Published online Feb 26, 2013. doi: 10.4331/wjbc.v4.i1.1
Revised: January 26, 2013
Accepted: February 25, 2013
Published online: February 26, 2013
Processing time: 118 Days and 24 Hours
The Ca2+-binding protein of the EF-hand type, S100B, exerts both intracellular and extracellular regulatory activities. As an intracellular regulator, S100B is involved in the regulation of energy metabolism, transcription, protein phosphorylation, cell proliferation, survival, differentiation and motility, and Ca2+ homeostasis, by interacting with a wide array of proteins (i.e., enzymes, enzyme substrates, cytoskeletal subunits, scaffold/adaptor proteins, transcription factors, ubiquitin E3 ligases, ion channels) in a restricted number of cell types. As an extracellular signal, S100B engages the pattern recognition receptor, receptor for advanced glycation end-products (RAGE), on immune cells as well as on neuronal, astrocytic and microglial cells, vascular smooth muscle cells, skeletal myoblasts and cardiomyocytes. However, RAGE may not be the sole receptor activated by S100B, the protein being able to enhance bFGF-FGFR1 signaling by interacting with FGFR1-bound bFGF in particular cell types. Moreover, extracellular effects of S100B vary depending on its local concentration. Increasing evidence suggests that at the concentration found in extracellular fluids in normal physiological conditions and locally upon acute tissue injury, which is up to a few nM levels, S100B exerts trophic effects in the central and peripheral nervous system and in skeletal muscle tissue thus participating in tissue homeostasis. The present commentary summarizes results implicating intracellular and extracellular S100B in tissue development, repair and regeneration.
- Citation: Sorci G, Riuzzi F, Arcuri C, Tubaro C, Bianchi R, Giambanco I, Donato R. S100B protein in tissue development, repair and regeneration. World J Biol Chem 2013; 4(1): 1-12
- URL: https://www.wjgnet.com/1949-8454/full/v4/i1/1.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v4.i1.1
S100B belongs to a multigenic family of small (mol. wt. between 9 kDa and 14 kDa) Ca2+-binding proteins of the EF-hand type comprising more than 20 members exclusively expressed in vertebrates[1,2]. Like other members of this protein family, S100B is expressed in a cell-specific manner; astrocytes, oligodendrocytes, neural progenitor cells, certain neuronal populations, ependymocytes, Schwann cells, enteric glial cells, melanocytes, kidney epithelial cells, adipocytes, chondrocytes, skin Langerhans cells, a subpopulation of lymphocytes, muscle satellite cells, skeletal myofibers, pituitary folliculo-stellate cells and Leydig cells in the testis are the cell types with the highest expression of S100B. However, at least cardiomyocytes, which normally do not express the protein, do express S100B post-infarction, and in several cell types S100B expression is upregulated in pathological conditions.
Within cells S100B exists in the form of a homodimer, sometime as an S100B-S100A1 heterodimer, in which the two subunits are arranged in an antiparallel fashion[1,3]. Like the majority of S100 members, S100B is a Ca2+ sensor protein that becomes activated by Ca2+ on the occasion of Ca2+ transients. Ca2+ induces a relatively large conformational changes in S100B C-terminal half resulting in the exposure of a hydrophobic patch through which the protein interacts with a wide array of target proteins (e.g., enzymes, enzyme substrates, cytoskeletal proteins, adaptor/scaffold proteins, transcription factors, ion channels and ubiquitin E3 ligases) thereby regulating their activities. Thus, S100B is involved in the regulation of energy metabolism, transcription, protein phosphorylation, cell proliferation, survival, differentiation and locomotion, and Ca2+ homeostasis.
However, S100B can also exert extracellular effects being secreted by certain cell types (e.g., astrocytes and adipocytes) or passively released by several cell types upon tissue injury. In this latter context S100B can be viewed as a damage-associated molecular pattern (DAMP) or alarmin, i.e., a danger signal capable of activating cells of the innate immune system[1,3-6]. Extracellular effects of S100B mostly have been studied in the context of the central nervous system likely because of its high abundance in the brain and the identification of neurons, astrocytes and microglia as its target cells. Indeed, extracellular S100B has long been implicated in the pathophysiology of Alzheimer’s disease and neuroinflammation largely via engagement of the receptor for advanced glycation end-products (RAGE). However, accumulating evidence suggests that effects of extracellular S100B are not restricted to the brain or to cells of the innate immune system, and that RAGE may not be the sole receptor transducing S100B effects. In the present commentary we shall discuss results implicating intracellular and extracellular S100B in tissue development, homeostasis, repair and regeneration.
S100B is involved in cell proliferation, survival and differentiation both as an intracellular regulator and an extracellular signal. Within cells S100B binds to and activates Ndr (nuclear Dbf2-related)[7], a serine/threonine protein kinase implicated in the regulation of cell division and morphology[8]. Regulation of Ndr by S100B involves a conformational change in the catalytic domain triggered by Ca2+/S100B binding to the junction region[9]. However, although S100B-dependent activation of Ndr in cell lines has been documented[7] and in non-dividing and dividing cells S100B localizes to centrosomes[10], which are Ndr targets[8], no evidence has been presented that S100B-dependent activation of Ndr results in stimulation of cell proliferation and/or changes in cell morphology.
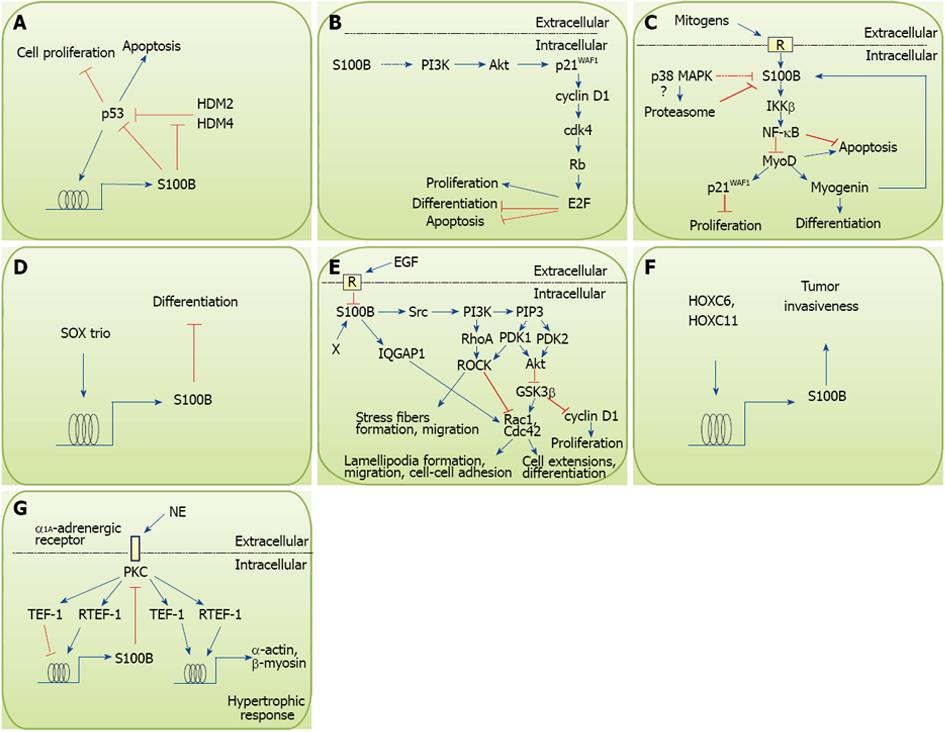
S100B also interacts with the tumor suppressor, p53, inhibiting its phosphorylation and tetramerization , i.e., its activation[11-14]. Also, S100B reduces p53 levels[15], and in turn, p53 upregulates S100B expression in melanoma cells[15]. In this scenario, p53 would reduce its own abundance by upregulating its inhibitor, S100B, which would result in uncontrolled proliferation[15] and reduced apoptosis[16] at least in melanoma cells (Figure 1A). However, phosphorylation of specific serine and/or threonine residues in p53 reduces the affinity of the S100B-p53 interaction by an order of magnitude, and is important for protecting p53 from S100B-dependent downregulation[17]. Thus, the S100B overall effect on p53 is likely to reflect a balance between inhibitory cues and intervening biochemical events (e.g., p53 phosphorylation). However, conflicting conclusions have been reported regarding functional implications of S100B/p53 interactions[15,18,19], and it is not known whether these interactions are relevant for tumor progression in other cancers and in non-neoplastic cells. In addition, it has been suggested that by interacting with the ubiquitin E3 ligases, MDM2 (HDM2) and MDM4 (HDM4)[17,20], that are central negative regulators of p53[21], S100B may actually promote p53 activities[20], which adds another layer of complexity to S100B-p53 interactions (Figure 1A). We have shown that forced expression of S100B in neuronal PC12 cells has no effects on p53 levels or nuclear translocation, and it results in enhanced proliferation and reduced differentiation and oxidative stress-induced apoptosis via activation of a PI3K/Akt/p21WAF1/cyclin D1/cdk4/Rb/E2F pathway in the absence of serum mitogens[22] (Figure 1B). S100B-dependent reduction of stress-induced apoptosis may also occur via interaction with and activation of the tetratricopeptide repeat protein, PP5, a member of the PPP family of serine/threonine phosphatases[23].
S100B is expressed in proliferating myoblast cell lines[24] and quiescent muscle satellite cells[25], the most relevant stem cell population in adult skeletal muscle tissue[26]. Increasing S100B levels in myoblast cell lines results in no effects on the proliferation rate of asynchronously proliferating myoblasts; however, S100B-overexpressing myoblasts are more resistant to basal and H2O2-induced apoptosis in an IκB kinase β (IKKβ)/ nuclear factor κB (NF-κB)-mediated manner[27] (Figure 1C). Thus, increasing S100B levels in myoblasts results in augmented cell numbers in consequence of their increased survival rate in stress conditions. Moreover, S100B-overexpressing myoblasts are less prone to acquire mitotic quiescence and proliferate faster than control cells upon re-exposure to serum mitogens after quiescence[27]. Proliferation of muscle satellite cells and their resistance to death-inducing stimuli are critical for efficient muscle regeneration as well as for successful cell therapy of muscular dystrophy[26,28-30]. Thus, intracellular S100B may contribute to muscle regeneration by reducing apoptosis and stimulating the expansion of activated satellite cells (Figure 1C). However, excess expression of S100B in activated satellite cells may be detrimental because its mitogenic effect might interfere with the reconstitution of the satellite cell reserve pool which normally occurs during muscle regeneration and requires that a fraction of cells stop proliferating and enter a quiescent state[26,28,29], and because myoblast proliferation and differentiation are mutually exclusive[26]. Considering that S100B is expressed in high abundance in several cancers[2,3], enhanced expression of S100B in activated muscle satellite cells, from which embryonal rhabdomyosarcomas are thought to originate[31], may also contribute to rhabdomyosarcomagenesis. Preliminary results show that embryonal rhabdomyosarcoma cells do indeed express elevated S100B levels (Riuzzi F, Sorci G, and Donato R, unpublished results).
Collectively, these data suggest that intracellular S100B may intervene in the regulation of proliferation, survival and apoptosis by mechanisms that vary depending on the cell type, the context and, probably, the cell’s normal or neoplastic condition. Further work is required to definitely establish the role of S100B in cell proliferation and survival in normal and neoplastic cells and the molecular mechanism(s) behind S100B overexpression in many cancers.
Intracellular S100B also functions as an inhibitor of differentiation. As mentioned above, expression of S100B in PC12 neuronal cells results in impaired NGF-induced differentiation via activation of a PI3K/Akt/p21WAF1/cyclin D1/cdk4/Rb/E2F pathway[22] (Figure 1B). However, induction of S100B expression in NGF-differentiated PC12 neuronal cells does not reverse the differentiated phenotype[22]. Also, S100B is induced in early-stage chondroblast differentiation by the SOX trio and negatively regulates chondrocyte terminal differentiation via an as yet undetermined mechanism[32] (Figure 1D). Interestingly, S100B expression in astrocytic cells is developmentally regulated albeit with different characteristics depending on whether subventricular or cortical astrocytic cells are considered[33]. These studies[33] have established that during the time interval between post-natal days 2 and 8 ramified, differentiating (i.e., GFAP filament-positive) astrocytes are S100B-negative. This suggests that during that time interval S100B may be downregulated and that the protein becomes re-expressed during the final phase(s) of astrocytic differentiation. S100B is expressed in radial glial precursors[34], in the ventricular zone of embryonic mouse cerebellum and progenitors of cerebellar granule cells[35], the protein being expressed in these latter cells as long as they are migrating. S100B interacts with the small GTPase Rac1 and Cdc42 effector, IQGAP1, at the polarized leading edge and areas of membrane ruffling in astrocytoma cell lines[36]. Hence, S100B has been proposed to regulate IQGAP1 activity in relation to cell migration (Figure 1E). In accordance with this view, reduction of S100B levels in astrocyte cell lines and primary astrocytes results in decreased proliferation and migration and acquisition of a differentiated phenotype (i.e., stellation) consequent to reduced activity of a Src/PI3K/RhoA/ROCK pathway and increased activity of the GSK3β/Rac1 module[37] (Figure 1E). These results are consistent with the possibility that repression of S100B expression at certain phases of development of astrocytes and certain neuronal populations may be functionally linked to their differentiation. Thus, S100B may contribute to expand the population of progenitors of neural cells and confer migratory capacity on undifferentiated astrocytes and neuroblasts, and S100B expression has to be repressed for differentiation to take place. In this context, S100B may act to avoid premature differentiation besides promoting cell migration; however, deregulated S100B expression may contribute to gliomagenesis. Intriguingly, knockdown of S100B in the Müller cell line, MIO-M1, results in remarkably inhibited neurosphere formation and differentiation of these cells towards the astrocyte phenotype[37]. Because MIO-M1 neurospheres have been shown to be made of neural precursor cells differentiating towards a neuronal phenotype when cultivated in the presence of bFGF or retinoic acid[38], the results in[37] suggest that S100B may contribute to confer stem cell-like properties on MIO-M1 cells and to reduce their propensity to differentiate into astrocytes. The expression of S100B in the murine cerebellar ventricular zone including the embryonic cerebellar rhombic lip and in cells lining cerebral ventricles[33-35] adds to the possibility that intracellular S100B may contribute to confer pluripotency on precursors of neural cells. Incidentally, the studies in[36,37] highlight S100B’s ability to regulate F-actin-based cytoskeleton in an indirect manner, i.e., via stimulation of a Src/PI3K/RhoA/ROCK and an IQGAP1/Rac1 pathway, and reduction of the activity of the GSK3β/Rac1 module (Figure 1E), as opposed to the protein’s direct effects on microtubule- and intermediate filament-based cytoskeleton[39-43].
On the other hand, HOXC6 and HOXC11, members of homeobox genes that encode transcription factors driving morphogenesis and cell differentiation during embryogenesis[44,45], have been reported to increase transcription of s100b in neuroblastoma cells[46] and this was interpreted as indicative of HOXC6 and HOXC11 stimulating differentiation of neuroblastoma cells into Schwann cells through the transcriptional activation of s100b. However, in the absence of data on the expression of additional markers such as myelin basic protein or GFAP, the expression of s100b may not be itself a proof of cell differentiation towards Schwann cells, oligodendrocytes or astrocytes[22,35,37]. Also, interactions of HOXC11 with the steroid receptor coactivator protein SRC-1, which is a strong predictor of reduced disease-free survival in breast cancer patients, induce the expression of S100B in resistant breast cancer cells[47] (Figure 1F). This latter study supports the notion that expression of S100B in proliferating and/or tumor cells may interfere with differentiation and/or is mechanistically linked to tumor progression. This study[47] also highlights the fact that S100B can be induced in precursors of certain cell types (breast cells, in the present case) and becomes repressed at completion of differentiation; differentiated breast cells do not express the protein whereas persistence of S100B in breast cell precursors may concur to tumor progression and invasion.
S100B is induced in post-infarction cardiomyocytes under the action of norepinephrine and phenylephrine via protein kinase C activation thereby limiting the hypertrophic response through the inhibition of the expression of the fetal proteins, skeletal α-actin and β-myosin heavy chain[48-50] (Figure 1G). Accordingly, norepinephrine-induced cardiac hypertrophy is inhibited in S100B transgenic mice[51]. Thus, S100B, which is not expressed in cardiomyocytes in normal physiological conditions, participates in the regulation of cardiomyocyte remodeling after infarction. These results appear in line with the notion that S100B is expressed in cells exhibiting properties of immature cells (post-infarction cardiomyocytes, in the present case). However, similarly to the majority of neuronal cells, in which S100B becomes stably repressed before differentiation, and differently from astrocytes (see above) and myoblasts (see below), in which a transient downregulation of S100B occurs at the beginning of differentiation, full maturation of cardiomyocytes is accompanied by stable repression of S100B expression.
Intracellular S100B modulates the differentiation of myoblasts, the precursors of skeletal myofibers. Indeed, overexpression of S100B in myoblasts blocks myogenic differentiation via IKKβ/NF-κB-mediated inhibition of expression of the muscle-specific transcription factor, MyoD, and the MyoD-downstream effectors myogenin and p21WAF1, and conversely, reduction of S100B expression in myoblasts by siRNA techniques results in reduced NF-κB activity and enhanced myogenic differentiation[25] (Figure 1C). It is known that NF-κB is a negative regulator of myogenic differentiation via inhibition of expression and/or reduction of stability of MyoD[52-54]. Also, S100B binds to, and inhibits EAG1 potassium channels Ca2+-dependently[55]. Because these channels have been reported to play a role in myoblast fusion into myotubes[56] it is possible that S100B may negatively affect myoblast differentiation via inhibition of EAG1 potassium channels as well. Moreover, compared with young subjects, muscle satellite cells from aged human subjects, which are known to be proliferation and differentiation defective[29,57], express higher levels of S100B and knockdown of S100B in aged satellite cells rescues their myogenic potential in part[58]. Notably, despite their high S100B levels, aged muscle satellite cells show a low proliferation rate and a remarkably reduced ability to secrete S100B and bFGF[58]. However, treatment of aged satellite cells with S100B or bFGF rescues their proliferative potential in part[58]. These results suggest that physiological levels of S100B in activated satellite cells and the satellite cells’ ability to secrete the protein concur to optimize the expansion of activated satellite cells required for satellite cell homeostasis, the maintenance of optimal muscular mass and/or efficient skeletal muscle regeneration after acute injury. In this context it is noteworthy that aged human satellite cells also exhibit altered expression of RAGE[58] shown to exert promyogenic effects[59-61] and to be required for S100B secretion[62]. Because transient transfection of aged satellite cells with full-length RAGE rescues their myogenin potential in part[58], one may speculate that the combination of enhanced S100B expression and expression of an altered form of RAGE may contribute significantly to their reduced myogenic potential, hence to sarcopenia. The recent demonstration that levels of bFGF are high and are responsible for disrupted satellite cell quiescence in aged skeletal muscle in homeostatic conditions[63] lend support to the possibility that excess S100B in aged satellite cells, potentially caused by high bFGF[1,3] may ultimately lead to defective muscle regenerative capacity as observed in sarcopenia. Interestingly, levels of S100B decrease in non-fused myoblasts early after their transfer to differentiation medium and S100B becomes re-expressed in differentiating (i.e., myogenin-positive) myocytes[25,27], which supports the notion that S100B levels have to decrease transiently in certain cell types for they to differentiate. Both differentiation cues (namely the activation of the promyogenic p38 MAPK) and reduction of mitogens appear to determine the transient downregulation of S100B in myoblasts in differentiation medium via transcriptional and post-translational (proteasome-dependent) mechanisms[27] (Figure 1C). Collectively, these results suggest that S100B in myoblasts contributes to reduce their premature differentiation which would be detrimental to skeletal muscle regeneration after acute injury, and that levels of S100B should be kept within a certain range of abundance in order to avoid excessive expansion of activated satellite cells leading to defective reconstitution of the damaged tissue and the pool of quiescent satellite cells.
Whereas EGF has been reported to reduce S100B expression in developing astrocytes[33] (Figure 1E), the extracellular stimuli and intracellular mechanisms causing transient or stable downregulation of S100B expression during cell differentiation are not completely defined. Also, because mature astrocytes, chondrocytes, myocytes (i.e., differentiated myoblasts), skeletal myofibers and certain neuronal populations in the adult brain express S100B[3,25,64], mechanisms should exist that cause re-expression of the protein at later developmental stages without determining cell de-differentiation[22,27]. In the case of skeletal muscle cells, the muscle-specific transcription factor, myogenin, that is essential for myogenic differentiation[26], has been implicated in the re-expression of S100B in myocytes[27] (Figure 1C). Overall, these observations suggest that functions of S100B may be different in developing and mature cells and that S100B may regulate different signaling pathways and functions depending on the cell type and the cell’s status. Future work should dissect the molecular mechanism(s) responsible for the regulation of S100B expression in immature (proliferating) and fully differentiated cells.
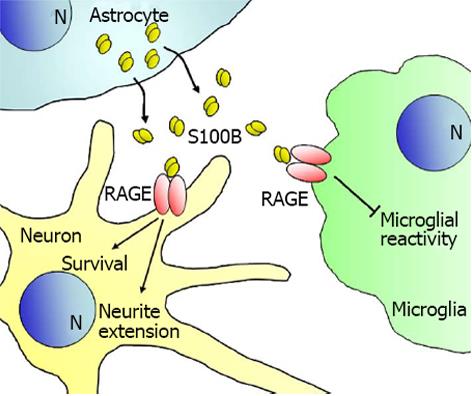
Extracellular S100B also regulates cell proliferation, survival and differentiation. Several factors/conditions regulate either positively or negatively S100B secretion by astrocytes, among which are interleukin-1β, extracellular levels of Ca2+ and K+, inhibitors of gap junctions, antioxidants, lipopolysaccharide, apomorphine and certain antipsychotic drugs[65-69]. At the low nM concentration found in the brain extracellular space in normal physiological conditions[3], S100B exerts pro-survival effects on neurons[70-73], stimulate astrocyte proliferation[74] and reduce microglial reactivity[75,76], via RAGE engagement in most cases (Figure 2). However, at low nM levels S100B synergizes with proinflammatory cytokines to activate microglia[77] suggesting that S100B may switch from antiinflammatory to proinflammatory at early phases of neuroinflammation (i.e., in the presence of low levels of inflammatory cytokines). Yet, attenuation of microglia activity by low concentrations of S100B may contribute to local tumor immunosuppression[76].
S100B has been implicated in the activity of antidepressants. The selective serotonin reuptake inhibitor, fluoxetine, increases S100B content in the hippocampus[78] and stimulates S100B secretion from astrocytes[79] and serotoninergic neurons[80]. It has been shown that secreted S100B downregulates microRNA-16 in noradrenergic neurons, which consequently acquire properties of serotoninergic neurons[80]. Although no information is available regarding the mechanism whereby fluoxetine induces serotoninergic neurons to express and secrete S100B, the mechanism whereby secreted S100B reduces microRNA-16 levels in noradrenergic neurons or the S100B-serotoninergic neuron relationships in S100B-null or transgenic mice, these results point to an important role of extracellular S100B in fluoxetine-dependent neurogenesis and neuronal plasticity[81,82].
Serum levels of S100B increase remarkably following an intense physical exercise[83,84], the source of the protein reasonably being skeletal myofibers in these circumstances. Indeed, intense physical exercise is associated with reversible skeletal muscle tissue damage and release of intracellular proteins[26], and the local concentration of S100B may be even higher than in serum thus allowing paracrine S100B effects on activated muscle stem (satellite) cells. In fact, at picomolar to low nanomolar concentrations S100B inhibits myoblast differentiation and stimulates myoblast proliferation[85-87] raising the possibility that the protein may participate in the process of skeletal muscle regeneration by expanding the myoblast population (see below).
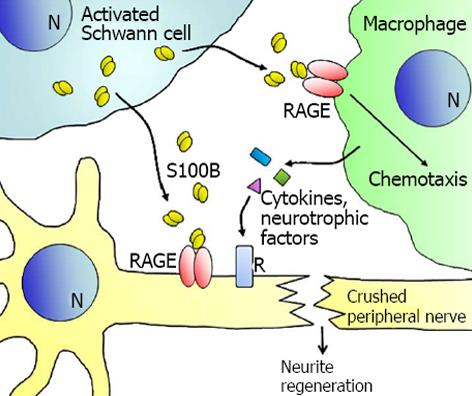
Since the discovery that a protein factor purified from brain and endowed with neurite extension activity was a disulfide cross-linked form of S100B[88] and the demonstration that S100B is found in the brain extracellular space[89] and is actively secreted by astrocytes[90], a mess of information has been provided over time on the protective and trophic role of S100B on neurons[70-73,91-98](Figure 2). S100B is found expressed in Schwann cells in uninjured peripheral nerves as well as in activated Schwann cells during the degeneration period of crushed nerves, i.e., up to day 7 post-injury, and in normal Schwann cells reappearing during the regeneration period, i.e., after day 7 post-injury, in the zone of the crush and proximal and distal to it[99]. In similar conditions, RAGE becomes expressed in axons and in infiltrating mononuclear phagocytes and reduction of RAGE expression and/or activity results in suppression of anatomical regeneration and functional recovery[100,101]. Upon acute peripheral nerve injury, S100B released from Schwann cells in damaged nerves activates RAGE in infiltrating macrophages[100,101] and in activated Schwann cells[102]; infiltrating macrophages exert beneficial effects by clearing cell debris and dead neutrophils and releasing cytokines and trophic factors, whereas activated Schwann cells release cytokines and neurotrophic factors shown to be crucial for the repair of injured nerves (Figure 3). S100B-activated RAGE promotes Schwann cell migration during the course of repair of injured peripheral nerves through the induction of thioredoxin interacting protein and activation of p38 MAPK, CREB and NF-κB[102]. S100B also stimulates proliferation and differentiation of neural progenitor cells from the subventricular zone of the adult mouse brain via RAGE activation[103]. These results complement the long-standing notion that S100B stimulates neuronal cell survival and differentiation via RAGE engagement[71,72].
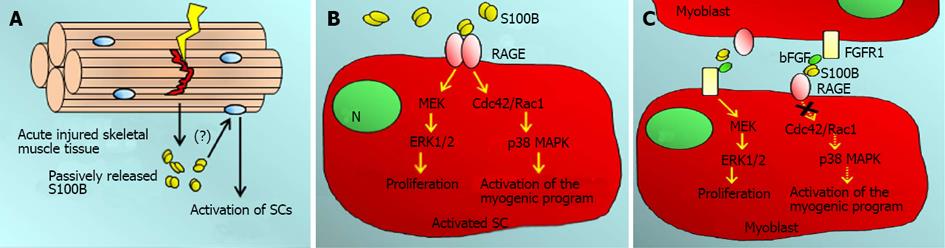
S100B also is expressed in skeletal myofibers[25,64] from which it is massively released early upon acute injury with declining release during the regeneration phase[104] (Figure 4A). Released S100B stimulates myoblast proliferation and concomitantly activates the myogenic differentiation program via RAGE engagement early after injury (Figure 4B), i.e., at a time when myoblast density and the level of released bFGF are low[104], thereby contributing to the timely and limited expansion of the myoblast population required for efficient muscle regeneration. Indeed, acutely injured Rage−/− muscles show delayed regeneration[61]. However, persistence of extracellular S100B in the damaged tissue is likely to prolong the myoblast proliferation phase at the expense of differentiation and reconstitution of the pool of quiescent satellite cells via enhancement of bFGF/FGFR1 signaling and blockade of RAGE signaling[87,104] (Figure 4C). The switch of S100B from a RAGE-activating factor to a bFGF/FGFR1 activating factor depends on the S100B concentration, the presence of bFGF and myoblast density[87,104]. Current findings indicate that neutralization of released S100B in acutely injured wild-type skeletal muscles results in defective regeneration as a consequence of reduced expansion of the population of activated satellite cells, reduced infiltration of the injured tissue with macrophages and delayed transition of macrophages from the M1 (proinflammatory) to the M2 (antiinflammatory) phase (Riuzzi F, Sorci G, Beccafico S and Donato R, in preparation). These results indicate that released S100B participates in the regeneration of acutely injured muscles by stimulating myoblast proliferation, macrophage infiltration and macrophage transition from a proinflammatory phenotype to an antiinflammatory phenotype. Our ongoing results also show that these effects of S100B are strictly RAGE-dependent, because neutralization of released S100B in acutely injured Rage−/− muscles does not change the muscle regeneration pattern described in[61]. However, one may anticipate that chronic release of S100B from skeletal myofibers in, e.g., muscular dystrophies and chronic inflammatory muscle diseases may translate into high local S100B concentrations amplifying or perpetuating muscle damage, a situation reminiscent of what occurs in the brain where low S100B levels are beneficial whereas chronically high S100B levels are detrimental, via RAGE engagement in both cases[1,3,71,72].
Also, cell/tissue identity appears to profoundly condition S100B’s extracellular effects. For example, whereas at concentrations ≤ 50 nmol/L S100B exerts trophic effects on neuronal and astrocytic cells and skeletal myoblasts[3,105,106], at doses ≥ 50 nmol/L the protein causes RAGE-dependent cardiomyocyte apoptosis[107]. However, a short-term (1 h) treatment of cardiomyocytes with S100B (100 nmol/L) (a condition insufficient to cause apoptosis) results in a RAGE-dependent secretion of vascular endothelial growth factor (VEGF) which in turn induces myofibroblast proliferation[108]. By this mechanism S100B might contribute to post-infarction scar formation, a kind of tissue reparative process. Whether S100B also causes VEGF-dependent post-infarction neoangiogenesis remains to be investigated. Intriguingly, whereas S100B is induced in the heart of diabetic mice as well, S100B mRNA and protein expression levels decrease in diabetes post-infarction by a mechanism that remains to be identified, and deletion of s100b has a deleterious effect on cardiac function in this condition partly attributed to increased ventricular dilation associated with increased AGE formation and reduced GLUT4 expression, i.e., reduced cardiac glucose metabolism[109]. Whether these changes are due to reduced intracellular or extracellular effects of S100B is not known. Yet, these results point to a protective role of S100B in post-infarction heart.
The role of extracellular S100B as a DAMP involved in inflammation is an accepted notion (see Refs.[1,3-6,106] for pertinent literature). However, for S100B to sustain inflammation via activation of macrophages/microglia it has to be present at relatively high concentration at injury sites[1,3-6,106,110], as it reasonably occurs during the course of chronic tissue damage as a result of a continuous release of the protein from injured cells, cell necrosis and/or defective clearance. However, recent evidence points to a novel role of S100B in resolution of inflammation in Aspergillus fumigatus infection in lung[111]. TLR2 activation on bronchial epithelial cells by the fungus results in upregulation of expression and release of S100B, that paracrinally binds to RAGE on polymorphonuclear neutrophils and mediates its association with TLR2 for subsequent inhibition. In addition, S100B upon binding to nucleic acids in bronchial epithelial cells, also activates an intracellular TLR3/TLR9/TRIF-dependent pathway leading to repression of s100b transcription. The transcriptional repression of s100b by the sequential action of downstream MyD88- and TRIF-dependent NF-κB signaling pathways[111] thus provides the molecular basis for a braking circuit in infection whereby the endogenous danger protects the host against pathogen-induced inflammation and a nucleic acid-sensing mechanism resolves danger-induced chronic inflammation. Whether this is a general mechanism of action of the S100B/RAGE axis in the course of infections remains to be determined. However, high local S100B concentrations exacerbates Aspergillus fumigatus-induced pulmonary inflammation[111] likely via sustained stimulation of RAGE signaling. Interestingly, the S100B (+427C/T) polymorphism results in S100B overexpression which associates with susceptibility to invasive aspergillosis in patients undergoing hematopoietic stem cell transplantation whenever the recipients show RAGE (-374T/A) polymorphism resulting in RAGE overexpression[112].
During the last decade there has been a burst of interest in S100B functions[3,6,106] following the seminal demonstration that S100B engages RAGE in immune cells and behaves like a DAMP[113]. Evidence has been provided shortly after that both the neurotrophic and neurotoxic effects of low and high S100B levels, respectively, on neuronal cells[1,3], are mediated by RAGE engagement[71]. However, S100B mostly has been viewed as a DAMP involved in the inflammatory response and S100B often has been used as a generic RAGE activator in the context of the inflammatory response[3,6,113].
Yet, a large body of information indicates that S100B protein is involved in cell proliferation, survival, motility and differentiation by acting as an intracellular regulator and an extracellular signal in normal physiological conditions and during the acute phase of tissue damage. In so doing, S100B may play a role in tissue development and repair after acute injury, through the refinement or fine tuning of enzyme activities, the dynamics of the cytoskeleton and cell-specific gene expression, and responses to external stimuli. Moreover, S100B exerts anti-infection effects in the bronchial epithelium where a tight regulation of its expression and release is mechanistically linked to the resolution of inflammation after fungal infection. Future work should assess the molecular mechanism(s) regulating S100B expression in developing and mature cells and during tissue repair/regeneration.
P- Reviewer Scatena R S- Editor Song XX L- Editor A E- Editor Lu YJ
| 1. | Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1140] [Cited by in RCA: 1184] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 2. | Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem Biophys Res Commun. 2004;322:1111-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 645] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 3. | Donato R, Sorci G, Riuzzi F, Arcuri C, Bianchi R, Brozzi F, Tubaro C, Giambanco I. S100B’s double life: intracellular regulator and extracellular signal. Biochim Biophys Acta. 2009;1793:1008-1022. [PubMed] [DOI] [Full Text] |
| 4. | Donato R. RAGE: a single receptor for several ligands and different cellular responses: the case of certain S100 proteins. Curr Mol Med. 2007;7:711-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 218] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 5. | Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta. 2009;1793:993-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 389] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 6. | Sorci G, Bianchi R, Riuzzi F, Tubaro C, Arcuri C, Giambanco I, Donato R. S100B Protein, A Damage-Associated Molecular Pattern Protein in the Brain and Heart, and Beyond. Cardiovasc Psychiatry Neurol. 2010;2010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 125] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 7. | Millward TA, Heizmann CW, Schäfer BW, Hemmings BA. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J. 1998;17:5913-5922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 122] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Hergovich A, Cornils H, Hemmings BA. Mammalian NDR protein kinases: from regulation to a role in centrosome duplication. Biochim Biophys Acta. 2008;1784:3-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416-14426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Sorci G, Agneletti AL, Bianchi R, Donato R. Association of S100B with intermediate filaments and microtubules in glial cells. Biochim Biophys Acta. 1998;1448:277-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence JJ. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc Natl Acad Sci USA. 1992;89:11627-11631. [PubMed] |
| 12. | Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca(2+)-dependent interaction of S100B(beta beta) with a peptide derived from p53. Biochemistry. 1998;37:1951-1960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Lin J, Blake M, Tang C, Zimmer D, Rustandi RR, Weber DJ, Carrier F. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J Biol Chem. 2001;276:35037-35041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 115] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 14. | van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J Biol Chem. 2009;284:13804-13811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 15. | Lin J, Yang Q, Yan Z, Markowitz J, Wilder PT, Carrier F, Weber DJ. Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. J Biol Chem. 2004;279:34071-34077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 104] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Lin J, Yang Q, Wilder PT, Carrier F, Weber DJ. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. J Biol Chem. 2010;285:27487-27498. [RCA] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Wilder PT, Lin J, Bair CL, Charpentier TH, Yang D, Liriano M, Varney KM, Lee A, Oppenheim AB, Adhya S. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim Biophys Acta. 2006;1763:1284-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Scotto C, Delphin C, Deloulme JC, Baudier J. Concerted regulation of wild-type p53 nuclear accumulation and activation by S100B and calcium-dependent protein kinase C. Mol Cell Biol. 1999;19:7168-7180. [PubMed] |
| 19. | Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc Natl Acad Sci USA. 2005;102:4735-4740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 147] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 20. | van Dieck J, Lum JK, Teufel DP, Fersht AR. S100 proteins interact with the N-terminal domain of MDM2. FEBS Lett. 2010;584:3269-3274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA. 2006;103:3232-3237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 204] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 22. | Arcuri C, Bianchi R, Brozzi F, Donato R. S100B increases proliferation in PC12 neuronal cells and reduces their responsiveness to nerve growth factor via Akt activation. J Biol Chem. 2005;280:4402-4414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Yamaguchi F, Umeda Y, Shimamoto S, Tsuchiya M, Tokumitsu H, Tokuda M, Kobayashi R. S100 proteins modulate protein phosphatase 5 function: a link between CA2+ signal transduction and protein dephosphorylation. J Biol Chem. 2012;287:13787-13798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 24. | Sorci G, Bianchi R, Giambanco I, Rambotti MG, Donato R. Replicating myoblasts and fused myotubes express the calcium-regulated proteins S100A1 and S100B. Cell Calcium. 1999;25:93-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Tubaro C, Arcuri C, Giambanco I, Donato R. S100B protein in myoblasts modulates myogenic differentiation via NF-kappaB-dependent inhibition of MyoD expression. J Cell Physiol. 2010;223:270-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Chargé SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1801] [Cited by in RCA: 1886] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 27. | Tubaro C, Arcuri C, Giambanco I, Donato R. S100B in myoblasts regulates the transition from activation to quiescence and from quiescence to activation and reduces apoptosis. Biochim Biophys Acta. 2011;1813:1092-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem. 2006;54:1177-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 466] [Cited by in RCA: 465] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 29. | Brack AS, Rando TA. Intrinsic changes and extrinsic influences of myogenic stem cell function during aging. Stem Cell Rev. 2007;3:226-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 30. | Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol. 2007;19:628-633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 403] [Cited by in RCA: 395] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 31. | Hettmer S, Wagers AJ. Muscling in: Uncovering the origins of rhabdomyosarcoma. Nat Med. 2010;16:171-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 101] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 32. | Saito T, Ikeda T, Nakamura K, Chung UI, Kawaguchi H. S100A1 and S100B, transcriptional targets of SOX trio, inhibit terminal differentiation of chondrocytes. EMBO Rep. 2007;8:504-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 93] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 33. | Raponi E, Agenes F, Delphin C, Assard N, Baudier J, Legraverend C, Deloulme JC. S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia. 2007;55:165-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 287] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 34. | Landry CF, Ivy GO, Dunn RJ, Marks A, Brown IR. Expression of the gene encoding the beta-subunit of S-100 protein in the developing rat brain analyzed by in situ hybridization. Brain Res Mol Brain Res. 1989;6:251-262. [PubMed] |
| 35. | Hachem S, Laurenson AS, Hugnot JP, Legraverend C. Expression of S100B during embryonic development of the mouse cerebellum. BMC Dev Biol. 2007;7:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 36. | Mbele GO, Deloulme JC, Gentil BJ, Delphin C, Ferro M, Garin J, Takahashi M, Baudier J. The zinc- and calcium-binding S100B interacts and co-localizes with IQGAP1 during dynamic rearrangement of cell membranes. J Biol Chem. 2002;277:49998-50007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 37. | Brozzi F, Arcuri C, Giambanco I, Donato R. S100B Protein Regulates Astrocyte Shape and Migration via Interaction with Src Kinase: Implications for astrocyte development, activation, and tumor growth. J Biol Chem. 2009;284:8797-8811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 38. | Lawrence JM, Singhal S, Bhatia B, Keegan DJ, Reh TA, Luthert PJ, Khaw PT, Limb GA. MIO-M1 cells and similar muller glial cell lines derived from adult human retina exhibit neural stem cell characteristics. Stem Cells. 2007;25:2033-2043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 196] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 39. | Donato R. Effect of S-100 protein on assembly of brain microtubule proteins in vitro. FEBS Lett. 1983;162:310-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 40. | Donato R. Mechanism of action of S-100 protein(s) on brain microtubule protein assembly. Biochem Biophys Res Commun. 1984;124:850-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 41. | Donato R. Calcium-independent, pH-regulated effects of S-100 proteins on assembly-disassembly of brain microtubule protein in vitro. J Biol Chem. 1988;263:106-110. [PubMed] |
| 42. | Garbuglia M, Verzini M, Giambanco I, Spreca A, Donato R. Effects of calcium-binding proteins (S-100a(o), S-100a, S-100b) on desmin assembly in vitro. FASEB J. 1996;10:317-324. [PubMed] |
| 43. | Sorci G, Agneletti AL, Donato R. Effects of S100A1 and S100B on microtubule stability. An in vitro study using triton-cytoskeletons from astrocyte and myoblast cell lines. Neuroscience. 2000;99:773-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 68] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 302] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 45. | McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1984] [Cited by in RCA: 1979] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 46. | Zhang X, Hamada J, Nishimoto A, Takahashi Y, Murai T, Tada M, Moriuchi T. HOXC6 and HOXC11 increase transcription of S100beta gene in BrdU-induced in vitro differentiation of GOTO neuroblastoma cells into Schwannian cells. J Cell Mol Med. 2007;11:299-306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 47. | McIlroy M, McCartan D, Early S, O Gaora P, Pennington S, Hill AD, Young LS. Interaction of developmental transcription factor HOXC11 with steroid receptor coactivator SRC-1 mediates resistance to endocrine therapy in breast cancer. Cancer Res. 2010;70:1585-1594. [RCA] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 48. | Tsoporis JN, Marks A, Kahn HJ, Butany JW, Liu PP, O’Hanlon D, Parker TG. S100beta inhibits alpha1-adrenergic induction of the hypertrophic phenotype in cardiac myocytes. J Biol Chem. 1997;272:31915-31921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 49. | Tsoporis JN, Marks A, Van Eldik LJ, O’Hanlon D, Parker TG. Regulation of the S100B gene by alpha 1-adrenergic stimulation in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2003;284:H193-H203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 50. | Tsoporis JN, Marks A, Haddad A, Dawood F, Liu PP, Parker TG. S100B expression modulates left ventricular remodeling after myocardial infarction in mice. Circulation. 2005;111:598-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 51. | Tsoporis JN, Marks A, Kahn HJ, Butany JW, Liu PP, O’Hanlon D, Parker TG. Inhibition of norepinephrine-induced cardiac hypertrophy in s100beta transgenic mice. J Clin Invest. 1998;102:1609-1616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 52. | Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS. NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363-2366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 701] [Cited by in RCA: 708] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 53. | Mourkioti F, Kratsios P, Luedde T, Song YH, Delafontaine P, Adami R, Parente V, Bottinelli R, Pasparakis M, Rosenthal N. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J Clin Invest. 2006;116:2945-2954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 258] [Cited by in RCA: 254] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 54. | Bakkar N, Wang J, Ladner KJ, Wang H, Dahlman JM, Carathers M, Acharyya S, Rudnicki MA, Hollenbach AD, Guttridge DC. IKK/NF-kappaB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol. 2008;180:787-802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 184] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 55. | Sahoo N, Tröger J, Heinemann SH, Schönherr R. Current inhibition of human EAG1 potassium channels by the Ca2+ binding protein S100B. FEBS Lett. 2010;584:3896-3900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 56. | Bijlenga P, Occhiodoro T, Liu JH, Bader CR, Bernheim L, Fischer-Lougheed J. An ether -à-go-go K+ current, Ih-eag, contributes to the hyperpolarization of human fusion-competent myoblasts. J Physiol. 1998;512:317-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 57. | Gopinath SD, Rando TA. Stem cell review series: aging of the skeletal muscle stem cell niche. Aging Cell. 2008;7:590-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 188] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 58. | Beccafico S, Riuzzi F, Puglielli C, Mancinelli R, Fulle S, Sorci G, Donato R. Human muscle satellite cells show age-related differential expression of S100B protein and RAGE. Age (Dordr). 2011;33:523-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 59. | Sorci G, Riuzzi F, Arcuri C, Giambanco I, Donato R. Amphoterin stimulates myogenesis and counteracts the antimyogenic factors basic fibroblast growth factor and S100B via RAGE binding. Mol Cell Biol. 2004;24:4880-4894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 60. | Riuzzi F, Sorci G, Donato R. The amphoterin (HMGB1)/receptor for advanced glycation end products (RAGE) pair modulates myoblast proliferation, apoptosis, adhesiveness, migration, and invasiveness. Functional inactivation of RAGE in L6 myoblasts results in tumor formation in vivo. J Biol Chem. 2006;281:8242-8253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 61. | Riuzzi F, Sorci G, Sagheddu R, Donato R. HMGB1-RAGE regulates muscle satellite cell homeostasis through p38-MAPK- and myogenin-dependent repression of Pax7 transcription. J Cell Sci. 2012;125:1440-1454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 62. | Perrone L, Peluso G, Melone MA. RAGE recycles at the plasma membrane in S100B secretory vesicles and promotes Schwann cells morphological changes. J Cell Physiol. 2008;217:60-71. [RCA] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 63. | Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355-360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 621] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 64. | Arcuri C, Giambanco I, Bianchi R, Donato R. Annexin V, annexin VI, S100A1 and S100B in developing and adult avian skeletal muscles. Neuroscience. 2002;109:371-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 65. | de Souza DF, Leite MC, Quincozes-Santos A, Nardin P, Tortorelli LS, Rigo MM, Gottfried C, Leal RB, Gonçalves CA. S100B secretion is stimulated by IL-1beta in glial cultures and hippocampal slices of rats: Likely involvement of MAPK pathway. J Neuroimmunol. 2009;206:52-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 66. | Nardin P, Tortorelli L, Quincozes-Santos A, de Almeida LM, Leite MC, Thomazi AP, Gottfried C, Wofchuk ST, Donato R, Gonçalves CA. S100B secretion in acute brain slices: modulation by extracellular levels of Ca(2+) and K (+). Neurochem Res. 2009;34:1603-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 67. | Leite MC, Galland F, de Souza DF, Guerra MC, Bobermin L, Biasibetti R, Gottfried C, Gonçalves CA. Gap junction inhibitors modulate S100B secretion in astrocyte cultures and acute hippocampal slices. J Neurosci Res. 2009;87:2439-2446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 68. | Nardin P, Tramontina AC, Quincozes-Santos A, Tortorelli LS, Lunardi P, Klein PR, Wartchow KM, Bobermin LD, Gottfried C, Elisabetsky E. In vitro S100B secretion is reduced by apomorphine: effects of antipsychotics and antioxidants. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1291-1296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 69. | Guerra MC, Tortorelli LS, Galland F, Da Ré C, Negri E, Engelke DS, Rodrigues L, Leite MC, Gonçalves CA. Lipopolysaccharide modulates astrocytic S100B secretion: a study in cerebrospinal fluid and astrocyte cultures from rats. J Neuroinflammation. 2011;8:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 70. | Barger SW, Van Eldik LJ, Mattson MP. S100 beta protects hippocampal neurons from damage induced by glucose deprivation. Brain Res. 1995;677:167-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 107] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 71. | Huttunen HJ, Kuja-Panula J, Sorci G, Agneletti AL, Donato R, Rauvala H. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. J Biol Chem. 2000;275:40096-40105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 475] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 72. | Businaro R, Leone S, Fabrizi C, Sorci G, Donato R, Lauro GM, Fumagalli L. S100B protects LAN-5 neuroblastoma cells against Abeta amyloid-induced neurotoxicity via RAGE engagement at low doses but increases Abeta amyloid neurotoxicity at high doses. J Neurosci Res. 2006;83:897-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 73. | Ellis EF, Willoughby KA, Sparks SA, Chen T. S100B protein is released from rat neonatal neurons, astrocytes, and microglia by in vitro trauma and anti-S100 increases trauma-induced delayed neuronal injury and negates the protective effect of exogenous S100B on neurons. J Neurochem. 2007;101:1463-1470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 74. | Selinfreund RH, Barger SW, Pledger WJ, Van Eldik LJ. Neurotrophic protein S100 beta stimulates glial cell proliferation. Proc Natl Acad Sci USA. 1991;88:3554-3558. [PubMed] |
| 75. | Reali C, Scintu F, Pillai R, Donato R, Michetti F, Sogos V. S100B counteracts effects of the neurotoxicant trimethyltin on astrocytes and microglia. J Neurosci Res. 2005;81:677-86. [RCA] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 76. | Zhang L, Liu W, Alizadeh D, Zhao D, Farrukh O, Lin J, Badie SA, Badie B. S100B attenuates microglia activation in gliomas: possible role of STAT3 pathway. Glia. 2011;59:486-498. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 77. | Bianchi R, Giambanco I, Donato R. S100B/RAGE-dependent activation of microglia via NF-kappaB and AP-1 Co-regulation of COX-2 expression by S100B, IL-1beta and TNF-alpha. Neurobiol Aging. 2010;31:665-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 205] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 78. | Akhisaroglu M, Manev R, Akhisaroglu E, Uz T, Manev H. Both aging and chronic fluoxetine increase S100B content in the mouse hippocampus. Neuroreport. 2003;14:1471-1473. [PubMed] |
| 79. | Tramontina AC, Tramontina F, Bobermin LD, Zanotto C, Souza DF, Leite MC, Nardin P, Gottfried C, Gonçalves CA. Secretion of S100B, an astrocyte-derived neurotrophic protein, is stimulated by fluoxetine via a mechanism independent of serotonin. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1580-1583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 80. | Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329:1537-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 381] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 81. | Maya Vetencourt JF, Sale A, Viegi A, Baroncelli L, De Pasquale R, O’Leary OF, Castrén E, Maffei L. The antidepressant fluoxetine restores plasticity in the adult visual cortex. Science. 2008;320:385-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 332] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 82. | David DJ, Samuels BA, Rainer Q, Wang JW, Marsteller D, Mendez I, Drew M, Craig DA, Guiard BP, Guilloux JP. Neurogenesis-dependent and -independent effects of fluoxetine in an animal model of anxiety/depression. Neuron. 2009;62:479-493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1033] [Cited by in RCA: 989] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 83. | Dietrich MO, Tort AB, Schaf DV, Farina M, Gonçalves CA, Souza DO, Portela LV. Increase in serum S100B protein level after a swimming race. Can J Appl Physiol. 2003;28:710-716. [PubMed] |
| 84. | Hasselblatt M, Mooren FC, von Ahsen N, Keyvani K, Fromme A, Schwarze-Eicker K, Senner V, Paulus W. Serum S100beta increases in marathon runners reflect extracranial release rather than glial damage. Neurology. 2004;62:1634-1636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 85. | Sorci G, Riuzzi F, Agneletti AL, Marchetti C, Donato R. S100B inhibits myogenic differentiation and myotube formation in a RAGE-independent manner. Mol Cell Biol. 2003;23:4870-4881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 86. | Riuzzi F, Sorci G, Donato R. S100B stimulates myoblast proliferation and inhibits myoblast differentiation by independently stimulating ERK1/2 and inhibiting p38 MAPK. J Cell Physiol. 2006;207:461-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 87. | Riuzzi F, Sorci G, Donato R. S100B protein regulates myoblast proliferation and differentiation by activating FGFR1 in a bFGF-dependent manner. J Cell Sci. 2011;124:2389-2400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 88. | Kligman D, Marshak DR. Purification and characterization of a neurite extension factor from bovine brain. Proc Natl Acad Sci USA. 1985;82:7136-7139. [PubMed] |
| 89. | Shashoua VE, Hesse GW, Moore BW. Proteins of the brain extracellular fluid: evidence for release of S-100 protein. J Neurochem. 1984;42:1536-1541. [PubMed] |
| 90. | Van Eldik LJ, Zimmer DB. Secretion of S-100 from rat C6 glioma cells. Brain Res. 1987;436:367-370. [PubMed] |
| 91. | Winningham-Major F, Staecker JL, Barger SW, Coats S, Van Eldik LJ. Neurite extension and neuronal survival activities of recombinant S100 beta proteins that differ in the content and position of cysteine residues. J Cell Biol. 1989;109:3063-3071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 226] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 92. | Azmitia EC, Dolan K, Whitaker-Azmitia PM. S-100B but not NGF, EGF, insulin or calmodulin is a CNS serotonergic growth factor. Brain Res. 1990;516:354-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 187] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 93. | Van Eldik LJ, Christie-Pope B, Bolin LM, Shooter EM, Whetsell WO. Neurotrophic activity of S-100 beta in cultures of dorsal root ganglia from embryonic chick and fetal rat. Brain Res. 1991;542:280-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 90] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 94. | Bhattacharyya A, Oppenheim RW, Prevette D, Moore BW, Brackenbury R, Ratner N. S100 is present in developing chicken neurons and Schwann cells and promotes motor neuron survival in vivo. J Neurobiol. 1992;23:451-466. [PubMed] |
| 95. | Alexanian AR, Bamburg JR. Neuronal survival activity of s100betabeta is enhanced by calcineurin inhibitors and requires activation of NF-kappaB. FASEB J. 1999;13:1611-1620. [PubMed] |
| 96. | Willoughby KA, Kleindienst A, Müller C, Chen T, Muir JK, Ellis EF. S100B protein is released by in vitro trauma and reduces delayed neuronal injury. J Neurochem. 2004;91:1284-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 97. | Kleindienst A, McGinn MJ, Harvey HB, Colello RJ, Hamm RJ, Bullock MR. Enhanced hippocampal neurogenesis by intraventricular S100B infusion is associated with improved cognitive recovery after traumatic brain injury. J Neurotrauma. 2005;22:645-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 145] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 98. | Pichiule P, Chavez JC, Schmidt AM, Vannucci SJ. Hypoxia-inducible factor-1 mediates neuronal expression of the receptor for advanced glycation end products following hypoxia/ischemia. J Biol Chem. 2007;282:36330-36340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 99. | Spreca A, Rambotti MG, Rende M, Saccardi C, Aisa MC, Giambanco I, Donato R. Immunocytochemical localization of S-100b protein in degenerating and regenerating rat sciatic nerves. J Histochem Cytochem. 1989;37:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 100. | Rong LL, Trojaborg W, Qu W, Kostov K, Yan SD, Gooch C, Szabolcs M, Hays AP, Schmidt AM. Antagonism of RAGE suppresses peripheral nerve regeneration. FASEB J. 2004;18:1812-1817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 85] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 101. | Rong LL, Yan SF, Wendt T, Hans D, Pachydaki S, Bucciarelli LG, Adebayo A, Qu W, Lu Y, Kostov K. RAGE modulates peripheral nerve regeneration via recruitment of both inflammatory and axonal outgrowth pathways. FASEB J. 2004;18:1818-1825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 105] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 102. | Sbai O, Devi TS, Melone MA, Feron F, Khrestchatisky M, Singh LP, Perrone L. RAGE-TXNIP axis is required for S100B-promoted Schwann cell migration, fibronectin expression and cytokine secretion. J Cell Sci. 2010;123:4332-4339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 103. | Meneghini V, Francese MT, Carraro L, Grilli M. A novel role for the Receptor for Advanced Glycation End-products in neural progenitor cells derived from adult SubVentricular Zone. Mol Cell Neurosci. 2010;45:139-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 104. | Riuzzi F, Sorci G, Beccafico S, Donato R. S100B engages RAGE or bFGF/FGFR1 in myoblasts depending on its own concentration and myoblast density. Implications for muscle regeneration. PLoS One. 2012;7:e28700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 105. | Van Eldik LJ, Wainwright MS. The Janus face of glial-derived S100B: beneficial and detrimental functions in the brain. Restor Neurol Neurosci. 2003;21:97-108. [PubMed] |
| 106. | Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of s100 proteins. Curr Mol Med. 2013;13:24-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 107. | Tsoporis JN, Izhar S, Leong-Poi H, Desjardins JF, Huttunen HJ, Parker TG. S100B interaction with the receptor for advanced glycation end products (RAGE): a novel receptor-mediated mechanism for myocyte apoptosis postinfarction. Circ Res. 2010;106:93-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 97] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 108. | Tsoporis JN, Izhar S, Proteau G, Slaughter G, Parker TG. S100B-RAGE dependent VEGF secretion by cardiac myocytes induces myofibroblast proliferation. J Mol Cell Cardiol. 2012;52:464-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 109. | Mohammadzadeh F, Desjardins JF, Tsoporis JN, Proteau G, Leong-Poi H, Parker TG. S100B: Role in cardiac remodeling and function following myocardial infarction in diabetes. Life Sci. 2013;92:639-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 110. | Chan JK, Roth J, Oppenheim JJ, Tracey KJ, Vogl T, Feldmann M, Horwood N, Nanchahal J. Alarmins: awaiting a clinical response. J Clin Invest. 2012;122:2711-2719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 382] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 111. | Sorci G, Giovannini G, Riuzzi F, Bonifazi P, Zelante T, Zagarella S, Bistoni F, Donato R, Romani L. The danger signal S100B integrates pathogen- and danger-sensing pathways to restrain inflammation. PLoS Pathog. 2011;7:e1001315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 79] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 112. | Cunha C, Giovannini G, Pierini A, Bell AS, Sorci G, Riuzzi F, Donato R, Rodrigues F, Velardi A, Aversa F. Genetically-determined hyperfunction of the S100B/RAGE axis is a risk factor for aspergillosis in stem cell transplant recipients. PLoS One. 2011;6:e27962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 113. | Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1392] [Cited by in RCA: 1484] [Article Influence: 57.1] [Reference Citation Analysis (0)] |