Published online Jul 26, 2012. doi: 10.4331/wjbc.v3.i7.159
Revised: February 10, 2012
Accepted: February 17, 2012
Published online: July 26, 2012
AIM: To report that Lpcat1 plays an important role in regulating lipopolysaccharide (LPS) inducible gene transcription.
METHODS: Gene expression in Murine Lung Epithelial MLE-12 cells with LPS treatment or Haemophilus influenza and Escherichia coli infection was analyzed by employing quantitative Reverse Transcription Polymerase Chain Reaction techniques. Nucleofection was used to deliver Lenti-viral system to express or knock down Lpcat1 in MLE cells. Subcellular protein fractionation and Western blotting were utilized to study Lpcat1 nuclear relocation.
RESULTS: Lpcat1 translocates into the nucleus from the cytoplasm in murine lung epithelia (MLE) after LPS treatment. Haemophilus influenza and Escherichia coli, two LPS-containing pathogens that cause pneumonia, triggered Lpcat1 nuclear translocation from the cytoplasm. The LPS inducible gene expression profile was determined by quantitative reverse transcription polymerase chain reaction after silencing Lpcat1 or overexpression of the enzyme in MLE cells. We detected that 17 out of a total 38 screened genes were upregulated, 14 genes were suppressed, and 7 genes remained unchanged in LPS treated cells in comparison to controls. Knockdown of Lpcat1 by shRNA dramatically changed the spectrum of the LPS inducible gene transcription, as 18 genes out of 38 genes were upregulated, of which 20 genes were suppressed or unchanged. Notably, in Lpcat1 overexpressed cells, 25 genes out of 38 genes were reduced in the setting of LPS treatment.
CONCLUSION: These observations suggest that Lpcat1 relocates into the nucleus in response to bacterial infection to differentially regulate gene transcriptional repression.
- Citation: Ellis B, Kaercher L, Snavely C, Zhao Y, Zou C. Lipopolysaccharide triggers nuclear import of Lpcat1 to regulate inducible gene expression in lung epithelia. World J Biol Chem 2012; 3(7): 159-166
- URL: https://www.wjgnet.com/1949-8454/full/v3/i7/159.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v3.i7.159
Lpcat1 was first cloned as an active lung surfactant phospholipid synthetic enzyme from lung type II epithelial cells[1,2]. This gene product expresses ubiquitously in almost all types of tissues, but is particularly high in surfactant-producing lung type II epithelial cells. The acyltransferase adds a palmitate group to the glycerol backbone of phospholipids with features of low selectivity to its acceptor substrates. Targeted disruption of this gene in mice increases newborn mortality[3]. Lpcat1 expresses at aberrantly high levels in colon tumor cells[4]. These observations suggest that Lpcat1 might possess a role distinct to its activity within the surfactant synthetic program. Lpcat1 is characterized as an ER protein with one trans-membrane domain and a carboxyl terminal ER localizing signal. Hence, Lpcat1 mainly localizes in the cytoplasm. A recent study showed that Lpcat1 co-localized with lipid droplets in the cytosol and might play a role in neutral lipid metabolism[5]. We have previously reported that Lpcat1 translocates into the nucleus after calcium (Ca2+) stimulation; nuclear Lpcat1 subsequently catalyzes histone palmitoylation through direct binding to histone protein. In these studies, nuclear Lpcat1 mediated histone palmitoylation results in increased total RNA synthesis suggesting that histone palmitoylation could be a novel epigenetic mark that controls pro-inflammatory gene transcription[6]. However, the nature of the genes that are regulated by Lpcat1-induced histone palmitoylation are unknown.
The host response to infectious lipopolysaccharide (LPS)-containing bacterial pathogens involves an initial modification of existed gene products[7]. In-turn, some of the modified effectors shift into the nucleus to modify histone proteins. Histone protein modification results in opening or closing of the chromatin structure, subsequently switching on or off gene transcriptional expression. Well characterized post-translational modifications of histone proteins include acetylation[8], methylation[9], phosphorylation[10], ubiquitination[11], and palmitoylation[6]. Of these, acetylation is believed to be one of the major known epigenetic pathways involved in LPS inducible inflammatory gene expression[12]. A recent study reported that about 139 genes are LPS inducible within 1 h of LPS exposure in macrophages[13]. Inhibition of acetylation by a pan-acetylation inhibitor suppressed 38 of the 139 LPS inducible inflammatory genes. Acetylation affects approximately one quarter of the total LPS inducible inflammatory genes indicating that the majority of the inflammatory-inducible genes are yet to be determined by possibly other post-translational histone modifications. It is also likely that gene transcriptional expression in response to histone modification is highly complex, controlled by dual- or multi-histone modification enzymes.
Pneumonia secondary to bacterial pathogens remains a major health concern and involves repetitive exposure of alveolar epithelial cells to LPS that results in pro-inflammatory gene activation. The inflammatory gene expression profile is vital for host immune responses, but exaggerated expression of these gene products is frequently deleterious to the host. In this study, we analyzed the expression of a subset of genes that are altered by modulating nuclear availability of the enzyme Lpcat1 during LPS exposure. We found that Lpcat1 shifts into the nucleus in response to both LPS treatment or gram-negative bacterial infection in lung epithelia. While knockdown of Lpcat1 modified somewhat LPS-inducible gene expression, overexpression of Lpcat1 suppressed approximately two thirds of the LPS inducible gene profile.
The murine lung epithelial cell line MLE-12 cells were maintained HITES medium complemented with 10% fetal bovine serum in a 37 °C incubator in the presence of 5% CO2. Human embryonic kidney (HEK) 293FT cells were cultured in modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 units/mL penicillin/streptomycin, and 2 mmol/L L-glutamine. Pneumonia causative pathogens Haemophilus influenza and Escherichia coli were from ATCC and inoculated into TSB (Tryptic Soy Broth) plates overnight before use. Bacteria were cultured in 20 mL of TSB broth up to a density of A = 0.6; MLE cells were infected with the bacteria at the indicated inoculums. Anti-Lamin A/C antibody and anti-histone H4 antibody were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). lipopolysaccharide (LPS) was purchased from from Sigma-Aldrich (St. Louis, MO). All other materials used in the study were the highest grades commercially available.
Cells in their exponential growth stage were treated with 10 μg/mL LPS for 2 h or the bacteria at the concentrations as indicated. Extraction of nuclear and cytosolic fraction proteins was performed as previously described[6]. Briefly, harvested cells were suspended in 1 mL of cytoplasmic membrane lysis buffer (10 mmol/L Hepes, pH 8.0, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 1 mmol/L DTT, and 1:1000 protease inhibitor mixture). The cell suspension was centrifuged at 2000 rpm for 10 min, and the supernatant was collected as the cytosolic fraction and condensed by Amicon Ultra filters. The pellet was lysed with 150 μL of nuclear envelope lysis buffer (20 mmol/L Hepes, pH 8.0, 1.5 mmol/L MgCl2, 420 mmol/L NaCl, 1mmol/L DTT, 0.2 mmol/L EDTA, 25% (v/v) glycerol, and 1:1000 protease inhibitor mixture) and cleared at 13 000 rpm for 10 min; the supernatant was collected as the nuclear fraction. The fractions were subjected to immunoblot analysis.
Nucleofection was carried out as previously described[14]. Briefly, one million log phase MLE cells in 0.1 mL of nucleofection buffer [1 × phosphate buffered solution (PBS), 20 mmol/L Hepes, pH 8.0] were mixed with 4 μg of lenti-viral vector, lenti-viral Lpcat1-shRNA, and lenti-viral Lpcat1 DNA respectively in nucleofection cuvette. Electroporation was performed with pre-set program T-013 in NucleofectionTM II system (Amaxa Biosystems, Gaithersburg, MD), and the cells were cultured in 2 mL of complete HITES medium for 48 h.
Immunoblotting was performed as previously described[6,14]. During exponential growth, cells were treated with a variety of reagents and pathogens for 2 h, and the cells were lysed with lysis buffer [0.3% Triton X-100 (v/v) in PBS and 1:1000 proteinase inhibitor mixture], or subjected to total RNA isolation or nuclear or cytosolic protein fractionation. The fractionated proteins or cell lysate were separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the proteins were transferred to nitrocellulose membranes. The membrane was blocked with 20% milk and incubated with primary and secondary antibodies respectively. The immunoblotting was visualized with enhanced chemiluminescence and the images were acquired under Kodak F in vivo carestream imaging system.
Total cellular RNA was isolated with Tri reagent (Invitrogen) following the directions of the manufacturer. Briefly, cells were washed twice with cold PBS and lysed with 0.6 mL of cold Tri reagents. The cell lysates were mixed thoroughly with 0.2 mL of chloroform, spun down and the upper fraction was collected. The fraction was mixed with same volume of isopropanol, and centrifuged. The pellet was washed with 0.8 mL of 70% ethanol, and dissolved in RNase free water. The dissolved solution was digested with DNase and stored at -80 °C freezer.
cDNA was synthesized from isolated total RNA with an iScript cDNA synthesis kit (Bio-Rad) following the directions of the manufacturer. A variety of quantitative-polymerase chain reaction (Q-PCR) primers were designed based on the NCBI mouse mRNA and genome DNA sequence database. DNA fragments of the PCR product were about 100 bp in length, the forward and reverse primers were primarily designed from different exons respectively to decrease the background noise from genomic DNA contamination. The sequences of the primers were listed in Table 1. Q-PCR reagents Ssofast evaGreen suppermix (were from Bio-Rad and the PCR was conducted following the directions of the manufacturer. Briefly, each reaction contains 2 μL of cDNA (from the reverse transcription of 0.1 μg of total RNA), 0.8 μL of primers (contains 100nM of forward and reverse primers), 2.2 μL of H2O, and 5 μL of Ssofast evaGreen suppermix. Q-PCR was conducted with the Bio-Rad C1000TM thermo cycler at a two-step program (95 °C for 3 s, and 55 °C for 3 s for 35 cycles). The quantitative reverse transcription-polymerase chain reaction (QRT-PCR) data were analyzed by the Q-PCR analyses software verion 3.4 from SABiosciences. The relative gene expression level (copy numbers) was obtained by the formula “2^-C.1000 000” (C here donates to the cycle number of the gene in Q-PCR). The gene expression fold change was obtained by dividing the treated group with the base expression level of the related gene in untreated MLE cells. In comparison to control, expression levels of genes that increased more than 2 folds represents genes that are defined as upregulated, whereas expression levels that decreased by 50% (or -2) represents genes that are suppressed.
| Gene symbol | Forward primer | Reverse primer |
| L19 | AGAGCTTGGAGTGCACAGGT | GGGACAGTCCAAACTGGGTA |
| TGF-b1 | GACTCTCCACCTGCAAGACC | GACTGGCGAGCCTTAGTTTG |
| IL-13Ra1 | CACAGAAGTTCAGCCACCTG | AATTTGGACTGGCTCCTTCA |
| ILRN | GTTGGAAGGCAGTGGAAGAC | GCATCTTGCAGGGTCTTTTC |
| IL4 | CCAAGGTGCTTCGCATATTT | ATCGAAAAGCCCGAAAGAGT |
| SPHK1 | GCTGTGAGGCTGGTGTTATG | ATATGCTTGCCCTTCTGCAT |
| EDG7 | ATTGCCTCTGCAACATCTCG | ATGAAGAAGGCCAGGAGGTT |
| IL-13Ra2 | TTGGTCTGCTCTTGGAAACC | CAGCACACTGTAAGGCATGA |
| Autotaxin | GAAACAGCACCTTCCCAAAC | AAGGTTTCCTTGCAACATGC |
| LPA4 | ACTGCGTTCCTCACCAACAT | CGATCGGAAGGGATAGACAA |
| sST2 | CATGGCATGATAAGGCACAC | GATGCAGTGCACAGCTGATT |
| LPCAT1 | CGACTGAGCGCCCTGCAGAA | AAGGGCCAGGCCAGCAGCAT |
| ST2L | GGAACGATGGCAAGCTCTAC | GGCAGAGTGTGGTGAACAAA |
| EDG1 | CTGTTAGATGTGGGCTGCAA | ATGATGGGGTTGGTACCTGA |
| MMP9 | CTTCGAGGGACGCTCCTATT | CAAATTTGCCGTCCTTATCG |
| IL-1β | CAGGCAGGCAGTATCACTCA | AGGTGCTCATGTCCTCATCC |
| IL33 | CCTTCTCGCTGATTTCCAAG | GCTGAACAGAACGTGTGCAT |
| IL-13 | CAGCAGCTTGAGCACATTTC | ATAGGCAGCAAACCATGTCC |
| EDG8 | ACACCAAATGCCCAGCTTAC | TGGAGCACTGTGCAAAAGTC |
| EDG2 | TCAACCTGGTGACCTTTGTG | GGTCCAGAACTATGCCGAGA |
| IL4R | TCACAGAGCAGCCTTCACAC | TTGTCTGCAAGGACAAGTGG |
| LPA5 | GCTCCAGTGCCCTGACTATC | CAGAGCGTTGAGAGGGAGAC |
| CCTa | GGAAACTCCGGGTAGTGGGTGC | TTCCCTAGCTGTCACTGGCCCA |
| COX2 | CCCCCACAGTCAAAGACACT | GGCACCAGACCAAAGACTTC |
| CTTN | AGCAGAGATGGGGTGCTAAA | TTGAGCGTCTGGTGTTCTTG |
| SGPP2 | CATCTCCTTCACCCTCCTCA | CCAGCGTAGAGAACACCACA |
| SGPL1 | AACTCTGCCTGCTCAGGGTA | CTCCTGAGGCTTTCCCTTCT |
| IL-6 | CCGGAGAGGAGACTTCACAG | TCCACGATTTCCCAGAGAAC |
| MET | CAGCCATCCCAATGTTCTCT | AATTTCGCAGATCTCCATGC |
| EDG3 | TGCCTTATGAACCCTGGAAG | AAGAGCAAATGCCATCAGGT |
| EDG6 | CTCCAAGGGCTATGTGCTCT | ATTGGCTCGGACCACTCTAA |
| CADHERIN | GCACTCTTCTCCTGGTCCTG | TATGAGGCTGTGGGTTCCTC |
| SGPP1 | GAGCAACTTGCCCGCTCTACT | AAGGGGTCGAGATTCCAGAT |
| SPHK2 | ACTGCTCGCTTCTTCTCTGC | GCCACTGACAGGAAGGAAAA |
All data were statistically analyzed and presented as means + SEM from 3 independent experiments.
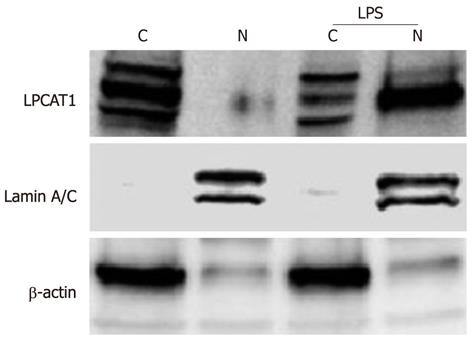
We have previously demonstrated that the cytosolic phospholipid synthetic enzyme Lpcat1 shifts into the nucleus and catalyzes histone protein palmitoylation after Ca2+ stimulation[6]. LPS-containing bacteria trigger Ca2+ cytosolic oscillations in alveolar epithelial cells and releases LPS to the cell surface, that in turn regulates expression of genes. To evaluate whether LPS induced gene expression might regulate Lpcat1 subcellular localization, we treated MLE cells with LPS (10 μg/mL) for 2 h and subjected the cells to nuclear and cytosolic protein extraction followed by immunoblotting. The cytosolic structural protein components β-actin and the nuclear structural protein lamin A/C were used as markers. Lpcat1 immunoblotting showed that Lpcat1 was mainly located in the cytosol of untreated (control) cells, existing as multiple species perhaps representing phosphorylation-modified variants. This is consistent with prior data[6], indicating that Lpcat1 itself is the subject of posttranslational modification. The majority of cytosolic Lpcat1 relocated into the nucleus after LPS treatment, with smaller amounts of Lpcat1 protein mass remaining in the cytosolic fraction (Figure 1). Among these modifications, only one form could enter into the nucleus. The data suggest that LPS containing intact bacteria might also elicit nuclear translocation of this surfactant enzyme.
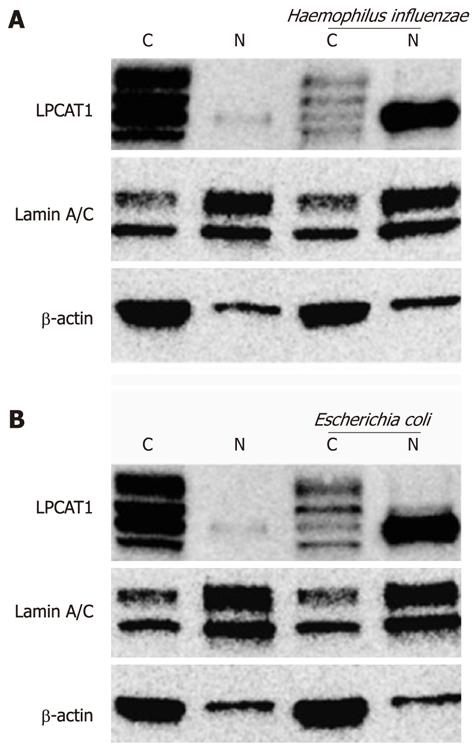
Haemophilus influenzae and Escherichia coli are gram-negative bacterial pathogens that release LPS eliciting biological responses. Immunoblotting analyses of the subcellular fractions revealed that both bacterial pathogens effectively caused Lpcat1 to shift from the cytosolic compartment to the nucleus (Figure 2). Similar to effects of LPS, only one form of Lpcat1 shifted into the nucleus, suggesting that Lpcat1 may be modified before entering into the nucleus after infection or LPS exposure.
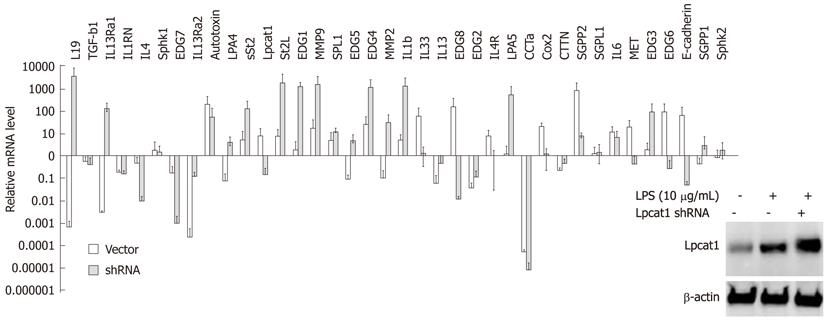
Since Lpcat1 shifted into the nucleus upon LPS treatment or bacterial infection, and nuclear Lpcat1 exhibits epigenetic activity, we hypothesized that Lpcat1 might participate in the regulation gene expression induced by LPS. To test this hypothesis, we treated MLE cells with LPS (10 μg/mL) for 2 h and isolated total cellular RNA for QRT-PCR analysis. The genes selected for QRT-PCR are diverse and include inflammatory cytokines, lipid signaling receptors, proteinases, and growth factors. β-actin was used as a systematic control to justify the variation among the different Q-PCR reactions. Lpcat1 itself was slightly upregulated but without detectable change in protein levels (Figure 3, inset). We found that 15 genes out of a total 38 screened genes were upregulated, 14 genes were suppressed, with 10 genes that remained unchanged in LPS treated cells in comparison to controls (Figure 3). To understand the potential regulatory role of Lpcat1 in LPS induced gene expression, we knocked down the LPCAT1 gene by hair-pin shRNA plasmid transfection. Lpcat1 was successfully knocked down by 48 h as shown by Lpcat1 immunoblotting (Figure 3). After silencing LPCAT1, cells were then challenged with LPS (10 μg/mL) for 2 h. QRT-PCR results showed that knockdown of LPCAT1 by shRNA changed the spectrum of LPS inducible gene expression patterns. As compared to the infected controls, 15 genes out of 38 genes were upregulated, of which 11 genes were LPS upregulated genes, 9 genes were LPS suppressed or unchanged genes compared to control. Notably, 14 LPS upregulated genes were downregulated in Lpcat1 knockdown cells, and 7 genes were downregulated in both groups. These results indicate that Lpcat1 may regulate LPS inducible gene expression.
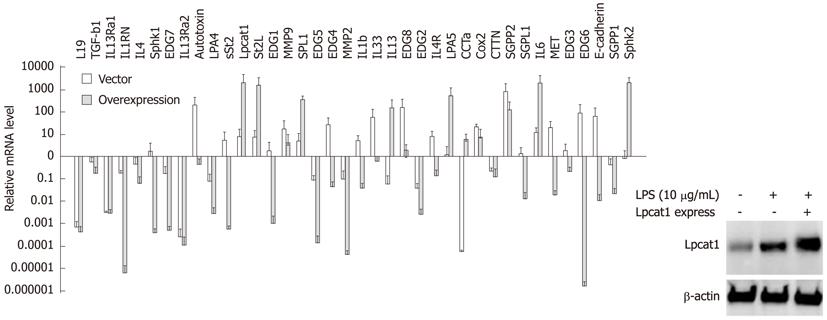
We next overexpressed LPCAT1 in cells by nucleofection for 24 h, and then challenged the cells with LPS (10 μg/mL) for 2 h. Overexpression of Lpcat1 was confirmed by Lpcat1 immunoblotting (Figure 4). Importantly, LPS inducible gene expression was suppressed after Lpcat1 overexpression, as 26 genes of 38 genes analyzed were reduced, with the exception of 10 genes that were upregulated, 3 genes remained unchanged. Within the 26 suppressed genes, 7 previously identified LPS-upregulated genes were downregulated, and 19 genes were LPS downregulated or unchanged genes. The majority of the upregulated genes in the Lpcat1 knockdown group were suppressed. Interestingly, a few genes were similarly regulated both in Lpcat1 knockdown and overexpressed cells. Of them, IL-13Ra2 and EDG2, expression were suppressed in both Lpcat1 knockdown and overexpressed cells, and another gene ST2L, was upregulated in both conditions. The fold regulation of each gene was listed in Table 2. Overall, these observations suggest that nuclear Lpcat1 may regulate early gene transcription in response to bacterial infection.
| Gene symbol | Vector | Lpcat1 shRNA | Lpcat1 overexpression |
| IL-1β | 2.29 | 119.70 | -29.79 |
| IL1RN | -1344.87 | 8.22 | -8579.43 |
| IL4 | -3029.24 | -2.65 | 1.86 |
| IL4R | 5.79 | 1.17 | -7.18 |
| IL6 | 7.38 | 10.88 | 998.30 |
| IL13 | -19.56 | -4.02 | 1237.60 |
| IL13Ra1 | -171.65 | 65.34 | -544.96 |
| IL13Ra2 | -2845.00 | 53.32 | 207.94 |
| IL33 | 20.48 | 1.38 | -1.46 |
| Autotaxin | 17.47 | 4.17 | -3.50 |
| sST2 | -4.11 | 1667.34 | 21.91 |
| ST2L | 2.42 | 138.14 | 781.44 |
| TGF-b1 | -1.07 | -6.35 | -14.69 |
| COX2 | 13.33 | 2.40 | 8.0 |
| MMP2 | -13.55 | 443.67 | -565.48 |
| MMP9 | 7.00 | 124.79 | -1.99 |
| EDG1 | -93.05 | 541.37 | -1231.90 |
| EDG2 | -6.85 | -1.26 | -48.50 |
| EDG3 | 2.07 | 30.06 | -3.70 |
| EDG4 | -1.27 | 83.48 | -36.25 |
| EDG5 | -1.59 | 50.21 | -198.09 |
| EDG6 | -70.20 | -1.56 | -343.30 |
| EDG7 | -180.60 | -1978.24 | 2189.93 |
| EDG8 | -441.62 | -2.52 | 19.79 |
| CCT | -17.71 | -227.54 | 6397.68 |
| CCTN | -1.49 | 1.10 | -22.63 |
| SPL1 | 5.62 | 7.11 | 186.54 |
| SGPP1 | -2.80 | -2.30 | -64.73 |
| SGPP2 | -26.85 | 67.18 | 305.14 |
| SGPL1 | -1.56 | -1.17 | -65.04 |
| SPHK1 | -8.28 | 22.37 | -133.74 |
| SPHK2 | 1.59 | -2.63 | 1223.39 |
| LPA4 | -21.96 | 15.63 | -110.15 |
| LPA5 | 1.15 | 275.01 | 3590.58 |
| MET | 1.72 | -8.00 | -38.49 |
| E-CADHERIN | -2.62 | -18.77 | -155.41 |
| L19 | - 667.15 | 6.09 | -2752.77 |
| LPCAT1 | 3.04 | -5.24 | 132.21 |
In this study, we found that Lpcat1 relocates into the nucleus in lung epithelia in response to LPS-containing bacterial pathogens that in turn differentially regulates the expression several genes involved in pulmonary homeostasis. We reported previously that elevated exogenous Ca2+ triggers Lpcat1 nuclear translocation, an effect that leads to histone modification that modulates global gene transcriptional activity[6]. However, the repertoire of specific genes that might be modulated by Lpcat1 was not investigated. In this study, we used a biologically relevant in vitro model system to assess the gene profiles that are modulated by relevant pulmonary bacterial pathogens and a major cell wall component that elicits alveolar injury and inflammation. Similar to Ca2+, LPS and bacterial pathogens were sufficient to trigger Lpcat1 nuclear entry in cells. Similar nuclear translocation behavior occurs with another surfactant synthetic enzyme, cholinephosphate cytidyltransferase, after stimulation with bacteria, high concentrations of Ca2+, or extracellular hypertonicity[15-17]. As gram-negative pathogens and LPS increase cytosolic Ca2+ levels in lung fluid and cytoplasm of epithelia, these factors may synergistically mediate nuclear translocation of such lipogenic enzymes. The biological relevance of our findings is that in gram negative bacterial infection caused sepsis or pneumonia, these pathogens may trigger modification of histones by nuclear import of Lpcat1, which could lead to attenuated levels of key genes that maintain lung balance or partake in lung host defense or repair.
There is a discrepancy in the immunoblotting bands of Lpcat1 in subcellular fractions and in whole cell lysates. We previously reported that Lpcat1 was susceptible to a variety of posttranslational modifications[14]. These modifications are, but not limited to, phosphorylation (at residues of S178-182) or ubiquitination (at K221). Therefore, it is possible that a range of modified Lpcat1 coexist in the settings of LPS treatment and bacterial infection. Furthermore, subcellular fractionation procedure takes several hours. Particularly, the subcellular cytosolic fraction has been concentrated and the cytosolic proteins were been enriched in Figures 1 and 2, so we see more bands in cytosol fractionations as compared to the one band in regular total cell lysates. In the bacterial infection experiments, the bacterial infected cells and control cells were maintained in bacterial incubator for 4 h without the supplement of 5% CO2 and 85% humidity. All the variation in experimental conditions may contribute to the discrepancy in Figures 1, 2, 3 and 4.
Both LPS or bacterial infection triggers only one form of Lpcat1 enters into the nucleus, a raised question is how this Lpcat1isoform is transported. We analyzed the sequence of the Lpcat1 and found that the NH-terminus contains a potential bipartite nuclear localization signal (NLS) peptide that is for nuclear translocation, deletion of the first 100 amino acids in its NH-terminus diminished its nuclear shifting with a relatively retained intact enzymatic function (unpublished data). This region also contains a Calmodulin binding domain and a possible serine phosphorylation site within the domain. We speculate that the Lpcat1 molecule should firstly bind to calcium and Calmodulin. Then, a kinase (potentially Calmodulin kinase I or Calmodulin kinase II) will phosphorylate the serine residue by bridge binding to calmodulin. The phosphorylated serine residue in the NLS signal peptide makes the NLS signal ready to associate with Importin and the Lpcat1 molecule migrates into the nucleus. This line of research works are undergoing.
The pattern of genes affected by Lpcat1 silencing or overexpression was complex and includes genes that encode expression of proteins involved in inflammation, signaling, biosynthetic proteins, matrix proteases, and membrane receptors. By gain of function and loss of function approaches, we found that Lpcat1 exhibits a tendency to be a transcriptional suppresser in the small population of genes we screened. This was evidenced by our findings that overexpression of Lpcat1 suppressed two third (26/38) of the genes screened. In contrast, knockdown of Lpcat1 by shRNA upregulated the expression levels in about half (20/38) of the genes analyzed. To obtain a more comprehensive analysis of genome-wide Lpcat1 regulated activity, further studies using microarray or ChIP-sequencing is necessary. One caveat in data interpretation of our profiles is that Lpcat1 itself appears upregulated by LPS stimulation. Nevertheless, as we confirmed that Lpcat1 protein decreased after Lpcat1 knockdown and increased after Lpcat1 overexpression indicating that these maneuvers were successful. Overall, our findings may open a new field of epigenetic study by a lipogenic enzyme that might shed light on the immunobiology of pulmonary infectious disease.
Post-translational modification of histones has emerged as a major regulatory mechanism that controls gene transcriptional activity by changing chromatin architecture. Histones package and organize DNA into structural subunits (nucleosomes) and regulate DNA accessibility, and are subject to extensive covalent post-translational modifications (methylation, acetylation, phosphorylation, etc.) These modifications, can in turn, selectively control gene expression that impact diverse processes such as development, carcinogenesis, and immunity. In the study of lung surfactant synthetic enzyme Lpcat1, the authors serendipitously found that this cytosolic resided enzyme migrates into the nucleus and O-palmitoylates histone protein by direct binding under certain conditions. The physio-pathological significance of this novel epigenetic code is illusive.
Histone modification has been the hottest research topic for the last two decades; it impacts significantly in both basic biological sciences and clinical applications. Lipidation of histone is a novel paradigm of these modifications. Fatty acid may not only be an energy source, a membrane component but also rigorously participate into gene expression regulation. The authors revealed histone O-palmitoylation and discovered the first enzyme Lpcat1 that modifies histone by O-palmitoylation. O-palmitoylation of histone results in gene expression. This study is the first attempt to linearize the Lpcat1 epigenetic function by using an acute lung injury model.
This study for the first time describes that a phospholipid enzyme Lpcat1 shifts into the nucleus under LPS treatment and bacterial infection. Nuclear Lpcat1 rigorously regulates inflammatory gene expression. Overexpression of Lpcat1 appears to repress the gene transcription while knockdown of Lpcat1 unleashes repression function which leads into robust gene expression.
This study could be expanded to other physio-pathological process such as development, immunity, cancer, etc.
Epigenetics is a rapidly emerging process that refers to non-DNA sequence changes that can significantly affect the clinical phenotype or expression of genes. One of the most important mechanisms of epigenetics is the modification of histone proteins. O-palmitoylation is a covalent attachment of a saturated free long fatty acid palmitoyl group (C16:0) to a phospholipid or a protein substrate via an oxy easter linkage.
This paper studied the role of Lpcat1 in regulating inducible gene expression in lung epithelia. The major finding is that Lpcat1 relocated into the nucleus in response to LPS. And LPS-containing bacterial pathogens has the similar effect on the expression of Lpcat1 in regulating the expression of several genes involved in pulmonary homeostasis, which provides some useful information for understanding this gene. However, there are some questions to be addressed.
Peer reviewer: Xiang-An Li, PhD, Assistant Professor, Department of Pediatrics, University of Kentucky, BBSRB B365, 741 South Limestone Street, Lexington, KY 40536, United States
S- Editor Cheng JX L- Editor A E- Editor Zhang DN
| 1. | Nakanishi H, Shindou H, Hishikawa D, Harayama T, Ogasawara R, Suwabe A, Taguchi R, Shimizu T. Cloning and characterization of mouse lung-type acyl-CoA: lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J Biol Chem. 2006;281:20140-20147. [PubMed] |
| 2. | Soupene E, Fyrst H, Kuypers FA. Mammalian acyl-CoA: lysophosphatidylcholine acyltransferase enzymes. Proc Natl Acad Sci USA. 2008;105:88-93. [PubMed] |
| 3. | Bridges JP, Ikegami M, Brilli LL, Chen X, Mason RJ, Shannon JM. LPCAT1 regulates surfactant phospholipid synthesis and is required for transitioning to air breathing in mice. J Clin Invest. 2010;120:1736-1748. [PubMed] |
| 4. | Mansilla F, da Costa KA, Wang S, Kruhøffer M, Lewin TM, Orntoft TF, Coleman RA, Birkenkamp-Demtröder K. Lysophosphatidylcholine acyltransferase 1 (LPCAT1) overexpression in human colorectal cancer. J Mol Med (. Berl). 2009;87:85-97. [PubMed] |
| 5. | Moessinger C, Kuerschner L, Spandl J, Shevchenko A, Thiele C. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J Biol Chem. 2011;286:21330-21339. [PubMed] |
| 6. | Zou C, Ellis BM, Smith RM, Chen BB, Zhao Y, Mallampalli RK. Acyl-CoA: lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J Biol Chem. 2011;286:28019-28025. [PubMed] |
| 7. | Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833-844. [PubMed] |
| 8. | Kawasaki H, Schiltz L, Chiu R, Itakura K, Taira K, Nakatani Y, Yokoyama KK. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature. 2000;405:195-200. [PubMed] |
| 9. | Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286-3295. [PubMed] |
| 10. | Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279-291. [PubMed] |
| 11. | Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873-878. [PubMed] |
| 12. | Cosío BG, Mann B, Ito K, Jazrawi E, Barnes PJ, Chung KF, Adcock IM. Histone acetylase and deacetylase activity in alveolar macrophages and blood mononocytes in asthma. Am J Respir Crit Care Med. 2004;170:141-147. [PubMed] |
| 13. | Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119-1123. [PubMed] |
| 14. | Zou C, Butler PL, Coon TA, Smith RM, Hammen G, Zhao Y, Chen BB, Mallampalli RK. LPS impairs phospholipid synthesis by triggering beta-transducin repeat-containing protein (beta-TrCP)-mediated polyubiquitination and degradation of the surfactant enzyme acyl-CoA: lysophosphatidylcholine acyltransferase I (LPCAT1). J Biol Chem. 2011;286:2719-2727. [PubMed] |
| 15. | Agassandian M, Chen BB, Schuster CC, Houtman JC, Mallampalli RK. 14-3-3zeta escorts CCTalpha for calcium-activated nuclear import in lung epithelia. FASEB J. 2010;24:1271-1283. [PubMed] |
| 16. | Chen BB, Coon TA, Glasser JR, Mallampalli RK. Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol Cell Biol. 2011;31:1905-1920. [PubMed] |
| 17. | Favale NO, Fernández-Tome MC, Pescio LG, Sterin-Speziale NB. The rate-limiting enzyme in phosphatidylcholine synthesis is associated with nuclear speckles under stress conditions. Biochim Biophys Acta. 2010;1801:1184-1194. [PubMed] |