Published online May 26, 2010. doi: 10.4331/wjbc.v1.i5.151
Revised: May 4, 2010
Accepted: May 17, 2010
Published online: May 26, 2010
The fruit fly Drosophila melanogaster has been successfully used to study numerous biological processes including immune response. Flies are naturally infected with more than twenty RNA viruses making it a valid model organism to study host-pathogen interactions during viral infections. The Drosophila antiviral immunity includes RNA interference, activation of the JAK/STAT and other signaling cascades and other mechanisms such as autophagy and interactions with other microorganisms. Here we review Drosophila as an immunological research model as well as recent advances in the field of Drosophila antiviral immunity.
-
Citation: Wang JH, Valanne S, Rämet M.
Drosophila as a model for antiviral immunity. World J Biol Chem 2010; 1(5): 151-159 - URL: https://www.wjgnet.com/1949-8454/full/v1/i5/151.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v1.i5.151
The fruitfly Drosophila melanogaster has been an important animal model in laboratory research since Thomas Hunt Morgan started using it to study chromosomes at the beginning of the 20th century. Drosophila is easy and inexpensive to rear in the laboratory, produces numerous progeny and has a short (about 10 d) generation time. As an invertebrate, Drosophila is considered an ethically acceptable animal model. Drosophila is an extremely useful tool for studying various biological processes, since many Drosophila external features such as compound eyes, wing veins and bristles can be affected by mutations and are easily visualized with a microscope[1].
The Drosophila genome is compact with low redundancy; single mutants are likely to reveal phenotypes of interest in contrast to mammals, whose genomes are more redundant and single mutation often does not produce a clear phenotype. Moreover, an analysis of the Drosophila euchromatin sequence revealed a high degree of similarity between flies and mammals[2-4] indicating a high degree of conservation of a very large number of biological processes. Moreover, 75% of known human genetic disease genes have homologues in the fly[5]. An online database named Homophila[5,6], which allows searching for human disease gene homologues in flies and vice versa, is available at http://superfly.ucsd.edu/homophila/.
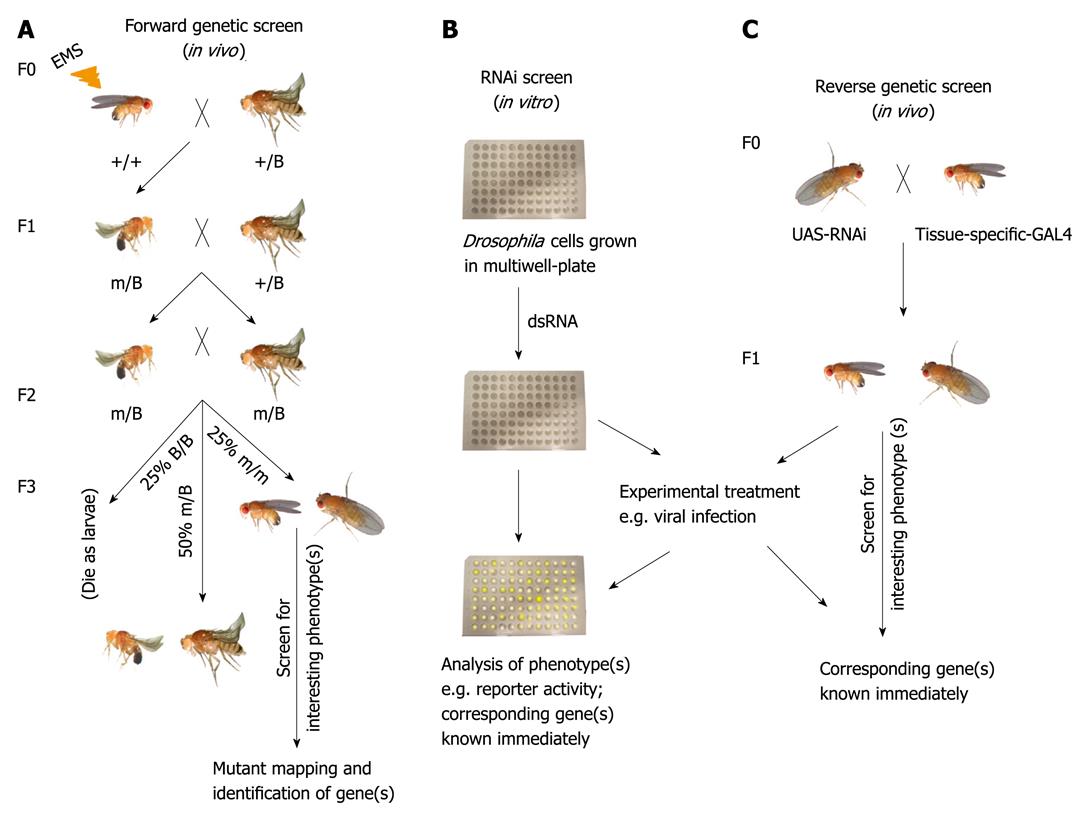
The major advantage of flies is the simplicity and scale for genetic analysis, which has been undertaken for a century with a large number of sophisticated genetic tools[7]. One of the most important tools in Drosophila, successfully used for decades for new gene discovery, is the possibility of carrying out genetic screens for mutations that affect chosen biological processes. The traditional forward genetic screen (Figure 1A) involves mutagenesis, which is most commonly carried out with ethyl methane sulphonate or X-rays[1]. The phenotype of interest is screened in the mutated population followed by mapping and identifying the gene(s) causing the phenotype[1] (Figure 1A). In addition, mutations caused by P-element insertions[8] may also be screened; the commonest approach is to screen existing P-element insertion collections from the Berkeley Genome project[9].
In addition to traditional screens, many molecular genetic techniques have been developed in the last few decades that allow e.g. germ-line transformation[10], homologous recombination[11], and RNA interference (RNAi)[12,13]. The discovery of RNAi in Drosophila[14], and the completion of the Drosophila genome sequence[2] boosted a large number of large-scale in vitro RNAi screenings[15-20] (Figure 1B) including microarray-based screens[21,22] in Drosophila macrophage-like S2 cells[23,24]. Recently, libraries of transgenic Drosophila based on the binary GAL4/UAS system[25], have been constructed, making large-scale in vivo RNAi screening studies feasible e.g.[26-28] (Figure 1C). The first generation of RNAi fly lines included the genome-wide VDRC lines[29] and lines from the NIG-FLY (National Institutes of Genetics, Japan). The second generation of RNAi lines from the TRiP (Transgenic RNAi project) collection at Harvard Medical School[30] and from the VDRC phiC31 RNAi library[31] contain improvements: for example, the insertion of the RNAi construct is site-specific and the efficacy of the RNAi phenotypes is improved[30,31]. It is likely that the availability of these GAL4/UAS RNAi-based libraries will spur publications in the field of genome-wide in vivo screens in the immediate future.
In the defense against pathogenic microorganisms, Drosophila relies on innate immunity including epithelial and systemic responses generating antimicrobial peptides (AMPs), a phenoloxidase reaction resulting in melanin deposition, and a cellular response leading to the encapsulation and phagocytosis of intruding microbes[32-34]. Several signaling pathways implicated in Drosophila immunity have been identified, such as Imd, Toll, JNK, JAK/STAT and RNAi pathways. Upon microbial invasion, corresponding pathway(s) are activated specifically or in combination, to mount anti-microbial responses. The evolutionarily conserved innate immunity between Drosophila and humans makes Drosophila a valuable model for deciphering the mechanisms underlying human immunity[35,36]. This is exemplified by the discovery of the Drosophila Toll receptor[37], which stimulated the identification of the human Toll-like receptors[38]. In addition, the discovery of Drosophila peptidoglycan recognition proteins PGRPs[39] and the immune functions of PGRP-SA[40] and PGRP-LC[15,41,42] prompted the identification of a human family of PGRPs[43].
In Drosophila innate immune signaling, it is well established that the Imd and Toll pathways respond to bacterial and fungal infections via production of AMPs[33]. AMPs are mostly cationic, small molecules with an activity range directed against a variety of microorganisms[34]. Drosophila JAK/STAT signaling is required to control immune and stress responses[34,44,45]. After septic injury, activation of the JAK/STAT pathway leads to the expression of a number of genes including Turandot stress genes in fat body[46-48]. The Drosophila cellular response involves Drosophila professional phagocytes i.e. plasmatocytes, which phagocytose pathogens and locally secrete extracellular matrix components, AMPs, clotting factors and signaling molecules[49]. Another cellular response mechanism at the larval stages is the encapsulation of microbes that are too large for phagocytosis; a specialized group of hemocytes called lamellocytes is responsible for this[33,50].
In comparison to bacteria and fungal pathogens, viral evoked immune response in Drosophila is less known. It is apparent that both local and systemic immune responses are involved in virus clearance. A detailed description of Drosophila viruses has recently been reviewed by Huszar et al[51]. In this article we will review the interactions of viruses with Drosophila at the molecular level.
Being the most abundant infectious agents, viruses cause diseases which represent a constant threat and cause significant mortality worldwide. They evolve rapidly to adapt to the changing environment of host cells and are a great challenge to their host as well as to the development of efficient therapies and vaccines. By 2005, more than 25 distinct Drosophila viruses had been identified and were all RNA viruses[33,51]. A large proportion of all flies are infected with viruses via horizontal transmission between any two individuals, and infection through vertical transmission from parent to offspring is also common in Drosophila[33]. The innate antiviral immunity in Drosophila has recently been reviewed by Sabin et al[52]. Drosophila can also be used as a model to study human pathogenic viruses, such as the human immunodeficiency virus 1 (HIV-1). It has been demonstrated that the HIV-1 gene Vpu in Drosophila fat body cells inhibits the activity of the Toll pathway[53]. In addition, the HIV-1 gene Nef in the wing disc inhibits the activity of the Imd pathway[54]. Bearing in mind the crucial contribution of the innate immune response both to fighting HIV infection and activating the adaptive immune response, it appears advantageous for HIV to produce proteins that interfere with innate immunity pathways[53]. Therefore, using a genetically tractable model, such as Drosophila, is very useful in investigating these important evolutionarily conserved processes.
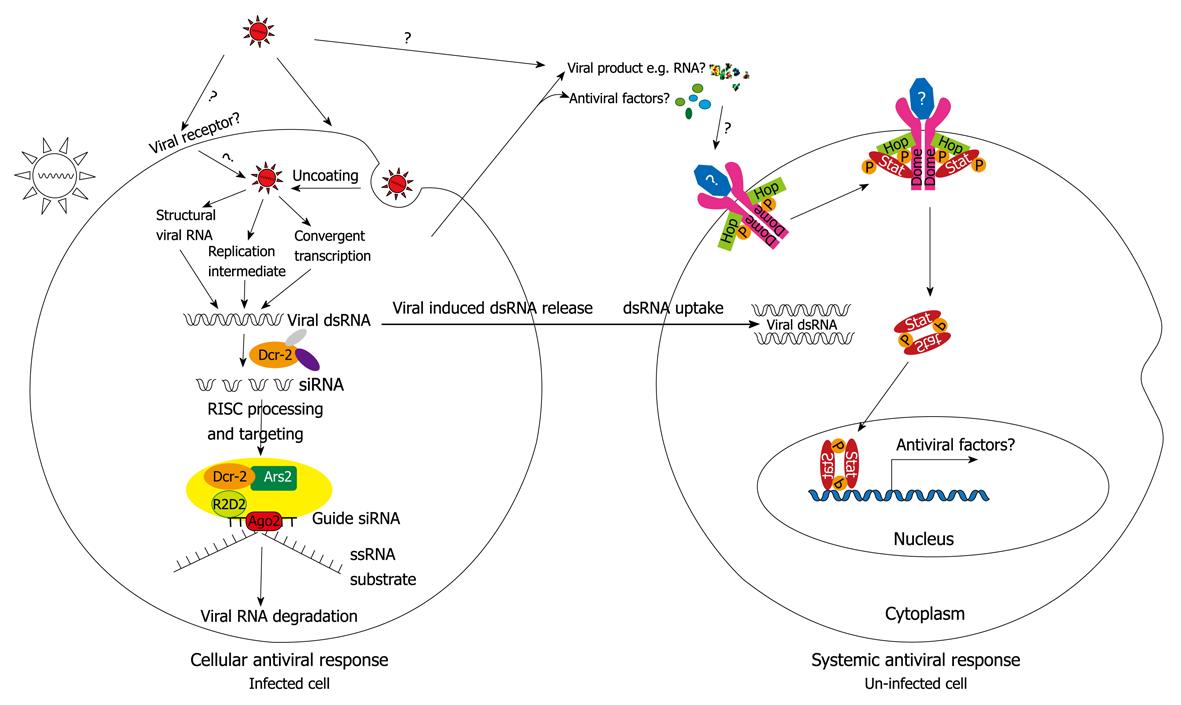
Initially described in plants[55,56], RNAi was later found in Caenorhabditis elegans with double-stranded RNA (dsRNA) as its initiating factor in silencing effects, and was named RNAi[57]. RNAi is an ancient, cell-intrinsic immune mechanism for the control of RNA viruses in plants and insects[58] as well as DNA viruses in mammals[59]. Three RNAi pathways have been identified in Drosophila[60], namely the small interfering RNA (siRNA) pathway, the micro-RNA (miRNA) pathway and the Piwi-interacting RNA (piRNA) pathway. The siRNA pathway utilizes AGO2 and Dicer2 and is activated by dsRNA. The miRNA pathway involves AGO1 and Dicer1 and regulates gene expression, particularly during development. The piRNA pathway involves three AGO proteins, is particularly active in the germline and seems to function in transposon silencing and epigenetic regulation[61,62]. In Drosophila, the exogenous part of the siRNA pathway is mainly responsible for the antiviral defense (Figure 2), whereas the endogenous siRNA pathway is involved in the regulation of transposons and transcripts[63].
The RNAi mediated antiviral response in Drosophila has been studied with evolutionarily diverse viruses including Drosophila X virus (DXV)[64], Drosophila C virus (DCV)[65], Cricket paralysis virus (CrPV)[65,66] and Flock house virus (FHV)[66,67]. Both single-stranded and dsRNA viruses can infect Drosophila, but to date, no Drosophila DNA viruses have been identified. Both RNA virus types produce dsRNA as a replication intermediate, and this dsRNA activates the host RNAi pathway. RNAi is currently considered the major antiviral immune defense mechanism in Drosophila and the above-mentioned studies point to the significant role of Dicer2, AGO2 and Ars2 in this process[64-68]. In addition, Zambon et al[64] indicated that piwi, vasa intronic gene, aubergine, armitage, Rm62, and r2d2 also have vital roles in anti-DXV response. It is worth noting that a low level of infection, which can be cleared, or persistent infection of Drosophila by Nora virus[69,70] are not affected by RNAi machinery, JAK/STAT pathway or Toll pathway[71]. Detailed RNAi in the antiviral innate immune defense has recently been reviewed by Ulvila et al[72].
One virulence mechanism for viruses is suppression of the host RNAi pathway. DCV encodes a suppressor of RNAi, DCV-1A, that binds to long dsRNA and inhibits Dicer2-mediated processing of dsRNA into siRNA, but does not bind to siRNAs nor disrupt the miRNA pathway[65]. On the other hand, successful infection and killing of Drosophila by FHV strictly depends on the expression of the viral protein B2[67], which binds to dsRNA regardless of the length and inhibits cleavage of dsRNA by Dicer2 as well as incorporation of FHV siRNAs into the RNA-induced silencing complex[73]. The N-terminal fragment of 140 amino acids of the CrPV-A was identified as a viral suppressor of RNAi (VSR) for CrPV[66].
Previously, it was believed that RNA silencing-mediated antiviral response is systemic in plants, Caenorhabditis elegans and fungi but not in Drosophila[58,74]. However, this view has been challenged. Saleh et al[75] showed that antiviral immunity against DCV and Sindbis virus (SINV) in Drosophila requires systemic RNAi spread through the endocytosis pathway, which had been identified previously[76,77]. The endocytic pathway is also used in viral entry. Flies heterozygous for mutations in components of the endocytic pathway were protected from DCV-induced lethality in vivo[78]. The virus is suggested to bind to its cell surface receptor, which is captured in clathrin-coated vesicles and trafficked through the endocytic pathway. When virus escapes from the endocytic pathway upon cell death or lysis, dsRNA is released and may be taken up by uninfected cells thus inducing the systemic RNAi-mediated antiviral mechanisms[75] (Figure 2).
JAK/STAT pathway in antiviral response: Known earlier as a regulator of multiple aspects of development, the JAK/STAT pathway was shown to partly contribute to the antiviral response in Drosophila, because DCV triggered STAT (a signal transducer and activator of transcription) DNA-binding activity in whole fly nuclear extract[79]. Moreover, loss-of-function mutation of the only Drosophila Jak kinase, Hopscotch resulted in increased viral replication upon DCV infection, and using a dominant negative form of the receptor Domeless, decreased the expression of a virus-specific target gene virus-induced RNA 1 (vir-1)[79]. It is suggested that DCV infection triggers the induction of an unidentified cytokine, possibly a member of the Upd family, which activates the Domeless receptor and Hopscotch, leading to activation of STAT and induction of a set of genes[80] (Figure 2). This set of induced genes appears to be dependent on the specific virus, at least in part, since the RNA level of a Drosophila JAK/STAT target gene Turandot M, although induced in FHV infection, was not induced upon DCV infection[79]. The role of JAK/STAT in the control of viral infection is further supported by a recent study[81] showing that flies heterozygous for a stat mutation displayed increased SINV replication. Together, these studies indicate an evolutionarily conserved involvement of the JAK/STAT pathway in antiviral response[79,82].
Toll and Imd pathways in antiviral response: Previously it was suggested that the fly nuclear factor (NF)-κB pathways, namely Toll and Imd pathways, have no role in antiviral defense, e.g.[80], but this view has been revised. The Toll pathway was shown to control the survival of and be required for the inhibition of the viral replication of DXV-infected flies[64,83]. Also, a role for the Toll pathway in the control of Dengue virus infection in mosquito has been shown[84]. Recently, two research groups demonstrated the involvement of the Imd pathway in antiviral defenses in Drosophila. Avadhanula et al[81] measured the SINV RNA change in transgenic flies expressing SINV replicon RNA and compared levels from flies with knock-down of different Imd pathway genes to that of wild-type flies. Their results showed that the Imd but not the Toll pathway plays a role in anti-SINV response. Later, Costa et al[85] demonstrated a role for hemocytes and the Imd pathway in combating against CrPV infection. Both studies showed increased viral loads in flies with knock-down of Imd pathway genes, which suggests a role for the Imd pathway at least in SINV and CrPV infections in Drosophila[81,85]. However, in all these studies it was agreed that AMPs are not involved, at least not directly, in the control of viral infection in Drosophila[79,81,83,85].
The importance of NF-κB signaling for antiviral defense was further highlighted by identification of viral proteins that suppress IMD and Toll pathway-mediated immune response. In Drosophila S2 cells, H4 and N5, two of the proteins encoded by Microplitis demolitor bracovirus (McBV), homologous to inhibitor κB (IκB) from insects and mammals, reduce the expression of Drosomycin and Attacin reporter constructs[86]. Moreover, H4 and N5 are able to bind to Dif and Relish and inhibit binding of Dif and Relish to κB sites in the promoters of the Drosomycin and Cecropin A1 genes[86]. Mimicking IκB factors thus appears to be one way for the virus to evade the insect immune system.
Other Drosophila antiviral defense: Induction of host antiviral responses via mechanisms independent of the RNAi, JAK/STAT, Toll and Imd pathways have also been found in Drosophila. One such inducible gene is Vago[80], identified in the screen for DCV-induced genes[79], which negatively controls DCV load in the fat body in a tissue-autonomous way. Vago induction by DCV requires Dicer2, but not AGO2 or r2d2[68,79]. This therefore, suggests a novel role for Dicer2 in sensing viral dsRNA in virus-infected cells, leading to induction of antiviral genes, in addition to its involvement in RNAi[80]. As well as DCV, Vago was also induced by SINV, but not FHV[80].
One other important factor in antiviral immunity appears to be dSUR, a homolog of the mammalian SUR2 protein, which was identified to be important for mammalian antiviral immunity in mapping the mice mayday phenotype[87]. SUR2 is a subunit of the ATP-sensitive potassium channel complex expressed in smooth muscle cells of coronary arteries. RNAi knock-down of Drosophila SUR (dSUR, CG5772) led to increased susceptibility to infection with FHV, but not for DCV, the bacterial species Enterococcus faecalis and Enterobacter cloacae, nor the fungus Beauveria bassiana[87]. Moreover, flies fed with tolbutamide, a member of the sulfonylurea class of channel blockers, showed increased susceptibility to FHV infection, indicating that the heart is one of the tissues maintaining homeostasis during the innate immune response[87].
Autophagy, an intrinsic mechanism that can degrade cytoplasmic components, was found to play a direct antiviral role against the mammalian viral pathogen, vesicular stomatitis virus (VSV), in Drosophila[88,89]. Autophagy decreased viral replication in a cell-autonomous manner and its activation does not require viral replication. Repression of autophagy led to increased viral replication and pathogenesis in cells and animals. Furthermore, the antiviral response of autophagy was controlled by the phosphatidylinositol 3-kinase (PI3K)-Akt-signaling pathway, which normally mediates autophagy in response to nutrient availability. On the other hand, flies depleted of Atg18, a component of the PI3K-Akt-signaling pathway, had a normal life span and were not more sensitive to infection with DCV, suggesting that the autophagy-mediated antiviral response is protective against VSV but not DCV infection. It is likely that the surface glycoprotein, VSV-G, which serves as the pathogen-associated molecular pattern initiated this cell-autonomous response. However, the intracellular sensor for VSV-G has not been identified in Drosophila.
Microarray studies have examined changes in gene expression in Drosophila infected with DCV[79,90], DXV[83], SIGMAV[91,92] and DCV/FHV/SINV[80]. It appears that Drosophila antiviral immune response is virus specific. Carpenter et al[92] also reported that there was a difference in gene regulation between male and female SIGMAV infected flies. On the other hand, although both studies from Tsai et al[91] and Carpenter et al[92] used SIGMAV, little overlap of regulated genes was observed. Therefore, it is apparent that the genetic background of Drosophila affects host-parasite interactions[93-95]. Genetic background may also explain, in part, the variable Nora virus titers observed between fly stocks in a recent study[70].
The genetic composition of alleles encoding a polymorphic gene ref(2)P appear important in Drosophila SIGMAV infection and have been intensively studied. Ref(2)P is evolutionarily conserved and its mammalian orthologue, p62, serves as an adaptor for the activation of the NF-κB pathway by aPKC (atypical protein kinase)[96-98]. A similar role for Ref(2)P has been proposed in Drosophila, where Ref(2)P/DaPKC activates the NF-κB proteins Dorsal and DIF in the Toll pathway[99]. In nature, there are ref(2)p alleles that are either permissive or restrictive for SIGMAV multiplication[51,100,101]. The mechanism by which the restrictive ref(2)p allele interacts with and blocks SIGMAV replication is not known, but it was shown that all allele encoded proteins can interact with SIGMAV P protein and share conformation-dependent epitopes with the N protein[102].
The interactions of Drosophila with other microorganisms also play a role in antiviral response. Drosophila infected with Wolbachia, a common bacterium in natural Drosophila populations, increases resistance to RNA viruses, namely DCV, Nora, FHV and CrPV, but not to a DNA virus insect iridescent virus 6[103,104]. Also, preinfection of flies with another virus, namely FHV, led to an increased induction of Vir-1 and more modest upregulation of Vago by DCV infection[80]. Antiviral silencing against FHV in S2 cells induced by FR1gfp, a construct defective in suppressing RNA silencing-based antiviral response in Drosophila cells, as described previously[105] was suppressed by CrPV superinfection[66].
In conclusion, the antiviral response in Drosophila is mainly mediated by the siRNA pathway, which functions against a broad range of viruses. Viruses have evolved ways to circumvent RNAi to survive in their host cells, exemplified by viral genome encoded VSRs. The JAK/STAT, and possibly Toll and Imd pathways are involved in the control of viral growth for specific virus(es) by mechanisms yet to be clarified. The viral infection could induce the expression of genes which are involved in decreasing the viral infection, and Dicer2 is needed for gene induction. Other aspects, including e.g. autophagy, also appear to play a role in the control of viral infection in Drosophila. Of note, the genetic background of Drosophila as well as infection with other microorganisms has effects on the antiviral responses.
Despite the progress in recent years in understanding fly antiviral immunity, there are still a lot of unanswered questions. Currently, dsRNA-triggered RNAi is considered the major antiviral immune response in Drosophila, and it has been intensively studied. What is the viral trigger for other pathways in antiviral response, such as the JAK/STAT pathway? What is the ligand of the virally-induced JAK/STAT pathway, and what do the activated genes do in combat against viruses at the molecular level? Do the fly NF-κB pathways mount a response specifically against some viruses, and what are the factors induced, since in all the studies it was agreed that AMPs are not directly involved[79,81,83,85]? What is the route of viral spreading in Drosophila[95]? What is the role of the newly identified Nora virus, which is present in large numbers in the intestine of infected flies[69-71]? In addition, it will be interesting to investigate the interactions between bacteria and viruses in the light of the new findings about Wolbachia conferring resistance against viral infections in Drosophila[103,104]. Further work is likely to uncover additional mechanisms, factors and pathways in the Drosophila antiviral immune response as well as insights into host-pathogen interactions.
Peer reviewers: Rongtuan Lin, PhD, Associate Professor, Lady Davis Institute, Department of Medicine, McGill University, 3755 Cote Ste-Catherine, Montreal, QC, H3T 1E2 Canada; Takashi Kuzuhara, PhD, Professor, Laboratory of Biochemistry, Faculty of Pharmaceutical Sciences, Tokushima Bunri University, Yamashiro-cho, Tokushima, 770-8514, Japan; Gianfranco Risuleo, PhD, Professor, Dipartimento di Genetica e Biologia Molecolare, Sapienza Università di Roma, P. Aldo Moro, 5 - 00185 Roma, Italy
S- Editor Cheng JX L- Editor Webster JR E- Editor Zheng XM
| 1. | St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176-188. |
| 2. | Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, WoodageT, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185-2195. |
| 3. | Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W. Comparative genomics of the eukaryotes. Science. 2000;287:2204-2215. |
| 4. | Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA. A whole-genome assembly of Drosophila. Science. 2000;287:2196-2204. |
| 5. | Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;11:1114-1125. |
| 6. | Chien S, Reiter LT, Bier E, Gribskov M. Homophila: human disease gene cognates in Drosophila. Nucleic Acids Res. 2002;30:149-151. |
| 7. | Rubin GM, Lewis EB. A brief history of Drosophila's contributions to genome research. Science. 2000;287:2216-2218. |
| 8. | Engels WR. The P family of transposable elements in Drosophila. Annu Rev Genet. 1983;17:315-344. |
| 9. | Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135-177. |
| 10. | Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348-353. |
| 11. | Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013-2018. |
| 12. | Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293-296. |
| 15. | Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644-648. |
| 16. | Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832-835. |
| 17. | Kleino A, Valanne S, Ulvila J, Kallio J, Myllymäki H, Enwald H, Stöven S, Poidevin M, Ueda R, Hultmark D. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 2005;24:3423-3434. |
| 18. | Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251-1253. |
| 19. | Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248-1251. |
| 20. | Cherry S. Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions? Curr Opin Microbiol. 2008;11:262-270. |
| 21. | Kallio J, Leinonen A, Ulvila J, Valanne S, Ezekowitz RA, Rämet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 2005;7:811-819. |
| 22. | Valanne S, Kleino A, Myllymäki H, Vuoristo J, Rämet M. Iap2 is required for a sustained response in the Drosophila Imd pathway. Dev Comp Immunol. 2007;31:991-1001. |
| 23. | Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353-365. |
| 24. | Rämet M, Pearson A, Manfruelli P, Li X, Koziel H, Göbel V, Chung E, Krieger M, Ezekowitz RA. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity. 2001;15:1027-1038. |
| 25. | Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401-415. |
| 26. | Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, Leibbrandt A, Simoes Rde M, Gruber S, Puc U. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science. 2009;325:340-343. |
| 27. | Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010;140:148-160. |
| 28. | Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987-992. |
| 29. | Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151-156. |
| 30. | Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089-1100. |
| 31. | Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312-3317. |
| 32. | Hultmark D. Drosophila immunity: paths and patterns. Curr Opin Immunol. 2003;15:12-19. |
| 33. | Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697-743. |
| 34. | Wang L, Ligoxygakis P. Pathogen recognition and signalling in the Drosophila innate immune response. Immunobiology. 2006;211:251-261. |
| 35. | Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457-483. |
| 36. | Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862-874. |
| 37. | Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973-983. |
| 38. | Medzhitov R, Preston-Hurlburt P, Janeway CA Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394-397. |
| 39. | Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:13772-13777. |
| 40. | Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756-759. |
| 41. | Choe KM, Werner T, Stöven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359-362. |
| 42. | Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640-644. |
| 43. | Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J Biol Chem. 2001;276:34686-34694. |
| 44. | Hombría JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12:R569-R575. |
| 45. | Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605-2616. |
| 46. | Ekengren S, Hultmark D. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem Biophys Res Commun. 2001;284:998-1003. |
| 47. | Ekengren S, Tryselius Y, Dushay MS, Liu G, Steiner H, Hultmark D. A humoral stress response in Drosophila. Curr Biol. 2001;11:714-718. |
| 48. | Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. 2003;5:441-450. |
| 49. | Stuart LM, Ezekowitz RA. Phagocytosis and comparative innate immunity: learning on the fly. Nat Rev Immunol. 2008;8:131-141. |
| 50. | Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230:243-257. |
| 51. | Huszar T, Imler JL. Drosophila viruses and the study of antiviral host-defense. Adv Virus Res. 2008;72:227-265. |
| 52. | Sabin LR, Hanna SL, Cherry S. Innate antiviral immunity in Drosophila. Curr Opin Immunol. 2010;22:4-9. |
| 53. | Leulier F, Marchal C, Miletich I, Limbourg-Bouchon B, Benarous R, Lemaitre B. Directed expression of the HIV-1 accessory protein Vpu in Drosophila fat-body cells inhibits Toll-dependent immune responses. EMBO Rep. 2003;4:976-981. |
| 54. | Lee SB, Park J, Jung JU, Chung J. Nef induces apoptosis by activating JNK signaling pathway and inhibits NF-kappaB-dependent immune responses in Drosophila. J Cell Sci. 2005;118:1851-1859. |
| 55. | van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291-299. |
| 56. | Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG. Induction of a Highly Specific Antiviral State in Transgenic Plants: Implications for Regulation of Gene Expression and Virus Resistance. Plant Cell. 1993;5:1749-1759. |
| 57. | Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358-363. |
| 59. | Aliyari R, Ding SW. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol Rev. 2009;227:176-188. |
| 60. | Kemp C, Imler JL. Antiviral immunity in drosophila. Curr Opin Immunol. 2009;21:3-9. |
| 61. | Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116-125. |
| 62. | Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761-764. |
| 63. | van Rij RP, Berezikov E. Small RNAs and the control of transposons and viruses in Drosophila. Trends Microbiol. 2009;17:163-171. |
| 64. | Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880-889. |
| 65. | van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985-2995. |
| 66. | Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452-454. |
| 67. | Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590-597. |
| 68. | Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340-351. |
| 69. | Habayeb MS, Ekengren SK, Hultmark D. Nora virus, a persistent virus in Drosophila, defines a new picorna-like virus family. J Gen Virol. 2006;87:3045-3051. |
| 70. | Habayeb MS, Cantera R, Casanova G, Ekström JO, Albright S, Hultmark D. The Drosophila Nora virus is an enteric virus, transmitted via feces. J Invertebr Pathol. 2009;101:29-33. |
| 71. | Habayeb MS, Ekström JO, Hultmark D. Nora virus persistent infections are not affected by the RNAi machinery. PLoS One. 2009;4:e5731. |
| 72. | Ulvila J, Hultmark D, Rämet M. RNA silencing in the antiviral innate immune defence--role of DEAD-box RNA helicases. Scand J Immunol. 2010;71:146-158. |
| 73. | Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952-957. |
| 74. | Reichhart JM, Ligoxygakis P, Naitza S, Woerfel G, Imler JL, Gubb D. Splice-activated UAS hairpin vector gives complete RNAi knockout of single or double target transcripts in Drosophila melanogaster. Genesis. 2002;34:160-164. |
| 75. | Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346-350. |
| 76. | Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, Rämet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370-14375. |
| 77. | Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O'Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793-802. |
| 78. | Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol. 2004;5:81-87. |
| 79. | Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6:946-953. |
| 80. | Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler JL. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425-1432. |
| 81. | Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5:e1000582. |
| 82. | Beutler B, Eidenschenk C, Crozat K, Imler JL, Takeuchi O, Hoffmann JA, Akira S. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7:753-766. |
| 83. | Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci USA. 2005;102:7257-7262. |
| 84. | Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. |
| 85. | Costa A, Jan E, Sarnow P, Schneider D. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS One. 2009;4:e7436. |
| 86. | Thoetkiattikul H, Beck MH, Strand MR. Inhibitor kappaB-like proteins from a polydnavirus inhibit NF-kappaB activation and suppress the insect immune response. Proc Natl Acad Sci USA. 2005;102:11426-11431. |
| 87. | Croker B, Crozat K, Berger M, Xia Y, Sovath S, Schaffer L, Eleftherianos I, Imler JL, Beutler B. ATP-sensitive potassium channels mediate survival during infection in mammals and insects. Nat Genet. 2007;39:1453-1460. |
| 88. | Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588-598. |
| 89. | Cherry S. VSV infection is sensed by Drosophila, attenuates nutrient signaling, and thereby activates antiviral autophagy. Autophagy. 2009;5:1062-1063. |
| 90. | Roxström-Lindquist K, Terenius O, Faye I. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. 2004;5:207-212. |
| 91. | Tsai CW, McGraw EA, Ammar ED, Dietzgen RG, Hogenhout SA. Drosophila melanogaster mounts a unique immune response to the Rhabdovirus sigma virus. Appl Environ Microbiol. 2008;74:3251-3256. |
| 92. | Carpenter J, Hutter S, Baines JF, Roller J, Saminadin-Peter SS, Parsch J, Jiggins FM. The transcriptional response of Drosophila melanogaster to infection with the sigma virus (Rhabdoviridae). PLoS One. 2009;4:e6838. |
| 93. | Bangham J, Knott SA, Kim KW, Young RS, Jiggins FM. Genetic variation affecting host-parasite interactions: major-effect quantitative trait loci affect the transmission of sigma virus in Drosophila melanogaster. Mol Ecol. 2008;17:3800-3807. |
| 94. | Bangham J, Kim KW, Webster CL, Jiggins FM. Genetic variation affecting host-parasite interactions: different genes affect different aspects of sigma virus replication and transmission in Drosophila melanogaster. Genetics. 2008;178:2191-2199. |
| 95. | Ammar el-D, Tsai CW, Whitfield AE, Redinbaugh MG, Hogenhout SA. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu Rev Entomol. 2009;54:447-468. |
| 96. | Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95-100. |
| 97. | Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18:3069-3080. |
| 98. | Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 1999;18:3044-3053. |
| 99. | Avila A, Silverman N, Diaz-Meco MT, Moscat J. The Drosophila atypical protein kinase C-ref(2)p complex constitutes a conserved module for signaling in the toll pathway. Mol Cell Biol. 2002;22:8787-8795. |
| 100. | Dru P, Bras F, Dezélée S, Gay P, Petitjean AM, Pierre-Deneubourg A, Teninges D, Contamine D. Unusual variability of the Drosophila melanogaster ref(2)P protein which controls the multiplication of sigma rhabdovirus. Genetics. 1993;133:943-954. |
| 101. | Carré-Mlouka A, Gaumer S, Gay P, Petitjean AM, Coulondre C, Dru P, Bras F, Dezélée S, Contamine D. Control of sigma virus multiplication by the ref(2)P gene of Drosophila melanogaster: an in vivo study of the PB1 domain of Ref(2)P. Genetics. 2007;176:409-419. |
| 102. | Wyers F, Dru P, Simonet B, Contamine D. Immunological cross-reactions and interactions between the Drosophila melanogaster ref(2)P protein and sigma rhabdovirus proteins. J Virol. 1993;67:3208-3216. |
| 103. | Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322:702. |
| 104. | Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. |
| 105. | Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, García-Sastre A, Ball LA. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA. 2004;101:1350-1355. |