancer and the benefits of targeted therapy. J Hematol Oncol. 2012;5:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 38. | Liao D, Johnson RS. Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 2007;26:281-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 544] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 39. | Heusschen R, van Gink M, Griffioen AW, Thijssen VL. MicroRNAs in the tumor endothelium: novel controls on the angioregulatory switchboard. Biochim Biophys Acta. 2010;1805:87-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Sundaram P, Hultine S, Smith LM, Dews M, Fox JL, Biyashev D, Schelter JM, Huang Q, Cleary MA, Volpert OV. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res. 2011;71:7490-7501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 41. | Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631-3641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 247] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 42. | Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, Lai L, Jiang BH. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012;40:761-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 237] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 43. | Lawler J. Thrombospondin-1 as an endogenous inhibitor of angiogenesis and tumor growth. J Cell Mol Med. 2002;6:1-12. [PubMed] |
| 44. | Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060-1065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 838] [Cited by in RCA: 821] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 45. | Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348-1354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 364] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 46. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4471] [Cited by in RCA: 4523] [Article Influence: 238.1] [Reference Citation Analysis (0)] |
| 47. | Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010;70:8233-8246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 223] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 48. | Ichikawa D, Komatsu S, Konishi H, Otsuji E. Circulating microRNA in digestive tract cancers. Gastroenterology. 2012;142:1074-1078.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 49. | Moertel CG, Fleming TR, Macdonald JS, Haller DG, Laurie JA, Tangen C. An evaluation of the carcinoembryonic antigen (CEA) test for monitoring patients with resected colon cancer. JAMA. 1993;270:943-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 249] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 50. | Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 750] [Cited by in RCA: 750] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 51. | Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 227] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 52. | Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 855] [Cited by in RCA: 901] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 53. | Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 54. | Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A, Galandiuk S. A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg. 2013;258:400-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 55. | Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36:e61-e67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 56. | Hofsli E, Sjursen W, Prestvik WS, Johansen J, Rye M, Tranø G, Wasmuth HH, Hatlevoll I, Thommesen L. Identification of serum microRNA profiles in colon cancer. Br J Cancer. 2013;108:1712-1719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |